Abstract
The high-mobility-group A2 protein (HMGA2) plays important functional roles in transcriptional regulation, DNA replication and chromatin structure. In this study, the effect of histone deacetylase inhibition on the transcriptional activity of the Hmga2 gene was investigated in vivo both at the endogenous gene level and in a variety of cell lines using transiently transfected promoter constructs. Trichostatin A (TSA) repressed both transfected murine and human Hmga2 promoter constructs 3–8-fold in NIH3T3, F9 and HeLa cells. Steady-state Hmga2 mRNA levels in NIH3T3 cells decreased 4–5-fold following TSA treatment, while pre- treatment of NIH3T3 cells with the transcriptional inhibitor, actinomycin D, completely blocked TSA mediated repression of the Hmga2 gene. Cross-linked chromatin immunoprecipitation (X-ChIP) analysis revealed a 5–6-fold decrease in endogenous Hmga2 promoter bound Sp1 and Sp3 proteins following TSA treatment in parallel with observed loss of acetylated histone H3 and H4. In addition, the poly-pyrimidine-tract-binding protein (PTB) was observed to bind to the Hmga2 promoter in both TSA treated and untreated NIH3T3 cells. Together, these results suggest TSA treatment leads to a decrease in Hmga2 gene transcription, and a significant decrease in promoter bound Sp1, Sp3 and acetylated histones H3 and H4.
INTRODUCTION
The high-mobility-group (HMG) non-histone chromosomal proteins HMGA1a, A1b and A2 proteins comprise a subgroup of HMG chromatin accessory factors, often referred to as architectural proteins (1–3). These proteins are abundant low molecular mass nuclear proteins of ∼100 amino acids which each possess three copies of a nine amino acid motif (AT-hook) that interacts with the minor groove of many AT-rich promoter and enhancer DNA regulatory elements (4). These HMG proteins possess no intrinsic transcriptional activity, but function to orchestrate the assembly of nucleoprotein structures involved in gene transcription, replication and overall chromatin structure through a complex network of protein–DNA and protein–protein interactions (5–7).
Hmga2 mRNA and protein is detected at high levels during the early stages of murine embryonic development (i.e. 10–14 d.p.c.), but not at 15 d.p.c., or in newborn tissues, as expression decreases rapidly with the beginning of organogenesis and becomes extinguished in both murine and human adult somatic tissues (8,9). Hmga2 becomes re-expressed, however, in many transformed cells and in experimental tumors, and is thought to contribute to the overall transformation process (10–12). A role for HMGA2 in mouse development is underscored by the finding that inactivation of the Hmga2 gene results in the pygmy mouse phenotype, which exhibits growth retardation, and a significant reduction of overall body adipose tissue (13).
Obesity-induced conditions in normal mice have been shown to increase adipocyte number due to an expansion of pre-adipocyte cells, and this results in measurable Hmga2 mRNA expression in mature adipocyte tissues under these conditions (14). Mice with a partial or complete deficiency of Hmga2 expression have been shown to resist diet-induced obesity, while disruption of the Hmga2 gene in Lepob/Lepob mice results in a reduction in obesity observed in leptin deficient ob/ob mice. Recently, the testes of Hmga2 null mice have been observed to contain few spermatids, completely lack mature spermatozoa and are not fertile at the homozygous state (15). Together, these results suggest HMGA2 plays an important role in normal embryonic development, pre-adipocyte cell proliferation and in mouse spermatogenesis.
The mouse and human Hmga2 proximal promoter DNA regions are highly conserved (16). These promoters contain no consensus TATA-box, but possess DNA-binding sites for the Sp1, Sp3, CTF/NF1 transcription factors as measured using mobility-shift and footprinting assays. In addition to these elements, an ∼60 bp polypyrimidine/polypurine (ppyr/ppur) region consisting of TCC repeats has been identified as an important functional element in vivo (17–19). The molecular events which serve to activate the Hmga2 gene transcription during normal embryonic development, and to repress expression in adult tissues are not well understood, nor have any distant enhancer elements been identified which may be involved in its developmental and tissue-specific transcriptional regulation.
In this study, we have utilized the histone deacetylase (HDAC) inhibitor, trichostatin A (TSA), to investigate the responsiveness of Hmga2 promoter constructs, and the endogenous Hmga2 gene, to changes in acetylation/deacetylation state, and to identify transcriptional regulatory proteins bound to the Hmga2 promoter in vivo. TSA treatment was found to reduce both transient Hmga2 promoter activity in NIH3T3, F9 and HeLa cells, and to significantly reduce the steady-state level of endogenous Hmga2 mRNA levels in NIH3T3 cells. Cross-linked chromatin immunoprecipitation (X-ChIP) analysis revealed poly-pyrimidine-tract-binding protein (PTB) occupancy levels on the murine Hmga2 promoter do not change significantly in response to TSA, whereas, the levels of Sp1, Sp3 and acetylated H3 and H4 levels on the Hmga2 promoter, decrease significantly following TSA treatment.
MATERIALS AND METHODS
Cell culture
NIH3T3, F9 and HeLa cells were obtained from the ATCC (Manassas, VA) and maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS) (Paragon Bioservices, Baltimore, MD) in a humidified incubator at 37°C with 5% CO2. TSA and actinomycin D (Act-D) were obtained from Calbiochem (San Diego, CA).
Reporter constructs
All plasmids were constructed by standard recombinant DNA techniques (20). DNA oligomers were synthesized by Invitrogen. Polymerase chain reactions (PCRs) were performed using normal murine or human genomic DNA (Promega), and the following specific promoter primer sets: mHMGA2, (5′ primer) 5′-GGGTCGCGAGGAATTCTTTC CCCGCCTAA-3′ and (3′ primer) 5′-GGGACGCGTGCC GCTACCACTGCCTCTAC-3′; mHMGA2 5′ deletion, (5′ primer) 5′-GGGTCGCGAACCTCCGCCACCCACTGCCC-3′ and (3′ primer) 5′-GGGACGCGTCGCTGCAGCCG CTCGGCCTC-3′; mHMGA2 3′ deletion, (5′ primer) 5′-GGGTCGCGAGGAATTCTTTCCCCGCCTAA-3′ and (3′ primer) 5′-GCTTAGGCTGCCGCCGCTG-3′; hHMGA2, (5′ primer) 5′-GGGTCGCGAGGAATTCTTTCCCCGCCTAA-3′ and (3′ primer) 5′-GCCTCCCGCCGCCGCTACCG-3′; hHMGA1, (5′ primer) 5′-GGGTCGCGAGGGCCCACAC GCCCTGGAAG-3′ and (3′ primer) 5′-GGGACGCGTGC TGGTAGCAAATGCGGATC-3′; hH2B, (5′ primer) 5′-CAATAGTAGTGCGTCTTCTG-3′ and (3′ primer) 5′-AGCACTGTGTAGCTATAAAGC-3′.
PCRs were performed for 30 cycles: 94°C for 1 min, 92°C for 30 s, 60°C for 30 s, 72°C for 1 min, followed by a final extension step at 72°C for 5 min using Taq polymerase (Qiagen). PCR products were gel-purified and ligated into the pCRII TA vector (Invitrogen). Promoter fragments were excised from pCRII by XhoI/HindIII digestion, and directionally cloned into the pGL3 Basic Luciferase reporter vector (Promega). The hH2B promoter was excised from pCRII with KpnI/XhoI, and cloned into these sites in pGL3 to create pGL3-hH2B. The pRL-TK vector (Promega) was digested with BglII/HindIII, and the 760 bp herpes simplex virus thymidine kinase (HSVTK) promoter was cloned into the corresponding sites in pGL3 to create pGL3-HSVTK. The 102 bp murine Hmga2 ppyr/ppur region (ppyr) (–109 to –8 bp) (17) was obtained from PCR of normal murine genomic DNA as described above using the following primers: (5′ primer) 5′-GGGTCGCGAACCTCCGCCACCCACTGCCC-3′ and (3′ primer) 5′-GGACGTCAGCGGCTGCCAA-3′, and cloned into the pCRII TA vector to create pCRII-ppyr. The KpnI/XhoI fragment derived from pCRII-ppyr was cloned into the corresponding sites in pGL3-HSVTK to create pGL3-ppyr-HSVTK. pGL3-H2B was digested with BamHI/HindIII, and the promoter fragment cloned into the BglII/HindIII sites in pGL3-ppyr-HSVTK, following gel-purification and removal of the HSVTK promoter, to create pGL3-ppyr-H2B. Promoter constructs were verified by DNA sequence analysis performed by Commonwealth Biotechnologies, Inc. (CBI; Richmond, VA) using an ABI Prism DNA sequencer.
Murine HMGA2 (mHMGA2) contains 480 bp of the murine Hmga2 promoter (8,16,17) from –298 to +182 bp (17), the mHMGA2 5′ deletion contains 274 bp of the proximal Hmga2 promoter from –115 to +158 bp inclusive of the TCC-rich ppyr/ppur region (17). The mHMGA2 3′deletion contains 325 bp of the Hmga2 promoter from –298 to +27 bp. Human HMGA2 (hHMGA2) contains 476 bp from –480 to –4 bp of the human Hmga2 promoter (18), and human HMGA1 (hHMGA1) contains 700 bp from –347 to +352 bp of the human Hmga1 promoter (21). Human H2B (hH2B) contains 200 bp of the human histone H2B promoter from –156 to +43 bp (22). Promoters are numbered according to their respective citations. Plasmid DNAs were transformed into the Escherichia coli strain, Top 10 (Invitrogen), and plasmid DNA was purified using Qiagen tip-100 columns (Qiagen) as described by the manufacturer.
Transient transfections and luciferase assays
NIH3T3 and HeLa cells were seeded in 12-well plates at 1 × 105 cells per well, and F9 at 2.5 × 105 per well. Prior to transfection, the cells were rinsed with serum-free DMEM, then cultured in 0.4 ml of serum-free DMEM. DNA constructs were incubated with Plus reagent (Invitrogen) in sterile 1× PBS (NIH3T3 and F9) or Opti-MEM (Invitrogen) (HeLa) for 15 min at room temperature. Lipofectamine (Invitrogen) (NIH3T3 and F9) or Lipofectin (Invitrogen) (HeLa cells, after 30 min incubation at room temperature in Opti-MEM) was then added, and the DNA mixture was incubated at room temperature for an additional 15 min. The DNA mixture was added to the cells, and incubated at 37°C for 3 h (NIH3T3, F9) or 5 h (HeLa). One milliliter of DMEM containing 15% FBS was then added to each well and the cells were incubated overnight at 37°C. The transfection media was replaced with fresh DMEM/10% FBS plus 200 ng/ml TSA or an equivalent volume of DMSO, and the cells were incubated for 24 h before harvest. Cells were processed using the Dual Luciferase Reporter Assay (Promega), and luciferase (LUC) activity was measured using a TD-20/20 Luminometer (Turner Designs). The pRL-null vector (Promega) was used as an internal control to normalize for transfection efficiency. TSA values were normalized to the DMSO control with or without the ppyr/ppur element, which was set to a value of 1.00, and are expressed as the standard error of the means. Duplicate or triplicate values of each construct were averaged from two to five experiments per cell line.
RNA isolation and semi-quantitative RT–PCR
Total RNA was extracted using RNAqueous (Ambion, Austin, TX) according to the manufacturer. RNA was quantified spectrophotometrically after treatment with DNA-free (Ambion). Extracted RNAs were additionally checked by analyzing aliquots on a 1% native agarose gel stained with ethidium bromide. RT–PCRs were generally performed with ∼1 µg of RNA in one-step RT–PCRs according to the manufacturer (Qiagen). Semi-quantitative RT–PCR analyses were performed using [α-32P]dCTP as described (23,24). Briefly, gene-specific PCRs were first titrated to determine linear ranges for product accumulation, then a fixed cycle number was used in subsequent 32P RT–PCRs. Negative controls were included that omitted the reverse transcription step to verify that PCR products were RNA dependent. Typical RT–PCRs were performed at 50°C for 30 min, 95°C for 15 min, 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, using 32 cycles for mHMGA2 and 25 cycles for GAPDH with a final extension step at 72°C for 5 min. The following specific RT–PCR primer sets were used: mHMGA2 primers (8), 5′-ATGGACTCATACACAGCAGCAG-3′ and 5′-AGGGAAT ATAGAGAGGAGAGAG-3′; and the murine GAPDH primers (8,25), 5′-TTAGCACCCCTGGCCAAGG-3′ and 5′-CTTACTCCTTGGAGGCCATG-3′.
RT–PCRs using mHMGA2 primers yield a 342 bp product (8) and using GAPDH primers a 540 bp PCR product (25). Amplified RT–PCR products were electrophoresed through a non-denaturing 5% polyacrylamide gel, dried and analyzed using a Molecular Dynamics Phosphoimager (Amersham-Pharmacia) and ImageQuant (Amersham-Pharmacia) software.
Trichostatin A and actinomycin D treatment
Two 100 mm cell culture dishes were seeded with 2 × 106 NIH3T3 cells and allowed to grow overnight to 70% confluency. The next day, TSA was added directly to the cells to a final concentration of 200 ng/ml. An equivalent volume of vehicle (DMSO) was added to the second cell culture dish. After 24 h incubation, cells were harvested and total RNA was prepared as described for RT–PCR analysis. In Act-D experiments, cells were pretreated with 4 µM Act-D for 30 min prior to TSA addition.
Cross-linked chromatin immunoprecipitation assay
We determined conditions to produce sheared chromatin of average lengths between 0.5 and 1.5 kb using a Branson Sonifer with a 1.5 mm micro-tip. Four cell culture plates of 70% confluent NIH3T3 cells were treated with TSA or DMSO as described above. Chromatin was cross-linked by the addition of formaldehyde to a final concentration of 1% and incubated for 30 min at 37°C. Cells were washed twice with PBS containing protease inhibitor cocktail (Sigma). Washed cells were counted and ∼1 × 106 cells were resuspended in 200 µl of SDS lysis buffer (26).
X-ChIP assays were performed essentially as described by Upstate Biotechnology (Lake Placid, NY). Each sample was sonicated using a Branson Sonifer at 30% intensity for 6 × 10 s bursts with a gap of 30 s between each burst. Cells were maintained on ice throughout. Sheared chromatin was centrifuged for 10 min at 14 000 g at 4°C, after which the supernatant was diluted 10-fold in dilution buffer (26). As input controls, 1% of each diluted supernatant was retained at this step. Sheared chromatin was pre-cleared for 1 h at 4°C with 50% Protein A Sepharose (Sigma), 20 µg of sonicated and sheared salmon sperm DNA and 50 µg of BSA. Sp1 (30 µg), Sp3 (10 µg), PTB (30 and 50 µg) antibodies (Santa Cruz) and acetylated histone H3 (5 µg) and acetylated histone H4 (5 µg) antibodies (Upstate Biotechnologies) were added to pre-cleared supernatant, and immunoprecipitates were allowed to form overnight at 4°C with end-over-end rocking. Immune complexes were incubated for 1 h at 4°C with 60 µl of Protein A Sepharose beads and 4 µg of sonicated and sheared salmon sperm DNA, then centrifuged gently for 5 min at 4°C. Beads were washed four times for 5 min, then eluted twice in 250 µl of fresh 1% SDS, 0.1 M NaHCO3. Samples were heated at 65°C for 4 h to reverse cross-links, then the DNA samples were phenol/chloroform extracted before ethanol precipitation. DNA was resuspended in 50 µl of H2O. X-ChIP samples including input controls were assayed by semi-quantitative 32P-PCR using the following primers: mHMGA2 promoter primers (16) to amplify a product of 477 bp, 5′-GGA ATTCTTTCCCCGCCTAA-3′ and 5′-GCCGCTACCACTG CCTCTAC-3′; murine H-rev107 promoter primers (27) to amplify a product of 280 bp, 5′-GAGGAGTGTG TATAGGCTG-3′ and 5′-CTTTGCTTGGGTCCCAGAA-3′; murine PO gene primers (28) to amplify a product of 552 bp, 5′-GTGATGCCCAGGGAAGACAGG-3′ and 5′-GTCTCCA GTCTTTATCAGCTG-3′; murine c-Ki-ras promoter primers (29) to amplify a product of 500 bp, 5′-GTCGGGG TCCGCGTTGGCCG-3′ and 5′-GAGCCGGCTTCATTC AGCGCCG-3′.
PCR conditions included one cycle of 3 min at 94°C; then 30 s at 94°C, 30 s at 60°C, 40 s at 72°C, for 30, 32, 36, 38 or 44 cycles, with a final extension step at 72°C for 5 min. X-ChIP 32P-PCR products were analyzed by phosphoimager analysis as described above for semi-quantitative RT–PCR.
RESULTS
TSA represses Hmga2 promoter activity
The murine and human Hmga2 promoters, and several control promoters were cloned into the pGL3 Basic LUC vector (Fig. 1), and tested in transient transfection assays to assess the effect of TSA on promoter activity in NIH3T3, F9 and HeLa cell lines (Fig. 2). Both the full-length murine and human Hmga2 promoters were down-regulated from 50 to 88%, in response to TSA in these cell lines (Fig. 2A–C, lanes 1 and 4). Of particular note, the murine Hmga2 promoter deletions which retain the TCC-rich ppyr/ppur region (17,19) were also down-regulated by TSA (Fig. 2A–C, lanes 2 and 3), suggesting Hmga2 promoter TSA responsiveness may require this ppyr/ppur region. TSA had no, or little, effect on the activity of the control human Hmga1 promoter construct in NIH3T3 cells (Fig. 2A, lane 5) and HeLa cells (Fig. 2C, lane 5), respectively, and resulted in an ∼2-fold increase in the human Hmga1 promoter in F9 cells (Fig. 2B, lane 5). TSA increased histone H2B promoter LUC activity in both NIH3T3 and F9 cells (Fig. 2A and B, lane 6). The Hmga1, and H2B promoters serve as controls in these TSA experiments, as these promoters share no known common DNA elements with the Hmga2 promoter (21,22). The additional positive control promoter, HSVTK, exhibited a 5–27-fold induction in promoter activity in response to TSA (Fig. 2A–C, lane 7) (30). These results suggest TSA down-regulates the Hmga2 promoter at the transcriptional level.
Figure 1.
Schematic representation of promoter regions cloned into the pGL3 Basic vector and tested in vivo. The 5′ and 3′ base pair positions of each promoter are denoted below at the left and right. The location of each promoter initiation site is denoted by a bent arrow. The TCC-rich ppyr region is denoted by a hatched box and its base pair position denoted. hH2B ppyr and HSVTK ppyr constructs contain the murine Hmga2 ppyr element. Promoter construction is described in Materials and Methods.
Figure 2.
TSA down-regulates Hmga2 promoter activity. Promoter constructs were transiently transfected into NIH3T3 (A), F9 (B) and HeLa (C) cells. Cells were treated with TSA or DMSO for 24 h, then LUC activity was measured. The fold change in promoter activity following TSA treatment is shown normalized to the internal control, pRL-null, and to the DMSO controls, which were set to a value of 1.00. Promoter constructs are labeled above each pair of comparisons (DMSO and TSA), which are numbered at the bottom. The mean fold change values are shown below each numbered comparison.
To examine the possible role of the murine Hmga2 ppyr/ppur element in TSA mediated transcriptional repression, the ppyr/ppur element was transferred to a 5′ position in the HSVTK and H2B promoters, and tested in NIH3T3 and F9 cells (Fig. 3). The ppyr/ppur element was observed to stimulate the HSVTK promoter ∼3-fold in both NIH3T3 and F9 cells, and stimulate the H2B promoter ∼5–6-fold in NIH3T3 cells in the absence of TSA. However, the ppyr/ppur element only slightly increased the activity of the H2B promoter in F9 cells in the absence of TSA (Fig. 3B). As observed previously, both the HSVTK and H2B promoters were activated by TSA in NIH3T3 and F9 cells. Transfer of the ppyr/ppur element to the HSVTK and H2B promoters, however, effectively blocked TSA mediated activation of these promoters in NIH3T3 cells (Fig. 3A). Similarly, in F9 cells, the ppyr/ppur element repressed the HSVTK promoter response 68% (∼3-fold) in TSA treated cells, however, in F9 cells the ppyr/ppur H2B promoter activity was increased ∼2-fold in response to TSA (Fig. 3B). These results suggest the murine Hmga2 ppyr/ppur element is capable of transferring TSA mediated repression to heterologous promoters, however, cell-type and promoter-specific differences were observed in one case. In F9 cells, and only in the case of the H2B promoter, a small increase in promoter activity was observed with the ppyr element following TSA treatment. These results suggest TSA mediated repression of Hmga2, and heterologous promoters containing the ppyr element, may depend on additional cell-specific factors and overall promoter-specific architecture.
Figure 3.
Effect of TSA on heterologous promoters containing the Hmga2 ppyr/ppur element. The murine Hmga2 ppyr/ppur element was positioned 5′ of the HSVTK and H2B promoters and compared to control promoter LUC activity following treatment with TSA or DMSO for 24 h in NIH3T3 (A) and F9 (B) cells. LUC activities were normalized to the internal control, pRL-null, and to the HSVTK and H2B promoter constructs with or without the ppyr/ppur element treated with DMSO which were set to a value of 1.00. Promoter constructs are denoted above each bar, and the mean fold change values are shown below for each construct.
TSA reduces steady-state Hmga2 mRNA levels at the transcriptional level
NIH3T3 cells are known to express high steady-state levels of Hmga2 mRNA (31). To determine if TSA is capable of regulating endogenous Hmga2 gene expression, Hmga2 mRNA levels in NIH3T3 cells were quantitated by RT–PCR analysis following TSA or DMSO treatment (Fig. 4). From two independent experiments we observed a 4.7-fold reduction in steady-state Hmga2 mRNA levels in TSA treated cells, suggesting TSA may affect the Hmga2 gene transcription rate, Hmga2 mRNA stability, or both. In further support of RT–PCR analysis of Hmga2 mRNA levels, northern blot analysis also revealed a significant decrease in the ∼3.7 kb Hmga2 mRNA species in TSA treated NIH3T3 cells (data not shown). To evaluate possible effects of TSA at the level of Hmga2 gene transcription, NIH3T3 cells were pre-treated with the transcriptional inhibitor, Act-D (27,32), 30 min prior to addition of either TSA or DMSO (27,33). From two independent experiments, RT–PCR analysis showed repressive effects of TSA on steady-state Hmga2 mRNA levels were blocked by pre-treatment with Act-D (Fig. 4). These results suggest TSA repression occurs at the Hmga2 gene transcriptional level in vivo.
Figure 4.
TSA represses transcriptional activity of the endogenous murine Hmga2 gene. (A) Semi-quantitative RT–PCR was performed on RNA derived from NIH3T3 cells treated with 200 ng/ml TSA or DMSO for 24 h. In addition, NIH3T3 cells were pre-treated for 30 min with or without 4 µM Act-D (lanes 3, 4, 7, 8). RNA samples were analyzed for Hmga2 (lanes 1–4) and GAPDH mRNA levels which served as an internal control (lanes 5–8). 32P-PCR products corresponding to Hmga2 and GAPDH were quantified, and Hmga2 levels were normalized to GAPDH. (B) Quantitated results are expressed as the average fold change of two experiments relative to the DMSO controls.
TSA reduces occupancy of acetylated histones H3 and H4 on the endogenous murine Hmga2 promoter
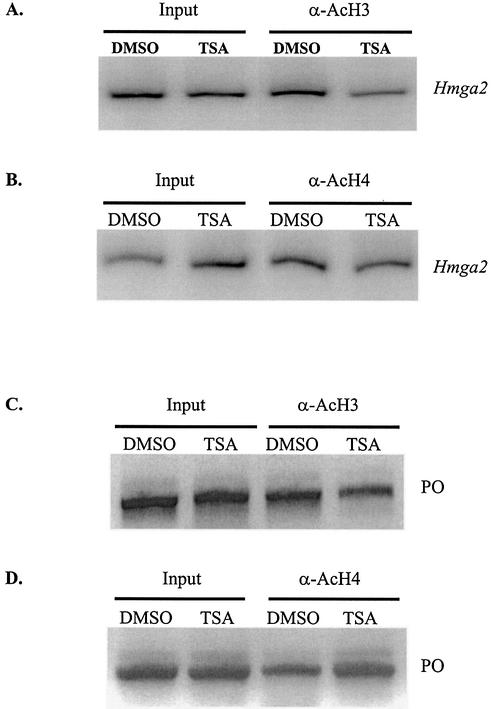
Histone acetylation is thought to be a crucial event in the remodeling of chromatin to allow greater accessibility of transcription factors to their DNA-binding sites resulting in an increased level of gene transcription. Localized acetylation of histones H3 and H4 in gene promoters have frequently been observed to be associated with active gene expression (34–36). The effect of TSA was further analyzed in the context of Hmga2 promoter localized histone H3 and H4 acetylation. Antibodies that specifically immunoprecipitate acetylated histones H3 and H4 were used in the X-ChIP assay (Fig. 5). An ∼500 bp region of the murine PO gene spanning the protein translational start site (28) was PCR amplified from input and immunoprecipitated acetylated H3 and H4 antibody X-ChIP samples at 38 and 44 PCR cycles, respectively, to verify sample integrity (Fig. 5C and D). X-ChIP analysis of DMSO treated cells revealed the presence of acetylated H3 and H4 histones (30 PCR cycles), on the Hmga2 promoter, commonly found associated with actively transcribed genes in vivo. Consistent with TSA induced Hmga2 gene transcriptional repression, X-ChIP analysis of TSA treated NIH3T3 cells showed a reduction of acetylated H3 and H4 histones (30 PCR cycles) by 2.2- and 2.5-fold, respectively, on the Hmga2 promoter (Fig. 5A and B).
Figure 5.
TSA treatment of NIH3T3 cells results in a decreased level of acetylated histone H3 and H4 on the murine Hmga2 promoter. X-ChIP analysis was performed on NIH3T3 cells treated with 200 ng/ml TSA or DMSO for 24 h using acetylated H3 and H4 antibodies. Cross-linked and sheared genomic DNA (input) serve as positive controls, and were used to normalize α-H3 (A) and α-H4 (B) IP results. In addition to the Hmga2 promoter (A and B) the PO gene (C and D) was detected by PCR amplification from input and IP samples to serve as a control for sample integrity.
Sp1 and Sp3 are present on the endogenous murine Hmga2 promoter
The ubiquitous transcription factors Sp1 and Sp3 have been implicated as positive transcriptional activators of the murine (17) and human Hmga2 genes (18), from in vitro DNA-binding assays, in vivo reporter assays and from in vivo transfection experiments performed in Drosophlia melanogaster SL-2 cells using Sp1 and Sp3 expression vectors. To determine if Sp1 and Sp3 proteins are indeed present on the endogenous Hmga2 promoter during active transcription in vivo, and to evaluate their potential status following TSA treatment, we performed X-ChIP analysis using NIH3T3 cells (Fig. 6). Using this approach, both Sp1 and Sp3 proteins were observed to be present on the murine Hmga2 promoter in both TSA treated and untreated cells (Fig. 6A and B, respectively). By normalizing each immunoprecipitation (IP) reaction to the corresponding input control we determined that the relative abundance of both Sp1 and Sp3 decreased 5–6-fold in TSA treated NIH3T3 cells as compared to control. However, 38 cycles were required to detect Sp3 bound to the Hmga2 promoter, whereas Sp1 was detected using 32 cycles in the X-ChIP assay in both control and TSA treated cells (Fig. 6C). These results suggest that both Sp1 and Sp3 proteins are bound to the murine Hmga2 promoter in vivo, and are both significantly reduced following TSA treatment. Both Sp1 and Sp3 have been identified on the murine H-rev107 promoter in NIH3T3 cells using the X-ChIP assay (27). As a positive control, we assayed our Sp1 and Sp3 X-ChIP samples, and likewise detected the murine H-rev107 promoter (Fig. 6D). Sp1 and Sp3 were not detected using 32 cycles in X-ChIP PCRs of the H-rev107 promoter, but required 36 cycles to detect.
Figure 6.

Sp1 and Sp3 are bound to the murine Hmga2 promoter in vivo. X-ChIP analysis was performed on NIH3T3 cells treated with 200 ng/ml TSA or DMSO for 24 h using Sp1 (A) and Sp3 (B) antibodies. Cross-linked and sheared genomic DNA (input) served as positive controls, and were used to normalize IP results. 32P-PCR bands corresponding to the Hmga2 promoter were quantified, and fold changes are expressed as a function of the X-ChIP results divided by the respective input control (C). As an additional control, input and IP samples from DMSO treated NIH3T3 cells were examined for the presence of the murine H-rev107 promoter (D).
PTB is bound to the murine Hmga2 and c-Ki-ras promoters in vivo
PTB is known to play a role in pre-mRNA splicing and mRNA translation (33,37,38), and is thought to participate in the control of gene transcription by binding to single-stand ppyr/ppur DNA elements in regulatory control regions (19,33). An ∼60 bp ppyr/ppur region consisting of TCC repeats has been identified in the Hmga2 promoter, and shown to function in promoter reporter assays (19). The murine c-Ki-ras promoter also possesses a ppyr/ppur region (19,29), whereas the murine acidic ribosomal phosphoprotein (PO; also known as 36B4) (28) gene lacks an identifiable ppyr/ppur element. The Hmga2 promoter together with these additional promoters were assayed for PTB binding using the X-ChIP assay, as additional controls were considered important, since to our knowledge, PTB has not been previously identified on any endogenous gene using the X-ChIP assay. To characterize the α-PTB reagent, two concentrations of α-PTB antibodies (30 and 50 µg) were tested to ensure complete IP, and both yielded identical results (data not shown). Results presented in Figure 7A demonstrate that PTB is bound to the endogenous murine Hmga2 promoter in vivo, with a 1.4-fold decrease in bound PTB in TSA treated NIH3T3 cells as compared to DMSO treated cells. To control for the specificity of the α-PTB X-ChIP assay a region of the murine PO gene spanning the protein translational start site (28) was PCR amplified from input X-ChIP samples at 38 PCR cycles. As expected, input samples yielded a PCR product corresponding to the PO gene, whereas α-PTB IP samples showed no PO product demonstrating the absence of PTB binding in this region of the PO gene in vivo (Fig. 7B). The murine c-Ki-ras promoter contains a shorter, but similar ppyr/ppur region as compared to Hmga2 promoter, and in vitro EMSA experiments demonstrate this ppyr/ppur element binds PTB (19,29). To provide a positive control for our α-PTB IP experiments, and to determine if PTB is actually bound to the endogenous murine c-Ki-ras promoter in vivo, we performed similar α-PTB X-ChIP assays using primers which span the c-Ki-ras ppyr/ppur element in this promoter (Fig. 7B). Our results suggest PTB is in fact bound to the c-Ki-ras promoter in vivo, and PTB levels decrease by a factor of 1.7-fold following TSA treatment in NIH3T3 cells. These results represent the first demonstration of PTB binding to ppyr/ppur containing promoters in vivo.
Figure 7.
PTB is bound to the murine Hmga2 and c-Ki-ras promoters in both TSA treated and untreated NIH3T3 cells. NIH3T3 cells were treated with 200 ng/ml TSA or DMSO for 24 h and analyzed using the X-ChIP assay employing α-PTB antibodies as described in Figures 5 and 6. Input and IP chromatin samples were analyzed using Hmga2 promoter-specific primers in 32P-PCRs, then quantified (A). In addition, input and IP samples were analyzed using murine PO gene-specific primers (B), and IP samples were analyzed using murine c-Ki-ras promoter-specific primers. The PO and c-Ki-ras IP reactions serve as negative and positive controls, respectively.
DISCUSSION
Hmga2 gene expression has been extensively characterized due to its important role in major cellular functions such as transcription, replication and control of chromatin structure. Overexpression of the Hmga2 gene is a common feature of many benign tumors of mesenchymal origin, and the role of Hmga2 in malignant tumor biology has been recently demonstrated where targeted anti-sense Hmga2 expression suppresses malignant transformation of thyroid cells by acute retroviruses (39).
Dynamic control of protein acetylation is now widely believed to be involved in regulating gene transcription through the interplay of histone acetyltransferase (HAT) and HDAC enzymes, which modify histones, transcription factors, and their associated cofactors and accessory chromatin factors. Specific HDAC inhibitors, such as TSA, have been employed to alter the level of histone acetylation, and to assess their functional impact on transcriptional activity of specific genes. Currently, over 10 HDAC inhibitors are in human clinical trials and represent a new anti-cancer therapeutic agent due to their ability to induce cell death and cell cycle arrest in tumor cells (40). HDAC inhibitors, such as TSA, may represent novel agents that decrease Hmga2 expression, thereby suppressing malignant cell transformation as has been shown using anti-sense methodologies (39). Recent microarray analysis of TSA effects on cellular gene expression in the human colon carcinoma cell line, SW620, suggests only a limited number of mammalian genes, ∼10% of >8000 genes examined, are significantly affected by HDAC inhibition (40,41). In the majority of genes examined using standard molecular biological assays, TSA treatment leads to an increase in gene expression (40). Of the few examples where inhibition of HDAC results in repression, TSA has been shown to decrease protein and mRNA levels of both the ETS domain transcription factor PU.1 (42) and the tetraspanin cell surface antigen CD9 (43) in murine macrophage cell lines.
To better understand the mechanisms of Hmga2 gene regulation we have investigated the effect of HDAC inhibition on the activity of the endogenous Hmga2 gene in vivo and Hmga2 promoter constructs in transfected cells. Transient transfection assays demonstrated both human and murine Hmga2 promoters were down-regulated following TSA treatment in both murine and human cell lines. Using both RT–PCR and northern analysis we observed a significant decrease in endogenous Hmga2 steady-state mRNA levels, while pretreatment of NIH3T3 with Act-D blocked this repression. Taken together, these results suggest repression of Hmga2 gene expression occurs at the transcriptional level.
Acetylated histones have been consistently observed associated with a more open and active chromatin environment leading to the hypothesis that HAT enzymes and histone acetylation are involved in transcriptional activation while HDAC enzymes and deacetylated histones are associated with gene repression. An increase in histone acetylation in promoters associated with TSA mediated gene induction has been observed in the majority of TSA inducible genes. A number of TSA inducible genes such as manganese superoxide dismutase (34), MKP-1 (35), HMG-CoA (36) and hLHR (26) have increased acetylated histone H3 and/or H4. Our data is consistent with this model in so far as acetylated histones H3 and H4 are present at the Hmga2 promoter under conditions favoring active gene transcription, but reduced in the repressed state. An interesting example of one of the few genes that are repressed by TSA, PU.1 associated histones H3 and H4 remained acetylated, and in the case of histone H4, TSA treatment actually increased its level of acetylation on the PU.1 promoter (42). It should be noted that TSA treatment not only leads to histone acetylation, but has also been shown to acetylate other transcription factors (44,45).
Recent studies have implicated a role for the Sp1 and Sp3 transcription factors in Hmga2 promoter activity and shown protein binding within the highly conserved TCC-rich ppyr/ppur region at position –83 to –25 bp (17–19). Ectopic expression of Sp1 in Drosophila SL-2 cells has also been shown to activate the murine Hmga2 promoter, while Sp3 enhances Sp1 functional activity but does not activate Hmga2 expression alone (17). In addition, a member of the hnRNP family, PTB, has been shown to bind in vitro within the Hmga2 ppyr/ppur tract when a single-stranded DNA conformation exists, suggesting PTB may play a functional role in Hmga2 transcription (19). Our results demonstrate that Sp1, Sp3 and PTB are bound to the Hmga2 promoter in vivo and suggest TSA mediated repression of the Hmga2 gene involves loss of bound Sp1 and Sp3 proteins, which is dependent on the Hmga2 ppyr/ppur region.
Sp1 and Sp3 are involved in transcriptional activation and repression of a large number of eukaryotic genes. In cases of TSA mediated activation, a number of genes, including p21waf1/cip1 (46), human telomerase reverse transcriptase (47) and manganese superoxide dismutase (41) depend on functional Sp1/Sp3 promoter sites. In general, HDAC enzymes are associated with Sp1 directly or present within these proximal promoter regions to maintain the repressed state, and dissociate following promoter activation by TSA (41,46–48). For both the human telomerase reverse transcriptase (47) and manganese superoxide dismutase (41) promoters, X-ChIP analyses show Sp1/Sp3 occupancy levels do not change with TSA mediated activation. These X-ChIP results suggest HDAC inhibition and gene activation do not involve any significant increase in promoter bound Sp1/Sp3 activator proteins, but may involve de-repression through the loss of bound HDAC enzymes. It is important to also note the levels of total nuclear extracted Sp1/Sp3 protein and its DNA-binding activity as measured using several distinct Sp1/Sp3 DNA-binding sites in these studies were unchanged following TSA treatment (26,27,41,46).
The X-ChIP assay provided an opportunity to identify proteins associated with the Hmga2 promoter in vivo. Our data provide the first evidence that both Sp1 and Sp3 transcription factors are bound to the Hmga2 promoter in vivo, and additionally that occupancy levels of Sp1 and Sp3 decrease significantly following TSA treatment. It is important to note, however, that the PCR cycle number was extended six cycles further to 38 cycles in order to detect and quantitate Sp3 levels. Loss of Hmga2 bound Sp1 and Sp3 proteins strongly suggests transcriptional repression is mediated through loss of these activator proteins, and to our knowledge this is the first demonstration of the loss of Sp1/Sp3 activator proteins following HDAC inhibition. It is also highly likely that the loss of Sp1/Sp3 in the presence of TSA is localized and specific to the Hmga2 promoter, as in other studies no TSA effects on total nuclear Sp1 protein abundance or DNA-binding activity have been observed (26,27). The presence of the ppyr/ppur element in the Hmga2 is known to provide a site for Sp1/Sp3 transcription factors and also for the PTB protein and may play a crucial role in overall repression of the Hmga2 promoter.
To further elucidate possible mechanisms of TSA mediated Hmga2 repression, the X-ChIP assay was performed with NIH3T3 cells to determine if PTB is bound in vivo to the endogenous Hmga2 and c-Ki-ras promoter ppyr/ppur elements, and if PTB protein occupancy levels change following HDAC inhibition. Previous studies have shown the murine Hmga2 promoter can adopt S1 nuclease sensitive structures in vitro which are capable of binding PTB (19), however, it is not known if single-stranded regions in the ppyr/ppur region exist in vivo. Our results demonstrate PTB is bound to the Hmga2 promoter in vivo, and show no significant decrease (1.4-fold) in the level of bound PTB in TSA treated cells as compared to untreated cells. These results suggest repression of Hmga2 gene transcription is not associated with increased levels of promoter bound PTB. PTB levels decreased 1.7-fold on the c-Ki-ras promoter following TSA treatment as compared to DMSO treatment. The significance of reduced PTB levels on the c-Ki-ras promoter in vivo are not known at present. Together, these X-ChIP analyses represent the first demonstration of PTB bound to ppyr/ppur element containing promoters in vivo, and suggest single-stranded H-DNA structures do indeed exist on these promoters in vivo. Although the precise role of PTB in both transcriptional activation and repression of the Hmga2 gene is not understood, TSA mediated repression of the Hmga2 gene may involve remodeling of PTB within the ppyr/ppur element such that double-stranded Sp1/Sp3 DNA-binding sites are converted into single-stranded sites following redistribution of PTB, which effectively reduces the level of bound Sp1/Sp3. Alternatively, TSA may stimulate covalent modification of Sp1/Sp3 proteins bound to the Hmga2 promoter that prevents their DNA-binding, leading to reduction in promoter bound Sp1/Sp3 (49). It is also possible that a repressor of Hmga2 gene transcription is induced by TSA and binds in the Hmga2 promoter region displacing Sp1 and Sp3. A bound repressor may result in histone deacetylation due to the loss of Sp1/Sp3 associated HAT activity and this may lead to remodeling of nucleosomes and a closed chromatin environment. An additional possibility is that TSA mediated transcriptional repression of the Hmga2 promoter is indirect where HDAC inhibition leads to acetylation of other non-histone chromosomal proteins such as p300 or CBP in addition to histones, which activates repressor expression (44,45).
Current studies are directed at determining if loss of histone acetylation is a result of deacetylation or remodeling of nucleosomes following TSA treatment and if known HAT enzymes, such as p300, CBP or pCAF, possibly associated with Sp1/Sp3, or within the Hmga2 ppyr/ppur region in vivo, may be displaced during HDAC inhibition. The inability of ectopically expressed p300 or CBP proteins (50) to activate the murine Hmga2 promoter in NIH3T3 cells, however, coupled with the inability of the adenovirus E1A 12S oncoprotein (51) to repress transient Hmga2 promoter activity, suggest these HAT enzymes and E1A 12S protein may not play a role in activation/repression of Hmga2 gene transcription (P.A.Henry, M.Ferguson and R.A.Currie, unpublished results). Further X-ChIP analysis of the endogenous Hmga2 gene for the presence of HAT activities and functional analysis of the Hmga2 promoter, should provide additional new insights into the mechanism of TSA mediated Hmga2 gene repression, and identification of basic components involved in Hmga2 transcriptional control.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by research grants, NIDDK 47272 and AI44111 from the National Institutes of Health to R.A.C.
REFERENCES
- 1.Wolffe A.P. (1996) Histone deacetylase: a regulator of transcription. Science, 272, 371–411. [DOI] [PubMed] [Google Scholar]
- 2.Grosschedl R., Giese,K. and Pagel J. (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet., 10, 38–42. [DOI] [PubMed] [Google Scholar]
- 3.Reeves R. (2000) Structure and function of the HMGI(Y) family of architectural transcription factors. Environ. Health Perspect., 108, 803–809. [DOI] [PubMed] [Google Scholar]
- 4.Reeves R. and Nissen,M.S. (1990) The A-T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem., 265, 8573–8582. [PubMed] [Google Scholar]
- 5.Thanos D. and Maniatis,T. (1995) Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell, 83, 1091–1100. [DOI] [PubMed] [Google Scholar]
- 6.John S., Reeves,R.B., Lin,J.-X., Child,R., Leiden,J.M., Thompson,C.B. and Leonard,W.J. (1995) Regulation of cell-type-specific interleukin-2 receptor α-chain gene expression. Potential role of physical interactions between Elf-1, HMG-I(Y) and NF-κB family proteins. Mol. Cell. Biol., 15, 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leger H., Sock,E., Renner,K., Grummt,F. and Wegner,M. (1995) Functional interaction between the POU domain Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol. Cell. Biol., 15, 3738–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X., Benson,K.F., Przybysz,K., Liu,J., Hou,Y., Cherath,L. and Chada,K. (1996) Genomic structure and expression of the murine Hmgi-c gene. Nucleic Acids Res., 24, 4071–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirning-Folz U., Wilda,M., Rippe,V., Bullerdiek,J. and Hameister,H. (1998) The expression pattern of the Hmgic gene during development. Genes Chromosomes Cancer, 23, 350–357. [PubMed] [Google Scholar]
- 10.Giancotti V., Buratti,E., Perissin,L., Zorzet,S., Balmain,A., Portella,G., Fusco,A. and Goodwin,G.H. (1989) Analysis of the HMGI nuclear proteins in mouse neoplastic cells induced by different procedures. Exp. Cell Res., 184, 538–545. [DOI] [PubMed] [Google Scholar]
- 11.Chiapetta G., Bandiera,A., Berlingiere,M.T., Visconti,R., Manfiolette,G., Battista,S., Martinez-Tello,F.J., Santoro,M., Giancotti,V. and Fusco,A. (1995) The expression of the high mobility group HMGI(Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene, 10, 1307–1314. [PubMed] [Google Scholar]
- 12.Vallone D., Battista,S., Pierantoni,G.,M., Fedele,M., Casalino,L., Santoro,M., Viglietto,G., Fusco,A. and Verde,P. (1997) Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J., 16, 5310–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X., Benson,K.F., Ashar,H.R. and Chada,K. (1995) Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature, 376, 771–774. [DOI] [PubMed] [Google Scholar]
- 14.Anand A. and Chada,K. (2000) In vivo modulation of Hmgic reduces obesity. Nature Genet., 24, 377–380. [DOI] [PubMed] [Google Scholar]
- 15.Chieffi P., Battista,S., Barchi, M, Di Agostino,S., Pierantoni,G.M., Federle,M., Chiariotti,L., Tramontano,D. and Fusco,A. (2002) HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene, 21, 3644–3650. [DOI] [PubMed] [Google Scholar]
- 16.Patel U.A., Bandiera,A., Manfiolette,G., Giancotti,V., Chau, K-Y. and Crane-Robinson,C. (1994) Expression and cDNA cloning of human HMGI-C phosphoprotein. Biochem. Biophys. Res. Commun., 201, 63–70. [DOI] [PubMed] [Google Scholar]
- 17.Rustighi A., Mantovani,F., Fusco,A., Giancotti,V. and Manfioletti,G. (1999) Sp1 and CTF/NF-1 transcription factors are involved in the basal expression of the Hmgi-c proximal promoter. Biophys. Res. Commun., 265, 439–447. [DOI] [PubMed] [Google Scholar]
- 18.Chau K.-Y., Arlotta,P., Patel,U.A., Crane-Robinson,C., Manfioletti,G. and Ono,S.J. (1999) A novel downstream positive regulatory element mediating transcription of the human high mobility group (HMG)I-C gene. FEBS Lett., 457, 429–436. [DOI] [PubMed] [Google Scholar]
- 19.Rustighi A., Tessari,M.A., Sgarra,R., Giancotti,V. and Manfioletti,G. (2002) A poly-pyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth related genes. Biochemistry, 41, 1229–1240. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J. and Russel,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Ogram S.A. and Reeves,R. (1995) Differential regulation of a multipromoter gene. Selective 12-o-tetradecanlylphorbol-13-acetate induction of a single transcription start site in the HMG-I/Y gene. J. Biol. Chem., 270, 14235–14242. [DOI] [PubMed] [Google Scholar]
- 22.Zhong R., Roeder,R.G. and Heintz,N. (1983) The primary structure and expression of four cloned human histone genes. Nucleic Acids Res., 11, 7409–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciolino H.P. and Yeh,G.C. (1999) The steroid hormone dehydroepiandrosterone inhibits CYP1A1 expression in vitro by a post-transcriptional mechanism. J. Biol. Chem., 274, 35186–35190. [DOI] [PubMed] [Google Scholar]
- 24.Ciolino H.P., Daschner,P.J. and Yeh,G.C. (1999) Dietary flavonols quercetin and kaempferol are ligands of the aryl hydrocarbon receptor that affect CYP1A1 transcription differentially. Biochem. J., 340, 715–722. [PMC free article] [PubMed] [Google Scholar]
- 25.Sabath D.E., Broome,H.E. and Prystowsky,M.B. (1990) Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene, 91, 185–191. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y. and Dufau,M.L. (2002) Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem., 277, 33431–33438. [DOI] [PubMed] [Google Scholar]
- 27.Roder K., Latasa,M.-J. and Sul,H.S. (2002) Murine H-rev107 gene encoding a class II tumor suppressor: gene organization and identification of an Sp1/Sp3-binding GC-box required for its transcription. Biochem. Biophys. Res. Commun., 293, 793–799. [DOI] [PubMed] [Google Scholar]
- 28.Krowczynska A.M., Coutts,M., Makrides,S. and Baweman,G. (1989) The mouse homologue of the human acidic ribosomal phosphoprotein PO: a highly conserved polypeptide that is under translational control. Nucleic Acids Res., 17, 6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman E.K., Trusko,S., Murphy,M. and George,D.L. (1990) An S1 nuclease-sensitive homopurine/homopyrimidine domain in the c-Ki-ras promoter interacts with a nuclear factor. Proc. Natl Acad. Sci. USA, 87, 2705–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safaya S., Ibrahim,A. and Rieder,R.F. (1994) Augmentation of γ-globin gene promoter activity by carboxylic acids and components of the human β-globin locus control region. Blood, 84, 3929–3935. [PubMed] [Google Scholar]
- 31.Battista S., Fidanza,V., Fedele,M., Klein-Szanto,A.J., Outwater,E., Brunner,H., Santoro,M., Croce,C. and Fusco,A. (1999) The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res., 59, 4793–4797. [PubMed] [Google Scholar]
- 32.Kloss S., Furneaux,H. and Mulsch,A. (2003) Posttranscriptional regulation of soluble guanylcyclase expression in rat aorta. J. Biol. Chem., 278, 2377–2383. [DOI] [PubMed] [Google Scholar]
- 33.Valcarcel J. and Gebauer,F. (1997) Post-transcriptional regulation: the dawn of PTB. Curr. Biol., 7, 705–708. [DOI] [PubMed] [Google Scholar]
- 34.Maehara K., Uedawa,N. and Isobe,K.-I. (2002) Effects of histone acetylation on transcriptional regulation of manganese superoxide dismutase gene. Biochem. Biophys. Res. Commun., 295, 187–192. [DOI] [PubMed] [Google Scholar]
- 35.Gorospe J.L., Hutter,D., Barnes,B., Keyse,S. and Liu,Y. (2001) Transcriptional induction of MKP-1 in response to stress is associated with histone H3 phosphorylation-acetylation. Mol. Cell. Biol., 21, 8213–8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camarero N., Nadal,A., Barrero,M.J., Haro,D. and Marrero,P.F. (2003) Histone deacetylase inhibitors stimulate mitochondrial HMG-CoA synthase gene expression via a promoter proximal Sp1 site. Nucleic Acids Res., 31, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunel F., Alzari,P.M, Ferrara,P. and Zakin,M.M. (1991) Cloning and sequencing of PYBP, a pyrimidine-rich specific single strand DNA-binding protein. Nucleic Acids Res., 19, 5237–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunel F., Zakin,M.M., Buc,H. and Buckle,M. (1996) The polypyrimidine tract binding (PTB) protein interacts with single-stranded DNA in a sequence-specific manner. Nucleic Acids Res., 24, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berlingieri M.T., Manfioletti,G., Santoro,M., Bandiera,A., Visconti,R., Giancotti,V. and Fusco,A. (1995) Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol. Cell. Biol., 15, 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Johnstone R.W. (2002) Novel deacetylase inhibitors: novel drugs for the treatment of cancer. Nature Rev. Drug Discov., 1, 287–299. [DOI] [PubMed] [Google Scholar]
- 41.Mariadason J.M., Corner,G.A. and Augenlicht,L.H (2000) Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac and curcumin and implications for chemoprevention of colon cancer. Cancer Res., 60, 4561–4572. [PubMed] [Google Scholar]
- 42.Laribee R.N. and Klemsz,M.J. (2001) Loss of PU.1 expression following inhibition of histone deacetylases. J. Immunol., 167, 5160–5166. [DOI] [PubMed] [Google Scholar]
- 43.Wang X.-Q., Alfaro,M.L., Evans,G.F. and Zuckerman,S.H. (2002) Histone deacetylase inhibition results in decreased macrophage CD9 expression. Biochem. Biophys. Res. Commun., 294, 660–666. [DOI] [PubMed] [Google Scholar]
- 44.Sterner D.E. and Berger,S. (2000) Acetylation of histones and transcription related factors. Micro. Mol. Biol. Rev., 64, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Q., Hutchins,A.E., Doyle,C.M., Lundblad,J.R. and Kwok,R.P.S. (2003) Acetylation of CREB by CBP enhances CREB-dependent transcription. J. Biol. Chem., 278, 15727–15734. [DOI] [PubMed] [Google Scholar]
- 46.Xiao H., Hasegawa,T. and Isobe,K.-I. (1999) Both Sp1 and Sp3 are responsible for p21waf1 promoter activity induced by histone deacetylase inhibitor in NIH3T3 cells. J. Cell. Biochem., 73, 291–302. [PubMed] [Google Scholar]
- 47.Won J., Yim,J. and Kim,T.K. (2002) Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transciptase (hTERT) promoter in normal human somatic cells. J. Biol. Chem., 277, 38230–38238. [DOI] [PubMed] [Google Scholar]
- 48.Doetzlhofer A., Rotheneder,H., Lagger,G., Koranda,M., Kurtev,V., Brosch,G., Winters-Berger,E. and Seiser,C. (1999) Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol., 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armstrong S.A., Barry,D.A., Leggett,R.W. and Mueller,C.R. (1997) Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA-binding activity. J. Biol. Chem., 272, 13489–13495. [DOI] [PubMed] [Google Scholar]
- 50.Yuan W., Condorelli,G., Caruso,M., Felsani,A. and Giordano,A. (1996) Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem., 271, 9009–9013. [DOI] [PubMed] [Google Scholar]
- 51.Chen Q., Dowhan,D.H., Liang,D., Moore,D.D. and Overbeek,P.A. (2002) CREB-binding protein/p300 co-activation of crystalline gene expression. J. Biol. Chem., 277, 24081–24089. [DOI] [PubMed] [Google Scholar]