Abstract
The BRCA1 tumor suppressor has been implicated in many cellular pathways, but the mechanisms by which it suppresses tumor formation are not fully understood. In vivo BRCA1 forms a heterodimeric complex with the related BARD1 protein, and its enzymatic activity as a ubiquitin ligase is largely dependent upon its interaction with BARD1. To explore the genetic relationship between BRCA1 and BARD1, we have examined the phenotype of Bard1-null mice. These mice become developmentally retarded and die between embryonic day 7.5 (E7.5) and E8.5. Embryonic lethality results from a severe impairment of cell proliferation that is not accompanied by increased apoptosis. In the absence of p53, the developmental defects associated with Bard1 deficiency are partly ameliorated, and the lethality of Bard1; p53-nullizygous mice is delayed until E9.5. This result, together with the increased chromosomal aneuploidy of Bard1 mutant cells, indicates a role for Bard1 in maintaining genomic stability. The striking similarities between the phenotypes of Bard1-null, Brca1-null, and double Bard1; Brca1-null mice provide strong genetic evidence that the developmental functions of Brca1 and Bard1 are mediated by the Brca1/Bard1 heterodimer.
Germ line mutations of the BRCA1 tumor suppressor gene are responsible for many cases of hereditary breast and ovarian carcinomas (34), and its protein product has been implicated in a broad spectrum of cellular processes that includes transcriptional regulation, chromatin remodeling, DNA repair, and cell cycle checkpoint control (for recent reviews, see references 2, 25, 35, 42, and 49). The major isoform of Brca1 has two recognizable amino acid motifs: a RING domain at the N terminus and two tandem copies of the BRCT domain at the C terminus (29, 34). In some patients, the predisposing BRCA1 lesion can be traced to missense mutations in the RING domain (8, 44), indicating that proper folding of this motif is essential for BRCA1-mediated tumor suppression. Many RING proteins are now known to function as ubiquitin E3 ligases, a family of enzymes that catalyze the final step in protein ubiquitination (22, 24). Recent studies have shown that the N-terminal RING sequence of BRCA1 can also catalyze the formation of polyubiquitin chains in vitro and that this activity is abolished by tumor-associated missense mutations (7, 17, 32, 39).
The in vivo functions of BRCA1 have been explored using genetically modified mice bearing either null Brca1 alleles, which are completely devoid of Brca1 activity and/or expression, or hypomorphic alleles that presumably retain some aspects of normal Brca1 activity (reviewed in references 4 and 19). Mice that are heterozygous for Brca1 mutations, whether null or hypomorphic, develop normally, but unlike human carriers of BRCA1 mutations, they are not predisposed to mammary carcinogenesis. On the other hand, mice that are homozygous for null Brca1 alleles die around the time of gastrulation, typically between days 6.5 and 7.5 of embryogenesis (15, 31, 33). Brca1-null embryos are not characterized by excessive apoptosis but instead display decreased cell proliferation and higher expression of the cyclin-dependent kinase inhibitor p21. Thus, it has been proposed that, in the absence of Brca1 function, DNA damage accumulates and ultimately elicits the activation of cell cycle checkpoints (5, 40). In this scenario, the embryonic lethality of Brca1-null mice is a direct consequence of the severe proliferation defect imposed by these checkpoints. In accord with this hypothesis, partial rescue of the Brca1-null phenotype is observed in mice that are also nullizygous for either p21 or its upstream transcriptional activator, the p53 tumor suppressor (15, 33).
BRCA1 exists primarily in the form of a heterodimer with BARD1, a protein that also harbors an N-terminal RING domain and two C-terminal BRCT motifs (23, 50). The association between BRCA1 and BARD1 is mediated by sequences encompassing their respective RING domains (50). Indeed, the molecular basis for heterodimerization was recently uncovered from the solution structure of a protein complex formed by the interacting sequences of BRCA1 and BARD1 (6). In this structure, the zinc-binding elements of both proteins are flanked by long α-helices that pair in an antiparallel fashion and promote heterodimerization by combining to form a stable four-helix bundle. Recent work has shown that the BRCA1/BARD1 interaction is essential for nuclear retention of Brca1 (10) as well as for suppression of mRNA processing during the DNA damage response (27, 28). The significance of the interaction has also been underscored by studies of its catalytic properties, which revealed that the ubiquitin E3 ligase activity of the heterodimer is dramatically higher than that of either BRCA1 or BARD1 alone (7, 17). These results imply that the BRCA1/BARD1 heterodimer is the primary mediator of the enzymatic activity attributed to BRCA1. Indeed, since mutations of the BARD1 gene are found in rare cases of breast, ovarian, and endometrial carcinoma (12, 48), BARD1 may itself serve as a target for tumor-associated lesions that disrupt the BRCA1 pathway. It has also been reported that BARD1 has proapoptotic functions independent of its association with BRCA1 (20).
If the biological activities of Brca1 are mediated primarily by the BRCA1/BARD1 heterodimer, then mutations of BARD1 should also serve to disrupt the BRCA1 pathway. To evaluate the developmental functions of Bard1 and to explore its genetic relationship to Brca1, we have characterized the phenotype of mice bearing a null Bard1 allele. These studies show that while heterozygous Bard1-null animals develop normally, homozygous Bard1-null embryos undergo proliferative arrest and suffer an early embryonic death that is essentially indistinguishable from that of Brca1-null mice and double Bard1; Brca1-null mice.
MATERIALS AND METHODS
Targeted mutagenesis.
The Bard1-hygromycin resistance gene and Bard1-neomycin resistance gene targeting vectors were constructed in several steps from subcloned fragments of overlapping λ clones isolated from a genomic 129/Sv library. The final Bard1 targeting constructs consisted of a 5′ homology fragment (2.0 kb), a selection marker gene cassette (hygromycin resistance-enhanced green fluorescent protein [EGFP] fusion gene [Clontech] or neomycin resistance gene) lacking both a promoter and a polyadenylation signal replacing the Bard1 open reading frame in exon 1, and a 3′ homology fragment (3.0 kb). A diphtheria toxin A gene cassette was included in the constructs as a negative selection marker against random integration (52). For gene targeting, linearized vector DNA was introduced into 129/Sv W9.5 embryonic stem (ES) cells by electroporation, and after drug selection, DNA of drug-resistant clones was analyzed by Southern blotting with a 5′ flanking probe. Male chimeras were generated by injection of ES cells into C57BL/6J blastocysts, and germ line transmission was verified by Southern analysis. Bard1 heterozygotes were intercrossed to generate homozygous mutants. Heterozygous Bard1 animals were also crossed with mice carrying null mutations of Brca1 (33) or p53 (21) to produce double heterozygous animals, which were subsequently intercrossed. The p53 mutant mice used in this genetic analysis were obtained from the Jackson Labs.
Southern blotting and PCR genotyping.
For genotyping by Southern analysis with the 5′ flanking probe, DNA was prepared from yolk sacs, whole embryos, or the tail tips of 10-day-old mice. PCR genotyping of whole embryonic day 6.5 (E6.5) and E7.5 embryos, as well as embryos from stained histological sections, was performed using a three-primer strategy: a common primer located in the first intron (5′-GTGCCGTTTGAGTCATCTTCGTTGC-3′) can pair with a wild-type allele-specific primer located within exon 1 (5′-GGCGTCCGACCAATTCAGAGACTCC-3′) or with a mutant allele-specific primer located at the 3′ end of the hygromycin resistance-EGFP fusion gene (5′-GGCACAAGCTGGAGTACAACTACAAC-3′). Amplification of the wild-type allele results in a 347-bp product, whereas the product of the mutant allele is 422 bp. Embryos from histological sections were captured onto CapSure LCM transfer film (Arcturus) by using an Arcturus PixCell Laser Capture Microdissection system according to the manufacturer's instructions as well as information available on the National Institutes of Health website (http://dir.nichd.nih.gov/lcm/lcm.htm). Samples were then digested for 20 h at 42°C in PicoPure digest buffer containing proteinase K (Arcturus), which was heat inactivated at 95°C for 10 min prior to PCR amplification.
Histological analyses.
Deciduae dissected at E6.5 and E7.5 were fixed overnight in 4% paraformaldehyde-0.1 M phosphate buffer (pH 7.3), washed for 24 h at 4°C in 0.25 M sucrose-0.2 M glycine-0.1 M phosphate buffer (pH 7.3), dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 μm and stained with hematoxylin and eosin.
BrdU labeling of embryos.
Labeling of embryonic cells in S phase with 5-bromo-2′-deoxyuridine (BrdU) was performed as previously described (18). BrdU (100 μg/g of body weight) was injected intraperitoneally into females pregnant from Bard1 heterozygous intercrosses at day E6.5. One hour after injection, the females were sacrificed. The deciduae were then dissected, fixed in fresh 4% paraformaldehyde, and processed for immunohistochemistry. Sections were incubated with a monoclonal anti-BrdU antibody (Becton-Dickinson) at a 1:20 dilution, and staining was visualized with a biotinylated antibody against mouse immunoglobulin G and avidin-conjugated peroxidase (Vectastain).
In vitro culture of blastocysts.
Blastocysts were collected by flushing the uteri of females pregnant from Bard1 heterozygous intercrosses at day 3.5 of gestation and individually cultured in 24-well plates in Dulbecco modified Eagle medium containing 20% fetal calf serum at 37°C in 5% CO2. The cultured blastocysts were examined and photographed daily for up to 6 days. At the end of the observation period, the tissue was scraped off the dish for DNA extraction and PCR genotyping.
Cytogenetic analysis.
Chromosomal spreads from E9.5 embryos were prepared as follows. Embryos were dissected and incubated in complete medium containing 0.1 μg of Colcemid (Invitrogen)/ml for 4 h at 37°C. Following hypotonic shock in 0.56% (wt/vol) KCl for 3 to 5 min, the embryos were fixed in fresh methanol-acetic acid (3:1), disaggregated in 60% acetic acid, and spread on glass slides. Giemsa-stained metaphase spreads were scored for numerical abnormalities.
Protein analysis.
Protein extracts were prepared from E9.5 embryos that were genotyped from yolk sac DNA as described above. The embryos were placed in lysis buffer (10 mM HEPES [pH 7.6], 250 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA) supplemented with 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors (Roche), homogenized, and incubated on ice for 30 min. The lysates were then centrifuged at 16,000 × g for 10 min, and the pellet was discarded. The protein concentrations of the extracts (supernatants) were determined by using a Bradford assay.
For Western blot analysis, equal amounts (30 μg) of protein extracts were boiled in sodium dodecyl sulfate sample buffer, electrophoresed on sodium dodecyl sulfate-6% polyacrylamide gels, and transferred onto nitrocellulose membranes (Amersham). Blots were then probed with primary antibodies against Bard1, Brca1 (kindly provided by S. Ganesan and D. Livingston), Ctip, and NaK-ATPase (Research Diagnostics), and the immunoreactive bands were visualized with ECL detection reagents (Amersham). The Bard1-specific rabbit antiserum was raised against a glutathione S-transferase fusion protein containing residues 86 to 268 of murine Bard1. The Ctip-specific rabbit antiserum was raised against a glutathione S-transferase fusion protein containing residues 133 to 370 of murine Ctip.
RESULTS
Targeted mutation of the mouse Bard1 gene.
To define the role of Bard1 in vivo, we generated a null Bard1 allele by using ES cell technology. The Bard1-hygromycin resistance gene targeting vector (Fig. 1A) was designed to delete the translation initiation site and coding sequence of exon 1 (Bard1 amino acids 1 to 46) and replace it with a hygromycin resistance gene cassette (Fig. 1A). To enrich for homologous recombination events, this cassette lacked both a promoter and a polyadenylation signal. Targeted ES clones carrying the mutant Bard1 allele were identified by Southern analysis (Fig. 1C), and three independently derived Bard1+/− clones were injected into blastocysts to establish the mutant allele in the mouse germ line. Mice heterozygous for the Bard1 mutation were indistinguishable from their wild-type littermates in viability, growth, development, and fertility. Thus far, none of the heterozygous Bard1+/− mutant mice has developed an overt tumor by 21 months of age. Therefore, Bard1+/− mice, like Brca1+/− heterozygous mice, do not appear to be predisposed to tumor development. However, it still remains possible that Bard1+/− mice will develop tumors at a more advanced age.
FIG. 1.
Targeted disruption of the mouse Bard1 gene. (A) A partial restriction map of the genomic region encompassing Bard1 exon 1 (Ex1; black box) is shown on top, followed by a diagram of the targeting vector used for insertion of the hygromycin resistance-EGFP gene cassette (HygEGFP) into exon 1 of the mouse Bard1 gene (bottom). A diphtheria toxin A gene (DT) was also included in the targeting construct for negative selection. The wavy lines represent plasmid sequences. Relevant restriction sites are EcoRI (E), HindIII (H), PstI (P), and SalI (S). The 5′ flanking probe used for Southern analyses, the sizes of the endogenous and expected DNA fragments, and the locations of the PCR primers a, b, and c are also shown. hyg, hygromycin resistance gene. (B) Schematics of the neomycin resistance gene (neo) targeting vector and the recombined Bard1 locus are shown. (C) Southern analysis of ES cell DNA digested with PstI to confirm gene targeting. Due to an additional PstI site in the hygromycin selection marker, the 7.2-kb PstI fragment detected in wild-type ES cells is reduced to 5.5 kb in properly targeted ES cells. (D) Southern analysis to identify the second targeting event in heterozygous ES cells. wt, wild type. (E) Representative PCR genotyping of E6.5 embryos with primers a, b, and c. PCR amplification was conducted on embryonic DNA from wild-type (lanes 1 and 2), Bard1+/− (lane 3), and Bard1−/− (lane 4) embryos. The PCR products derived from wild-type and mutant alleles are 347 and 422 bp, respectively.
Bard1 deficiency causes early embryonic death.
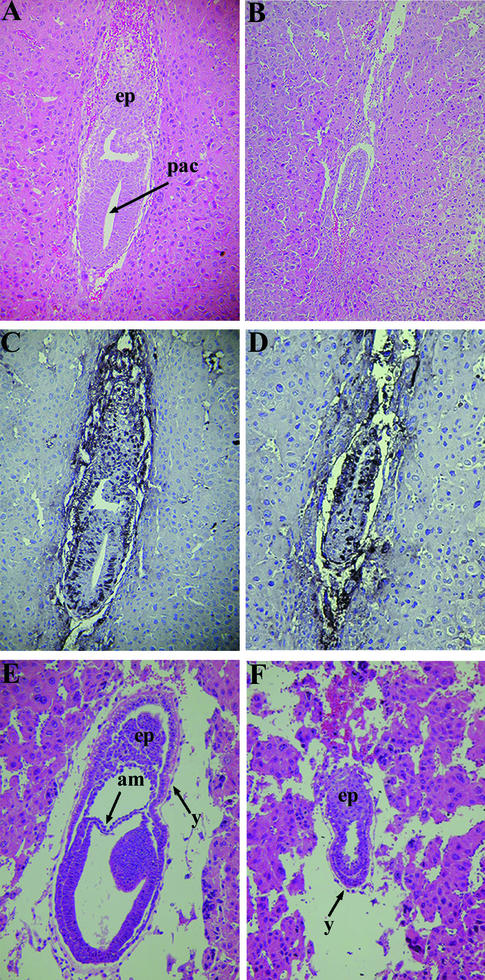
To evaluate the phenotype of Bard1-null animals, Bard1+/− mice were intercrossed and their progeny were genotyped. Of the 137 offspring, 47 were wild types (Bard1+/+) and 90 were heterozygotes (Bard1+/−). The complete absence of homozygous (Bard1−/−) offspring, while wild-type and heterozygous animals were obtained at the expected 1:2 ratio, indicates that Bard1 ablation results in embryonic lethality. To analyze the stage of lethality, embryos from Bard1+/− intercrosses were genotyped and the gross morphologies of the embryos and histological sections of dissected deciduae were examined at different times postcoitum (Table 1). At E6.5, the Bard1−/− mutant embryos were two-layered egg cylinders approximately half the size of normal embryos (Fig. 2A and B). The reduction in size was consistently more pronounced in the embryo proper than in the extraembryonic regions, a feature also observed in Brca1−/− null embryos (33). While all normal (wild-type and heterozygous) embryos examined at E7.5 had gastrulated and possessed a third (mesodermal) germ layer, the Bard1−/− mutant embryos did not develop past the egg cylinder stage (Fig. 2E and F). Six of the 22 deciduae dissected at E8.5 were significantly smaller and contained embryos that were undergoing resorption; the 16 remaining E8.5 embryos had head folds and up to nine somites and were genotyped by PCR as either wild types or heterozygotes (data not shown). Therefore, Bard1−/− mutant embryos display severe growth and morphogenic defects by the onset of gastrulation and die prior to E8.5. This phenotype is remarkably similar to that of Brca1−/− nullizygous embryos (15, 31, 33), consistent with the notion that normal Brca1 functions are mediated by the Brca1/Bard1 heterodimer (2).
TABLE 1.
Genotype and phenotype analysis of offspring and embryos from Bard1+/− intercrossesa
| Stage | No. of embryos with genotypeb:
|
Total no. | ||
|---|---|---|---|---|
| Bard1+/+ | Bard1+/− | Bard1−/− | ||
| E6.5 | 3 (0) | 7 (0) | 4 (4) | 14 (4) |
| E7.5 | 15 (0) | 26 (0) | 15 (15) | 56 (15) |
| E8.5 | 5 (0) | 11 (0) | 6 (6) | 22 (6) |
| E9.5 | 2 (0) | 6 (0) | 3 (3) | 11 (3) |
| Offspring | 47 (0) | 90 (0) | 0 | 137 (0) |
Live-born offspring from Bard1+/− intercrosses were genotyped by Southern analysis with the 5′ flanking probe indicated in Fig. 1A. Embryos were dissected at days 6.5, 7.5, 8.5, and 9.5 of gestation and genotyped by PCR amplification with the primers indicated in Fig. 1A.
The numbers of phenotypically abnormal embryos are indicated in parentheses.
FIG. 2.
Development of Bard1−/− embryos. Hematoxylin- and eosin-stained sagittal sections of wild-type (A and E) and Bard1−/− (B and F) embryos at E6.5 (A and B) and E7.5 (E and F) are shown. (A) Normal (Bard1+/−) egg cylinder with proamniotic cavity (pac) and ectoplacental cone (ep). (B) Developmentally retarded Bard1 mutant embryo. (C and D) The proliferating cells of wild-type (C) and Bard1 mutant (D) E6.5 embryos are highlighted by BrdU labeling. Strong BrdU staining can be detected throughout the wild-type embryo (C), whereas the Bard1 mutant embryo has fewer BrdU-labeled nuclei (D). (E) E7.5 wild-type embryo postgastrulation with three distinct germ layers. am, amnion; y, yolk sac. (F) Bard1 homozygous mutant at the egg cylinder stage with prominent proamniotic cavity.
Bard1 is required for early embryonic cell proliferation.
The growth deficiencies of Brca1-nullizygous embryos were previously shown to correlate with decreased cell proliferation rather than increased cell death (15, 31, 33, 43, 47). Similarly, the reduced size of Bard1 mutant embryos could not be attributable to excessive apoptosis, as terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays on sections from E6.5 embryos showed the same levels of apoptotic nuclei in wild-type and mutant embryos (data not shown). To ascertain whether the Bard1 mutation affects cell proliferation, we examined the incorporation of BrdU into DNA. Females pregnant from heterozygous intercrosses were injected with BrdU 1 h prior to sacrifice, and E6.5 deciduae were dissected, serially sectioned, and PCR genotyped. BrdU incorporation was assayed by counting labeled and unlabeled nuclei in the extraembryonic and embryonic regions of representative sagittal sections (Fig. 2C and D). Analysis of three wild-type and four mutant embryos revealed that 84.1% ± 1.9% and 63.0% ± 1.6%, respectively, of cells had incorporated BrdU, indicating that growth retardation of Bard1−/− nullizygous embryos correlates with relative hypoproliferation.
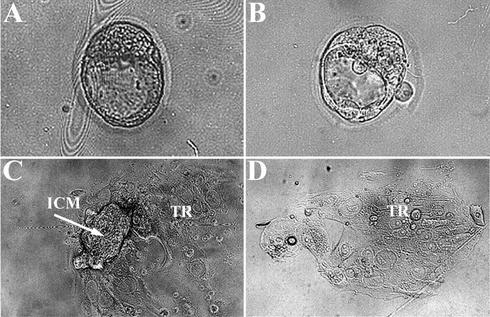
To determine whether proliferation is intrinsically impaired in Bard1−/− mutant embryos, we also examined the growth capabilities of preimplantation embryos in vitro. In this assay, E3.5 blastocysts from heterozygous matings were isolated, cultured individually for 6 days, and subsequently genotyped. After 1 day, all isolated blastocysts hatched from the zona pellucida, adhered to the tissue culture plastic, and began to grow out. For the first 2 days, the outgrowths of Bard1−/− blastocysts (n = 8) were indistinguishable from those of Bard1+/+ (n = 10) and Bard1+/− (n = 18) blastocysts (Fig. 3A and B). However, whereas the inner cell masses (ICMs) of Bard1+/+ and Bard1+/− embryos continued to proliferate and expand throughout the observation period (Fig. 3C), the ICMs of Bard1−/− blastocysts began to disintegrate within 2 days and were invariably lost after 4 days. After 6 days in culture, only the nondividing, endoreplicating, trophoblastic giant cells of Bard1−/− blastocysts appeared to be unaffected (Fig. 3D).
FIG. 3.
Impaired proliferation of Bard1−/− mutants in vitro. (A) Wild-type E3.5 blastocyst. (B) Bard1−/− E3.5 blastocysts. (C and D) Wild-type (C) and Bard1−/− (D) blastocyst outgrowths after 6 days in culture. Proliferation of both the ICM and the trophoblastic giant cells (TR) is evident in the wild-type embryo. In contrast, in the Bard1−/− mutant embryo, the ICM is completely missing and only the trophoblastic giant cells remain.
To delineate the biological function of Bard1 at the cellular level, we also attempted to generate homozygous mutant ES cells. One of the hygromycin-resistant Bard1+/− ES clones (1B1) was electroporated with a targeting vector similar to the original Bard1-hygromycin resistance gene vector, except that the neomycin resistance gene was used as a positive selection marker (Fig. 1B). A total of eight homologous recombinants were identified out of 32 Neor clones. However, Southern analysis revealed that none of these had targeted the remaining wild-type allele; instead, recombination involved the inactive Bard1-hygromycin resistance allele in each of the eight clones (Fig. 1D). In addition, multiple attempts to generate homozygous ES cells by increasing the hygromycin B concentration (up to 3.6 mg/ml) to force homogenotization of heterozygous ES cells were also unsuccessful (data not shown). The failure to obtain Bard1 homozygous mutant ES clones suggests that Bard1 function is essential for the viability and/or proliferation of ES cells.
Phenotypes of Bard1; Brca1 double-mutant embryos.
To assess the combined effect of Bard1 and Brca1 deficiency on embryonic development, we examined and genotyped 154 E9.5 embryos derived from intercrosses of Bard1+/−; Brca1+/− double heterozygous animals. Embryos of all nine possible genotypes were recovered at the expected Mendelian ratios (Table 2). As expected, all embryos that carried at least one wild-type allele of both Bard1 and Brca1 were normal and had 20 to 25 somites, while each embryo that was null for either Bard1 or Brca1 exhibited the characteristic phenotype of severe growth retardation and degeneration. Interestingly, the 10 embryos with a double-mutant Bard1−/−; Brca1−/− genotype were phenotypically indistinguishable from either single Bard1 or single Brca1 homozygous mutants (Fig. 4B).
TABLE 2.
Phenotype analysis of E9.5 embryos from intercrosses of Bard1+/−; Brca1+/− double heterozygotes
| Genotype | No. of E9.5 embryos
|
|
|---|---|---|
| Observed | Expected | |
| Bard1+/+; Brca1+/+ | 11 | 10 |
| Bard1+/−; Brca1+/+ | 16 | 19 |
| Bard1+/+; Brca1+/− | 23 | 19 |
| Bard1+/−; Brca1+/− | 39 | 38 |
| Bard1−/−; Brca1+/+ | 12 | 10 |
| Bard1−/−; Brca1+/− | 16 | 19 |
| Bard1+/+; Brca1−/− | 10 | 10 |
| Bard1+/−; Brca1−/− | 17 | 19 |
| Bard1−/−; Brca1−/− | 10 | 10 |
| Total no. of embryos | 154 | |
FIG. 4.
Gross morphologies of wild-type, single homozygous mutant, and double homozygous mutant embryos. (A) Normal (Bard1+/−) and mutant (Bard1−/−) embryos at E7.5. (B) A normal (Bard1+/−; Brca1+/−) E9.5 embryo is shown on the left. A Bard1; Brca1 (bd1/br1) double homozygous mutant embryo is shown in comparison to a single Bard1 (bd1) mutant (heterozygous for Brca1) and a single Brca1 (br1) nullizygote (wild type for Bard1) that exhibit similar extents of development. (C) A normal (Bard1+/+; p53−/−) E9.5 embryo on the right compared to a Bard1; p53 double homozygous mutant (Bard1−/−; p53−/−) E9.5 embryo on the left. Note that the Bard1; p53 double homozygous mutant E9.5 embryo has developed much further than any single Bard1−/− embryo. Scale bar, 300 μm (A) and 800 μm (B and C).
Partial rescue of Bard1−/− embryos by loss of p53.
To ascertain genetically whether the early embryonic lethality of the Bard1 mutation could be mitigated by altering p53 levels, as shown previously for Brca1 null mutations (15, 33), we placed the Bard1 null allele in a p53-deficient background by intercrossing Bard1+/−; p53−/− males and Bard1+/−; p53+/− females. No double homozygous (Bard1−/−; p53−/−) mutant mice were found among 74 genotyped offspring (the expected number was nine), indicating that the loss of p53 is not sufficient to sustain viability of Bard1−/− mutants. To investigate the possibility of a partial rescue during embryonic development, we dissected 101 embryos at E9.5 and genotyped each by Southern and/or PCR analysis. Embryos of all six genotypes were detected with the expected frequencies (Table 3). Wild-type Bard1+/+ and heterozygous Bard1+/− embryos, regardless of their p53 status, appeared to be normal, with 20 to 25 somites, while each of the Bard1−/−; p53+/− embryos (n = 12) was already in an advanced stage of resorption and contained only embryonic remnants in a small yolk sac. On the other hand, all 13 embryos genotyped as Bard1−/−; p53−/− double homozygotes were still alive at day 9.5 of gestation. However, complete rescue of the Bard1 mutation did not occur, as these mice were developmentally retarded compared to their wild-type and heterozygous littermates, as shown by their small sizes and open neural tubes (Fig. 4C). Thus, the embryonic lethality of Bard1-null mice, like that of Brca1-null animals, is partly—but not completely—mitigated in the absence of p53.
TABLE 3.
Partial rescue of Bard1−/− mutants by loss of p53
| Genotype | No. of E9.5 embryos
|
|
|---|---|---|
| Observed | Expected | |
| Bard1+/+; p53+/− | 14 | 13 |
| Bard1+/−; p53+/− | 27 | 25 |
| Bard1−/−; p53+/− | 8a | 13 |
| Bard1+/+; p53−/− | 12 | 13 |
| Bard1+/−; p53−/− | 28 | 25 |
| Bard1−/−; p53−/− | 13b | 13 |
| Total no. of embryos | 102 | |
Eight Bard1−/−; p53+/− embryos were severely retarded and undergoing resorption.
All Bard1−/−; p53−/− embryos were alive at E9.5.
Interdependence of BRCA1 and BARD1 protein levels.
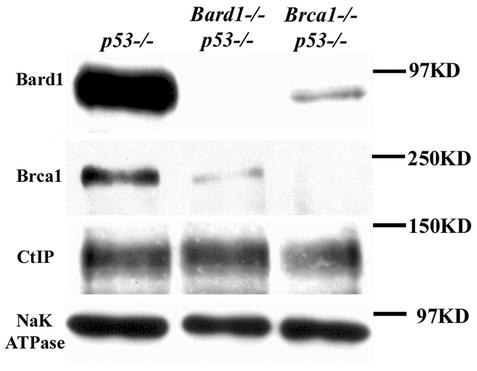
The partial rescue of some Bard1 mutant embryos in a p53-negative background to more advanced developmental stages allowed us to obtain sufficient embryonic protein for Western analysis. Therefore, to confirm that the null Bard1 mutation indeed results in loss of Bard1 expression, lysates of E9.5 Bard1−/−; p53−/− embryos were immunoblotted with an antiserum raised against murine Bard1 (Fig. 5). As expected, the Bard1 polypeptide was absent from Bard1−/−; p53−/− lysates. Notably, however, levels of Brca1 protein were dramatically reduced or absent in these lysates, while the levels of a control protein (NaK-ATPase) and another Brca1-associated protein (Ctip) were not affected. In a reciprocal manner, the levels of Bard1 polypeptides were substantially reduced or absent in Brca1−/−/ p53−/− embryos. Thus, there appears to be an obligate requirement for coexpression of both proteins in order to maintain normal endogenous levels of Bard1 and Brca1 in early mouse embryos.
FIG. 5.
Reciprocal stability control of Bard1 and Brca1 proteins in vivo. Protein lysates of Bard1+/+; p53−/−, Bard1−/−; p53−/−, and Brca1−/−; p53−/− embryos were analyzed by Western blotting. Anti-NaK-ATPase was used as a loading control.
Increased chromosomal instability in Bard1−/−; p53−/− embryos.
In keeping with the notion that Brca1 normally serves to promote genomic stability, aneuploidy is a common feature of chromosomal spreads from Brca1−/−/ p53−/− embryos bearing a hypomorphic Brca1 mutation (Brca1Δ11) (9). To determine whether Bard1 inactivation also causes aneuploidy, we compared mitotic spreads of Bard1+/+; p53−/− and Bard1−/−; p53−/− embryos at E9.5. In p53-deficient embryos that carry at least one wild-type Bard1 allele, we found that approximately 10% of the cells (9 of 94 metaphase spreads) had a reduced number of chromosomes (Table 4). In contrast, however, 44.7% of mitotic spreads from Bard1−/−; p53−/− embryos (42 out of 94) displayed numerical abnormalities (Table 4): 37 spreads (39.4%) had a reduced number of chromosomes, while 5 (5.3%) contained more than 40 chromosomes. The chromosome counts in those five spreads were 42, 44, 45, 56, and 83 chromosomes. The accumulation of chromosomal abnormalities in Bard1−/−; p53−/− embryos suggests a role for Bard1 in the maintenance of genomic integrity.
TABLE 4.
Cytogenetic abnormalities in Bard1−/−; p53−/− embryos
| Genotype | No. of embryos with the following no. of chromosomes/cell:
|
Total no. of spreads | % Abnormal | ||||
|---|---|---|---|---|---|---|---|
| ≤36 | 37-38 | 39-40 | 41-50 | ≥51 | |||
| Bard1+/+; p53−/− | 1 | 8 | 85 | 0 | 0 | 94 | 9.6 |
| Bard1−/−; p53−/− | 7 | 30 | 52 | 3 | 2 | 94 | 44.7 |
DISCUSSION
The only enzymatic property yet ascribed to BRCA1, its ubiquitin E3 ligase activity, is likely to play a central role in BRCA1 function (7, 17, 32, 39). It is noteworthy that this activity is dramatically enhanced in the presence of BARD1 (7, 17). This, together with the evidence that most cellular BRCA1 polypeptides exist in association with BARD1 (53), suggests that the BRCA1/BARD1 heterodimer is the natural mediator of BRCA1-dependent ubiquitination and as such may be responsible for most BRCA1 functions, including those required for tumor suppression (2). Consistent with this hypothesis, we have found that Bard1- and Brca1-null mice are phenocopies of one another, at least with respect to the parameters examined here. Hence, Bard1-null mice experience an early embryonic lethality (between E7.5 and E8.5) that is not associated with increased levels of cell death. Instead, two separate lines of evidence argue that the developmental retardation and death of Bard1-null embryos reflect an intrinsic defect in cell proliferation: (i) incorporation of BrdU is reduced in Bard1−/− nullizygous embryos relative to that in their wild-type (Bard1+/− or Bard1+/+) littermates, and (ii) the ICMs of Bard1−/− blastocysts, but not those of wild-type blastocysts, fail to proliferate in vitro. In addition, Bard1 targeting of heterozygous (Bard1+/−) ES cells by homologous recombination repeatedly disrupted the inactive, but not the active, Bard1 allele, suggesting that Bard1 function is also essential for the viability and/or proliferation of ES cells. Interestingly, as is the case for Brca1-null mice (33), the proliferation defect of Bard1-null mice appears to affect the embryo proper more profoundly than the extraembryonic tissues.
BRCA1-deficient cells are hypersensitive to a range of genotoxic agents and display defects in several distinct repair pathways, such as homology-directed DNA repair and nucleotide excision repair (1, 3, 11, 13, 16, 30, 36, 37, 41, 45, 46). These findings indicate an important role for BRCA1 in the cellular response to DNA damage and raise the possibility that BRCA1 normally suppresses tumor formation through its capacity to preserve genome integrity. Although the biochemical basis for BRCA1 function in the DNA damage response is not understood, BRCA1-deficient cells are known to accumulate DNA damage in the form of increased aneuploidy, chromosomal rearrangements, and double-strand DNA breaks (41, 51). The increased aneuploidy of Bard1-null embryonic cells indicates that Bard1 is also required for preservation of genomic stability, presumably through its participation in the Brca1/Bard1 heterodimer.
It has been proposed that the accumulating DNA damage associated with BRCA1 deficiency induces cell cycle checkpoints that are in turn responsible for the cell proliferation defects and embryonic lethality of Brca1-null mice (5, 40). This hypothesis is supported by the observation that p53 nullizygosity partly mitigates the developmental defects of Brca1-null embryos (14, 33). Here we found that the embryonic lethality of Bard1-null mice is also rescued to a similar extent in a p53-null background. Thus, the timings of embryonic deaths are indistinguishable between Bard1- and Brca1-null mice whether in the presence (lethality at E7.5 to E8.5) or absence (lethality at E9.5 to E10.5) of p53 function. These data, together with the observation of increased aneuploidy in Bard1-null cells, argue that the genomic lesions associated with Bard1 deficiency also retard cell proliferation and embryonic development by inducing cell cycle checkpoints—at least some of which are dependent on p53. That these phenotypic effects reflect functional inactivation of the Brca1/Bard1 heterodimer is supported by (i) the analysis of double homozygous Bard1−/−; Brca1−/− animals, which suffer embryonic lethality in the same manner as single homozygous Bard1- and Brca1-null mice, and (ii) the severe reduction or absence of Brca1 and Bard1 proteins in Bard1; p53 and Brca1/p53 mutant embryos, respectively. This observation also supports a model in which the functions attributed to Brca1 and Bard1, at least to the extent that they are manifested in early embryogenesis, are mediated by the Brca1/Bard1 heterodimer. Reciprocal stabilization of BRCA1 and BARD1 proteins has also been observed in frog embryos depleted of xBRCA1 or xBARD1 polypeptides by treatment with antisense oligonucleotides (26) and in mammalian cells that transiently overexpress exogenous BRCA1 or BARD1 polypeptides (17). We cannot exclude the possibility that these proteins also function independently of one another in some biological settings. However, the striking similarity between the phenotypes of single Brca1- and Bard1-null animals, together with the lack of additive effects in the Bard1−/−; Brca1−/− double mutants, suggests that Brca1 and Bard1 do not have essential independent functions in early embryonic development.
Interestingly, frog embryos depleted of xBRCA1 or xBARD1 polypeptides also display developmental defects and increased numbers of aneuploid nuclei (26). In these experiments, however, the developmental abnormalities appeared to arise during late embryogenesis, without overtly affecting germ layer formation or gastrulation. It is unclear why the impact of BRCA1 and BARD1 inactivation is less severe in frogs. On one hand, it may reflect phylogenetic differences in the functions of the BRCA1/BARD1 heterodimer in amphibians and mammals—a plausible possibility given the low level of amino acid sequence conservation among BRCA1 orthologs (26, 38). On the other hand, a simpler explanation might be that antisense treatment depletes, rather than abolishes, expression of the xBRCA1 and xBARD1 polypeptides. Regardless, the fact that aneuploidy is induced in both frogs and mice by either BRCA1 or BARD1 deficiency suggests that maintenance of chromosomal stability is a fundamental function of the BRCA1/BARD1 heterodimer that has been conserved throughout vertebrate evolution.
Acknowledgments
We thank Mian Su for expert technical assistance, M. Mendelsohn for blastocyst injections, S. Ganesan and D. Livingston for the Brca1 antibody, V. Murty for suggestions regarding the cytogenetic analysis, and A. Efstratiadis for helpful discussions and invaluable support.
This work was supported by the Avon Products Foundation Breast Cancer Research and Care Program (T.L.), NIH grant R01-CA76334 (R.B.), and NIH Cancer Biology Training Grant T32-CA09503 (E.E.M.).
REFERENCES
- 1.Abbott, D. W., M. E. Thompson, C. Robinson-Benion, G. Tomlinson, R. A. Jensen, and J. T. Holt. 1999. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J. Biol. Chem. 274:18808-18812. [DOI] [PubMed] [Google Scholar]
- 2.Baer, R., and T. Ludwig. 2002. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12:86-91. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya, A., U. S. Ear, B. H. Koller, R. R. Weichselbaum, and D. K. Bishop. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899-23903. [DOI] [PubMed] [Google Scholar]
- 4.Brodie, S. G., and C. X. Deng. 2001. BRCA1-associated tumorigenesis: what have we learned from knockout mice? Trends Genet. 17:S18-S22. [DOI] [PubMed] [Google Scholar]
- 5.Brugarolas, J., and T. Jacks. 1997. Double indemnity: p53, BRCA and cancer. p53 mutation partially rescues developmental arrest in Brca1 and Brca2 null mice, suggesting a role for familial breast cancer genes in DNA damage repair. Nat. Med. 3:721-722. [DOI] [PubMed] [Google Scholar]
- 6.Brzovic, P. S., P. Rajagopal, D. W. Hoyt, M. C. King, and R. E. Klevit. 2001. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8:833-837. [DOI] [PubMed] [Google Scholar]
- 7.Chen, A., F. E. Kleiman, J. L. Manley, T. Ouchi, and Z. Q. Pan. 2002. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem. 277:22085-22092. [DOI] [PubMed] [Google Scholar]
- 8.Couch, F. J., B. L. Weber, et al. 1996. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Hum. Mutat. 8:8-18. [DOI] [PubMed] [Google Scholar]
- 9.Deng, C. X. 2001. Tumorigenesis as a consequence of genetic instability in Brca1 mutant mice. Mutat. Res. 477:183-189. [DOI] [PubMed] [Google Scholar]
- 10.Fabbro, M., J. A. Rodriguez, R. Baer, and B. R. Henderson. 2002. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J. Biol. Chem. 277:21315-21324. [DOI] [PubMed] [Google Scholar]
- 11.Foray, N., V. Randrianarison, D. Marot, M. Perricaudet, G. Lenoir, and J. Feunteun. 1999. Gamma-rays-induced death of human cells carrying mutations of BRCA1 or BRCA2. Oncogene 18:7334-7342. [DOI] [PubMed] [Google Scholar]
- 12.Ghimenti, C., E. Sensi, S. Presciuttini, I. M. Brunetti, P. Conte, G. Bevilacqua, and M. A. Caligo. 2002. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer 33:235-242. [DOI] [PubMed] [Google Scholar]
- 13.Gowen, L. C., A. V. Avrutskaya, A. M. Latour, B. H. Koller, and S. A. Leadon. 1998. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science 281:1009-1012. [DOI] [PubMed] [Google Scholar]
- 14.Hakem, R., J. L. de la Pompa, A. Elia, J. Potter, and T. W. Mak. 1997. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat. Genet. 16:298-302. [DOI] [PubMed] [Google Scholar]
- 15.Hakem, R., J. L. de la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmair, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 16.Hartman, A. R., and J. M. Ford. 2002. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat. Genet. 32:180-184. [DOI] [PubMed] [Google Scholar]
- 17.Hashizume, R., M. Fukuda, I. Maeda, H. Nishikawa, D. Oyake, Y. Yabuki, H. Ogata, and T. Ohta. 2001. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276:14537-14540. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi, Y., M. Koike, M. Matsutani, and T. Hoshino. 1988. Effects of fixation time and enzymatic digestion on immunohistochemical demonstration of bromodeoxyuridine in formalin-fixed, paraffin-embedded tissue. J. Histochem. Cytochem. 36:511-514. [DOI] [PubMed] [Google Scholar]
- 19.Hohenstein, P., M. F. Kielman, C. Breukel, L. M. Bennett, R. Wiseman, P. Krimpenfort, C. Cornelisse, G. J. van Ommen, P. Devilee, and R. Fodde. 2001. A targeted mouse Brca1 mutation removing the last BRCT repeat results in apoptosis and embryonic lethality at the headfold stage. Oncogene 20:2544-2550. [DOI] [PubMed] [Google Scholar]
- 20.Irminger-Finger, I., W. C. Leung, J. Li, M. Dubois-Dauphin, J. Harb, A. Feki, C. E. Jefford, J. V. Soriano, M. Jaconi, R. Montesano, and K. H. Krause. 2001. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol. Cell 8:1255-1266. [DOI] [PubMed] [Google Scholar]
- 21.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Y., X. L. Xu, M. C. Yang, F. Wei, T. C. Ayi, A. M. Bowcock, and R. Baer. 1997. Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc. Natl. Acad. Sci. USA 94:12075-12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, R. D., and M. Jasin. 2001. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29:196-201. [DOI] [PubMed] [Google Scholar]
- 26.Joukov, V., J. Chen, E. A. Fox, J. B. Green, and D. M. Livingston. 2001. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. USA 98:12078-12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleiman, F. E., and J. L. Manley. 2001. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104:743-753. [DOI] [PubMed] [Google Scholar]
- 28.Kleiman, F. E., and J. L. Manley. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285:1576-1579. [DOI] [PubMed] [Google Scholar]
- 29.Koonin, E. V., S. F. Altschul, and P. Bork. 1996. BRCA1 protein products… functional motifs. Nat. Genet. 13:266-268. [DOI] [PubMed] [Google Scholar]
- 30.Le Page, F., V. Randrianarison, D. Marot, J. Cabannes, M. Perricaudet, J. Feunteun, and A. Sarasin. 2000. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. 60:5548-5552. [PubMed] [Google Scholar]
- 31.Liu, C. Y., A. Flesken-Nikitin, S. Li, Y. Zeng, and W. H. Lee. 1996. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 10:1835-1843. [DOI] [PubMed] [Google Scholar]
- 32.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig, T., D. L. Chapman, V. E. Papaioannou, and A. Efstratiadis. 1997. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11:1226-1241. [DOI] [PubMed] [Google Scholar]
- 34.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66-71. [DOI] [PubMed] [Google Scholar]
- 35.Monteiro, A. N. 2000. BRCA1: exploring the links to transcription. Trends Biochem. Sci. 25:469-474. [DOI] [PubMed] [Google Scholar]
- 36.Moynahan, M. E., J. W. Chiu, B. H. Koller, and M. Jasin. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4:511-518. [DOI] [PubMed] [Google Scholar]
- 37.Moynahan, M. E., T. Y. Cui, and M. Jasin. 2001. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61:4842-4850. [PubMed] [Google Scholar]
- 38.Orelli, B. J., J. M. Logsdon, Jr., and D. K. Bishop. 2001. Nine novel conserved motifs in BRCA1 identified by the chicken orthologue. Oncogene 20:4433-4438. [DOI] [PubMed] [Google Scholar]
- 39.Ruffner, H., C. A. Joazeiro, D. Hemmati, T. Hunter, and I. M. Verma. 2001. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA 98:5134-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scully, R., J. Chen, A. Plug, Y. Xiao, D. Weaver, J. Feunteun, T. Ashley, and D. M. Livingston. 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88:265-275. [DOI] [PubMed] [Google Scholar]
- 41.Scully, R., S. Ganesan, K. Vlasakova, J. Chen, M. Socolovsky, and D. M. Livingston. 1999. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell 4:1093-1099. [DOI] [PubMed] [Google Scholar]
- 42.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 44.Shattuck-Eidens, D., M. McClure, J. Simard, F. Labrie, S. Narod, F. Couch, K. Hoskins, B. Weber, L. Castilla, M. Erdos, et al. 1995. A collaborative survey of 80 mutations in the BRCA1 breast and ovarian cancer susceptibility gene. Implications for presymptomatic testing and screening. JAMA 273:535-541. [PubMed] [Google Scholar]
- 45.Shen, S. X., Z. Weaver, X. Xu, C. Li, M. Weinstein, L. Chen, X. Y. Guan, T. Ried, and C. X. Deng. 1998. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene 17:3115-3124. [DOI] [PubMed] [Google Scholar]
- 46.Snouwaert, J. N., L. C. Gowen, A. M. Latour, A. R. Mohn, A. Xiao, L. DiBiase, and B. H. Koller. 1999. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene 18:7900-7907. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, A., J. L. de la Pompa, R. Hakem, A. Elia, R. Yoshida, R. Mo, H. Nishina, T. Chuang, A. Wakeham, A. Itie, W. Koo, P. Billia, A. Ho, M. Fukumoto, C. C. Hui, and T. W. Mak. 1997. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 11:1242-1252. [DOI] [PubMed] [Google Scholar]
- 48.Thai, T. H., F. Du, J. T. Tsan, Y. Jin, A. Phung, M. A. Spillman, H. F. Massa, C. Y. Muller, R. Ashfaq, J. M. Mathis, D. S. Miller, B. J. Trask, R. Baer, and A. M. Bowcock. 1998. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum. Mol. Genet. 7:195-202. [DOI] [PubMed] [Google Scholar]
- 49.Welcsh, P. L., and M. C. King. 2001. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10:705-713. [DOI] [PubMed] [Google Scholar]
- 50.Wu, L. C., Z. W. Wang, J. T. Tsan, M. A. Spillman, A. Phung, X. L. Xu, M. C. Yang, L. Y. Hwang, A. M. Bowcock, and R. Baer. 1996. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 14:430-440. [DOI] [PubMed] [Google Scholar]
- 51.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 52.Yagi, T., Y. Ikawa, K. Yoshida, Y. Shigetani, N. Takeda, I. Mabuchi, T. Yamamoto, and S. Aizawa. 1990. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proc. Natl. Acad. Sci. USA 87:9918-9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, X., and R. Baer. 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 275:18541-18549. [DOI] [PubMed] [Google Scholar]