Abstract
The precursor terminal protein pTP is the primer for the initiation of adenovirus (Ad) DNA replication and forms a heterodimer with Ad DNA polymerase (pol). Pol can couple dCTP to pTP directed by the fourth nucleotide of the viral genome template strand in the absence of other replication proteins, which suggests that pTP/pol binding destabilizes the origin or stabilizes an unwound state. We analyzed the contribution of pTP to pTP/pol origin binding using various DNA oligonucleotides. We show that two pTP molecules bind cooperatively to short DNA duplexes, while longer DNA fragments are bound by single pTP molecules as well. Cooperative binding to short duplexes is DNA sequence independent and most likely mediated by protein/protein contacts. Furthermore, we observed that pTP binds single-stranded (ss)DNA with a minimal length of approximately 35 nt and that random ssDNA competed 25-fold more efficiently than random duplex DNA for origin binding by pTP. Remarkably, short DNA fragments with two opposing single strands supported monomeric pTP binding. pTP did not stimulate, but inhibited strand displacement by the Ad DNA binding and unwinding protein DBP. These observations suggest a mechanism in which the ssDNA affinity of pTP stabilizes Ad pol on partially unwound origin DNA.
INTRODUCTION
The linear, double-stranded adenovirus (Ad) genome has a size of ∼36 000 bp and contains terminal proteins covalently attached to the 5′ ends. The origins of DNA replication are located at the genome termini in the inverted terminal repeats, which are 103 bp long in the case of human Ad2/5. The Ad DNA polymerase (pol) initiates the replication of the viral genome with the covalent coupling of a dCTP to the protein primer pTP, the precursor terminal protein (for reviews see 1,2). Pol and pTP form a pTP/pol heterodimer in solution, which binds the terminal 18 bp of the viral genome, referred to as the core origin (3). The pTP/pol dimer can be recruited to the viral origins of DNA replication by the interaction with two cellular transcription factors, Oct-1 and NFI. NFI binds pol, while Oct-1 contacts pTP. Both transcription factors use their DNA binding domains for these protein/protein interactions and recognize binding sites in the viral origins, stimulating replication levels up to 200-fold.
After replication initiation, pol elongates the protein primer while the transcription factors are displaced from their downstream binding sites. Elongation depends on the action of the Ad encoded DNA binding protein DBP, which can unwind DNA because it multimerizes on single-stranded (ss)DNA by hooking a C-terminal flexible arm in a neighboring protein (4). Full length replication of the genome requires the presence of NFII/topoisomerase I (5).
The terminus of Ad genomes contains a sequence repeat that permits a jumping-back mechanism to start DNA synthesis. The coupling of dCTP to pTP by pol is directed by the fourth nucleotide in the template strand. After synthesis of the trinucleotide intermediate pTP-CAT, the template strand slides back along the pTP/pol complex to pair CAT with the three 3′ terminal genome nucleotides (6). Pol then elongates the pTP-CAT primer, from which it dissociates after the jumping-back step (7). This mechanism is highly reminiscent of the sliding back mechanism employed by protein primed bacteriophages, like phage φ29, which also start replication internally in a sequence repeat (8,9). This internal replication initiation does not depend on the unwinding capability of DBP, suggesting that the pTP/pol complex possesses at least limited unwinding activity. In this study, we investigate the potential contribution of DNA binding by pTP to destabilization of the origin structure.
As the initiator of replication, the 671 amino acid Ad5 pTP protein harbors a multitude of interactions important for replication. DNA and pol interact with N-terminal and more C-terminal pTP regions, respectively, while for the interaction with Oct-1, multiple elements throughout the primary sequence are important (10,11). The pivotal amino acid in pTP is Ser580, which is covalently coupled to the incoming dCTP. Ad5 pTP contains three protease cleavage sites that are recognized by a virally encoded protease. Proteolytic processing at the iTP sites takes place during the course of infection (12), while cleavage at the TP site most likely occurs after packaging into the viral particle (13,14), resulting in the formation of TP-DNA lacking the N-terminal 349 amino acids of pTP. The integrity of the TP cleavage site might determine the intranuclear localization of pTP and the viral genome (15). The efficient nuclear import of pTP is dependent on its nuclear localization sequence, which is capable of stimulating nuclear import of pol as well (16). The viral genome is attached to the nuclear matrix via the interaction between pTP and the nuclear CAD enzyme involved in nucleotide synthesis (17). The interaction with CAD could provide a scaffold for the recruitment of replication proteins that explains the observation of specific replication foci in the nucleus.
The function of the DNA binding capacity of pTP is uncertain. The association of pol with the origin does not depend on pTP, since pol can bind DNA independently. pTP fails to bind DNA when it has been cleaved by the Ad protease, but this proteolytic processing did not block DNA replication in vivo (15). The N-terminal pTP fragments might remain associated with pol during and after processing, which explains why the C-terminal proteolytic pTP fragments containing the priming Ser580 did not support DNA replication alone (18). pTP point mutants that failed to bind DNA were impaired in replication initiation, suggesting that DNA binding by pTP contributes to DNA replication. Some of these mutations caused disruption of the pol interaction as well (18), although these two interactions mapped to different pTP regions (10). Apparently, these regions cannot be separated without influencing both interactions simultaneously.
To obtain insight in the contribution of pTP DNA binding to Ad DNA replication, we analyzed the characteristics of pTP binding to various DNA fragments. We observe that pTP dimerizes on short DNA duplexes, most likely caused by a protein/protein interaction, and show that pTP binds ssDNA. These data support a model in which pTP stabilizes the pol complex on partially unwound origin DNA.
MATERIALS AND METHODS
Proteins and nucleic acids
In all experiments, baculovirally expressed Ad5 pTP was used unless stated otherwise. Baculoviral Ad5 pTP was expressed and purified to >90% purity as described (19). His-tagged pTP was expressed in bacteria and purified to >90% purity as described (19). Purified baculovirally expressed DBP was a gift of B. van Breukelen. The monoclonal antibody recognizing a penta-His epitope was obtained from Qiagen. The anti-pTP antibody used to detect pTP/DNA complexes is a rabbit polyclonal antiserum raised against the pTP/pol complex as described previously (20). Oligonucleotides were purchased from Amersham/Pharmacia Biotech. An aliquot of 10 pmol of oligonucleotide was 5′ end-labeled with T4 polynucleotide kinase (Pharmacia). After stopping the labeling reaction by boiling for 3 min, a 1.2 M surplus of unlabeled complementary oligonucleotide was added to generate duplex DNA probes, which were subsequently purified by 12% acrylamide PAGE. The sequences of oligonucleotides used were as follows: T(1–50), 5′-CATCATCAATAATATACCTTATTTTGGAT TGAAGCCAATATGATAATGAG-3′; D(1–50), 5′-CTCAT TATCATATTGGCTTCAATCCAAAATAAGGTATATTATTGATGATG-3′; T(15–50), 5′-TACCTTATTTTGGATT GAAGCCAATATGATAATGAG-3′; D(15–50), 5′-CTCA TTATCATATTGGCTTCAATCCAAAATAAGGTA-3′. Other Ad ori derived oligonucleotides are numbered accordingly.
DIM probes were generated by hybridization of two fully complementary oligonucleotides with the forward sequences: DIM-wt, 5′-CATCATCAATAATATACCTTAATATAC CTT-3′; DIM-AT, 5′-CATCATCAATTTATAATTAAAAT ATACCTT-3′; DIM-CG, 5′-CATCATCAATGGCGCCGG CCAATATACCTT-3′; DIM-Oct, 5′-CATCATCAATTAT GCAAATGAATATACCTT-3′.
YG:YT was formed by hybridization of 5′-GCGAAT TCGGGCGGCGGG-3′ and 5′-CCCGCCGCCCGAATCC GC-3′ as described previously (21). YGT:YTC contained the sequence 5′-YG-TTTTTTTT-3′ hybridized with 5′-TTTTTTTT-YC-3′. The 19mer DNA duplex used in Figure 7 is the unwinding probe described previously, but was labeled with Klenow DNA polymerase (4).
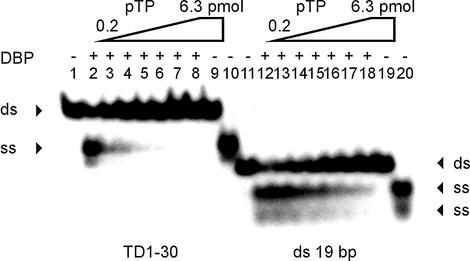
Figure 7.
pTP inhibits strand displacement by DBP in a DNA unwinding assay. TD(1–30) DNA (lanes 1–10, ds) or a probe unrelated to the origin (lanes 11–20, ds) were incubated with DBP (lanes 2 and 12), pTP (lanes 9 and 19) or DBP mixed with 0.2–6.3 pmol pTP in 2-fold increments (lanes 3–8 and 13–18). Displaced single strands are indicated by arrowheads (ss). Lanes 1 and 11, free probe; lanes 10 and 20, probe boiled to separate individual strands (ss).
DNA binding assay (EMSA)
DNA binding assays were performed essentially as described (22) with the following modifications. pTP was incubated with DNA probes for 10 min at 0°C in KB50 buffer (20 mM HEPES–KOH pH 7.5, 4.0 mM Mg2+, 50 mM NaCl, 50 µg ml–1 BSA, 1.0 mM dithiothreitol, 4.0% Ficoll-400). Protein/DNA complexes were separated at 4°C on 7% polyacrylamide in TBE for 14 h at 1 V cm–1. The amount of labeled DNA added (1000 c.p.m.) was ∼0.1–0.3 ng depending on the specific activity of the probe. Quantifications were performed using a Storm 820 phosphorimager; the percentage shift is defined as the intensity of the DNA/protein complex divided by the summed intensity of both free and bound DNA.
When indicated, DNA competitors were added preceding addition of the labeled DNA probe: pRP265NB DNA (19) as circular DNA or digested with AluI, Sau3A or CfoI to generate DNA fragments with blunt, 5′ or 3′ overhangs, respectively. Restriction enzymes were inactivated by heating for 15 min at 65°C. Digestions of competitor DNA were monitored on agarose.
Time course analysis of EMSA was performed by preincubating 8 nM pTP with DNA as indicated in the legend to Figure 6B for 10 min at 0°C in a volume of 100 µl and a 10 µl sample was loaded on a running gel (10 V cm–1). After the addition of 31 ng competitor pRP265NB double-stranded (ds)DNA, samples were mixed by pipetting and 10 µl aliquots were loaded on the running gel at the indicated time points. The intensities of the shifted pTP1 and pTP2 complexes were determined using a Storm 820 phosphorimager (Molecular Dynamics), summed and divided by the value after preincubation (= 100%).
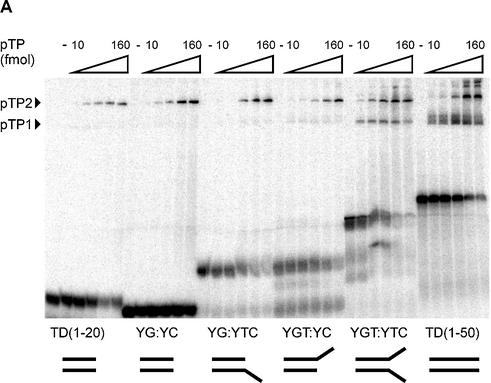
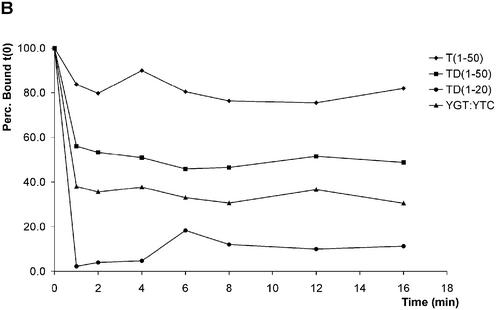
Figure 6.
pTP binds as a monomer to short DNA duplexes in the presence of two opposing single strands. At the bottom of (A), the structures of the DNA probes examined are schematically indicated. (A) pTP ranges from 0 to 160 fmol in 2-fold increments were incubated with the indicated DNA probes in an EMSA. YG:YC, CG-rich stem; YG:YTC, CG-stem with 5′ 8 nt T overhang; YGT:YC, CG-stem with 3′ 8 nt T overhang; YGT:YTC, CG-stem with two opposing 8 nt T overhangs. (B) Time course analysis of 8 nM pTP binding in EMSA before (0 min) and after addition of 31 ng competitor vector dsDNA (1–16 min). The fraction pTP bound to DNA was indexed at 100% to allow comparison of the relative stability of pTP binding.
Crosslinking assay
Chemical crosslinking of proteins was performed using N-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) supplied by Sigma in crosslinking buffer (20 mM MES–NaOH pH 6.0, 100 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA pH 8.0, 10% glycerol). Aliquots of 5 µM of each protein (or 7 µM DNA) were incubated for 1 h at room temperature, after which reactions were stopped by addition of 50 mM NH4CH3COO. pTP was preincubated with pol or DNA for 30 min in the absence of EDC when indicated. Proteins were separated by SDS–PAGE and analyzed by silver staining or western blotting using a rabbit polyclonal antiserum raised against pTP.
DNA unwinding assay
DNA unwinding assays were performed essentially as described (23). pTP and/or DBP were preincubated with DNA probes for 60 min at 30°C in unwinding buffer (25 mM HEPES–KOH pH 8.0, 1.0 mM dithiothreitol, 50 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 20% glycerol, 0.02% NP-40, 5 mM EDTA and 40 ng µl–1 BSA) in a volume of 25 µl. Unwinding reactions were stopped by adding 5.0 µl stop buffer (40% sucrose, 0.1% bromophenol blue, 0.1% xylene cyanol, 1.0% SDS) and unwound DNA was separated from dsDNA by SDS–PAGE (15%) at room temperature.
RESULTS
pTP binds both single- and double-stranded DNA
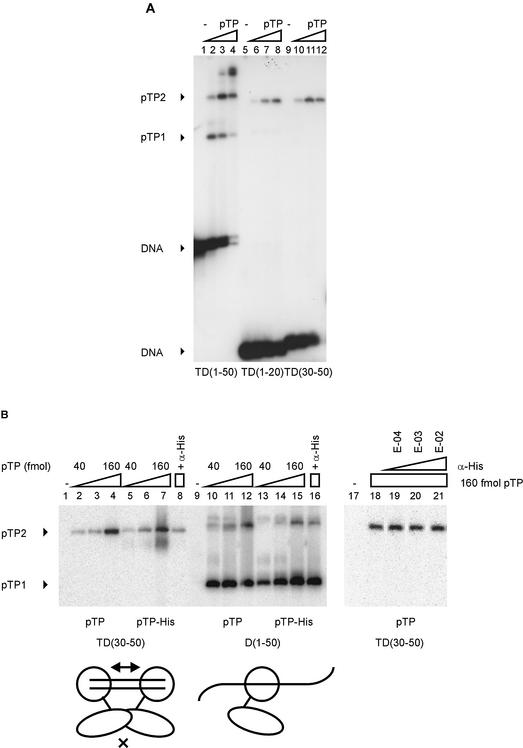
The pTP/pol heterodimer has been shown to bind both dsDNA and partially duplex DNA fragments derived from the origin of Ad DNA replication (3,22). To identify the contribution to this binding by pTP, we assayed the binding of purified Ad5 pTP to dsDNA and ssDNA probes derived from the Ad origin of DNA replication. The DNA probes in this study were designated D for displaced and T for template strand (Fig. 1). pTP was able to bind both to the double-stranded origin probes TD(1–20) and TD(1–50), but pTP binding gave rise to different complexes on these probes (Fig. 1A). On TD(1–50), pTP binding resulted first in the formation of the pTP1 complex, whereas the pTP2 complex was formed at higher pTP concentrations. On TD(1–20), the pTP1 complex was less prominent, while the pTP2 complex seemed to be formed preferentially. Both complexes contain pTP, since the addition of α-pTP polyclonal antibody inhibited the formation of both complexes (lanes 13–16). The pTP1 complex on TD(1–20) does not correspond to a breakdown product of pTP, since its intensity did not increase at higher pTP concentrations. The concentration dependence of pTP2 formation on TD(1–50) suggests that pTP2 corresponds to a pTP dimer or to two single pTP molecules on one DNA probe. The pTP2 complex is already formed on TD(1–20) while most DNA is still free; this indicates that pTP binds to TD(1–20) in a cooperative manner.
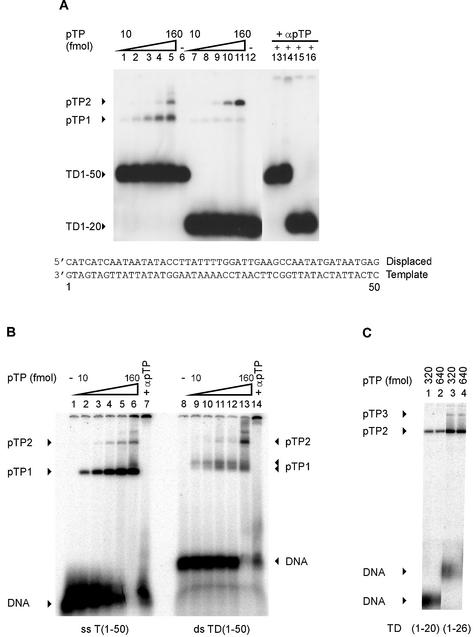
Figure 1.
pTP binds both ssDNA and dsDNA. Increasing amounts of pTP were incubated with radiolabeled DNA probes containing Ad5 origin sequences and analyzed using EMSA. T(emplate) or D(isplaced) strand oligonucleotides were hybridized to form the double-stranded TD probes as indicated at the bottom of (A). DNA probes and DNA/protein complexes are indicated by arrowheads. (A) pTP binding to double-stranded TD(1–50) (lanes 1–6 and 13–14) and TD(1–20) (lanes 7–12 and 15–16). Lanes 1–5 and 7–11, 10–160 fmol (0.5–8 nM) pTP in 2-fold increments; lanes 6 and 12, free probes; lanes 13–16, 160 fmol pTP with α-pTP polyclonal antibody at 1:1000 (lanes 13 and 15) or 1:100 dilutions (lanes 14 and 16). (B) pTP binding to single-stranded T(1–50) (lanes 1–7) and double-stranded TD(1–50) (8–14). Lanes 1 and 8, free probe; lanes 2–6 and 9–13, 10–160 fmol pTP in 2-fold increments; lanes 7 and 14, 160 fmol pTP with α-pTP polyclonal antibody at 1:100. The different percentage of bound DNA in (B) when compared to (A) is caused by a lower specific activity of the DNA probes in (A), which allows the visualization of the pTP1 complex at TD(1–20). (C) pTP binding to TD(1–20) (lanes 1 and 2) and TD(1–26) (lanes 3 and 4). pTP3 marks the formation of a third pTP complex (arrowhead). Lanes 1 and 3, 160 fmol pTP; lanes 2 and 4, 320 fmol pTP.
We also analyzed the capacity of pTP to bind ssDNA (Fig. 1B). We observed pTP complexes on TD(1–50) and T(1–50) with similar mobility, corresponding to the pTP1 complex observed in Figure 1A. pTP bound ssDNA and dsDNA with similar efficiency. The complex formed on single stranded T(1–50) contains pTP: addition of a polyclonal α-pTP antibody retarded part of the protein/DNA complexes and inhibited the formation of protein/DNA complexes as well (lanes 7 and 14). On TD(1–50), the pTP1 complex appeared as a diffuse double band, which was observed repeatedly. This observation might be explained by two conformations of the pTP/DNA complex running with different mobilities.
On TD(1–50), higher order complexes appeared with increasing pTP concentrations. This could be caused by additional pTP molecules binding to DNA directly or by pTP proteins binding via pTP (‘piggybacking’). When pTP was incubated with the small DNA duplexes TD(1–20) and TD(1–26), predominantly the pTP2 complex was formed on both duplexes. At higher concentrations of pTP modest loading of a third pTP molecule was observed, but only on the 26mer (Fig. 1C). Together with the observation of even higher order complexes on TD(1–50), this suggests that the non-specific loading of additional pTP molecules on DNA requires direct contacts with the DNA. It also indicates that pTP covers a binding site size of ∼8–10 bp.
Two molecules of pTP bind to short dsDNA fragments
If pTP dimerizes on short DNA duplexes, internal DNA sequence dependence does not explain the cooperativity of pTP binding (Fig. 1A, lanes 11 and 12): the sequence of TD(1–20) is present within TD(1–50). If the DNA termini contribute to pTP binding stability, the variant sequence of the downstream terminus could cause disruption of the pTP dimer. To exclude this possibility, we tested pTP binding to TD(30–50) (Fig. 2A, lanes 9–12). pTP readily formed the pTP2 complex on TD(30–50), showing that neither the internal sequence nor the sequence of the termini determines whether pTP binding is cooperative.
Figure 2.
pTP dimerizes specifically on small DNA duplexes. (A) pTP binding to TD(30–50) was studied using EMSA. Lanes 1–4, TD(1–50); lanes 5–8, TD(1–20); lanes 9–12, TD(30–50). Lanes 1, 5 and 9, free probe; lanes 2, 6 and 10, 40 fmol pTP; lanes 3, 7 and 11, 80 fmol pTP; lanes 4, 8 and 12, 160 fmol pTP. Free DNA, pTP1 and pTP2 complexes are indicated. (B) Comparison of pTP and pTP-His binding to TD(30–50) and D(1–50). Lanes 1–8 and 17–21, TD(30–50); lanes 9–16, D(1–50). Lanes 2–4 and 10–12, 40, 80 and 160 fmol pTP; lanes 5–7 and 13–15, 40, 80 and 160 fmol pTP-His. Lanes 4, 16 and 18–21, 160 fmol pTP + α-His antibody added during preincubation at E-04 (lane 19), E-03 (lane 20) or E-02 dilution (lanes 8, 16 and 21). pTP1 and pTP2 complexes are indicated; free DNA was run off the gel to obtain large separation of the pTP1 and pTP2 (and potential pTP-His/antibody) complexes. Schematically indicated at the bottom of the figure are two pTP-His (large circle with extending line)/α-His (small oval) complexes binding to the short DNA duplex TD(30–50) or a single complex bound to D(1–50). E-02, 1:100 dilution; E-03, 1:1000 dilution; E-04, 1:10 000 dilution.
The method of choice to demonstrate protein dimerization on DNA is forming heterodimers with differently sized proteins, but we were unable to obtain soluble expression of pTP with C-terminally fused thioredoxin or GST tags. Therefore we prepared pTP containing a C-terminal His tag and examined the effect of the addition of a monoclonal α-His antibody. When we incubated pTP containing a C-terminal His tag with TD(30–50) in the presence of α-His, the antibody inhibited pTP2 formation by 82% (Fig. 2B, compare lanes 7 and 8). The same antibody inhibited pTP1 formation on D(1–50) by only 18% (lanes 15 and 16) and the antibody did not interfere with the binding of pTP without His tag to TD(30–50) (lanes 18 and 21). This suggests that in solution the formation of pTP2, but not pTP1, is inhibited by the addition of α-His antibody. We did not observe complexes of α-His antibody with pTP-His that were stable under EMSA conditions, although pTP-His was recognized by the α-His antibody during western blotting (data not shown).
At the bottom of the figure, we have indicated how this might be explained: the size of the antibody could inhibit binding of two closely spaced pTP molecules on TD(30–50), while it does not affect the binding of a single pTP molecule to D(1–50). We consider cooperative binding of two pTP molecules to small DNA duplexes the most likely explanation for the observations in Figures 1 and 2, but realize that we observed only indirect evidence for pTP dimerization. Inhibition of pTP2 formation on TD(30–50) did not result in pTP1 binding, indicating that pTP only binds stably to small DNA duplexes in the presence of a second pTP.
Binding of two pTP molecules is mediated by protein/protein contacts
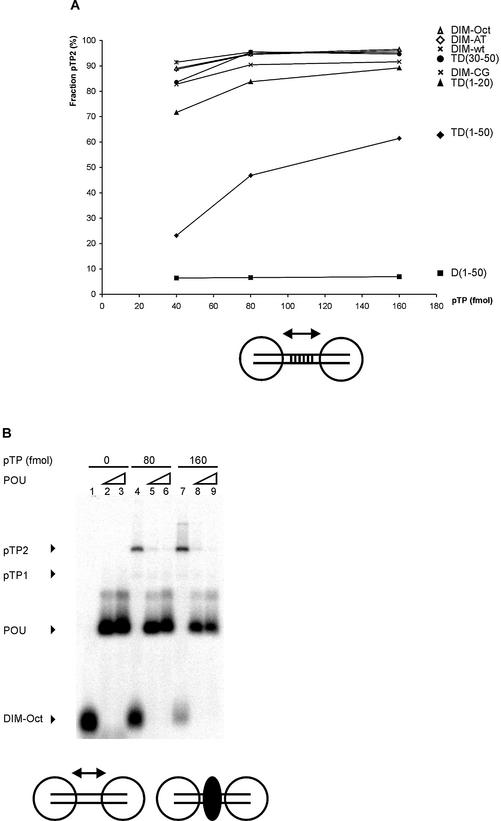
Cooperative binding of two pTP molecules on TD(1–20) could be mediated by two different mechanisms: protein/protein contacts once both pTP monomers are loaded on DNA or DNA structural changes caused by pTP binding that influence the binding stability of either monomer. When the oligomeric state of pTP in solution was monitored by gel filtration analysis in the presence of 400 mM KCl, pTP was predominantly monomeric (3). Therefore, we first assayed the influence of intervening base pairs using a 30 bp dimerization probe with varying base pairs 11–20 (Fig. 3A). The tested DNA fragments consisted of TD(1–20) with a 10 bp insertion between base pairs 10 and 11: Ad origin base pairs 11–20 (DIM-wt), AT-rich (DIM-AT), CG-rich (DIM-CG) or an octamer sequence (DIM-Oct). Origin, AT-rich, CG-rich and octamer-containing DNA probes supported dimerization with similar efficiency (Fig. 3A). This suggests that putative structural changes in the DNA caused by pTP binding are not the primary determinant of the formation of the pTP2 complex.
Figure 3.
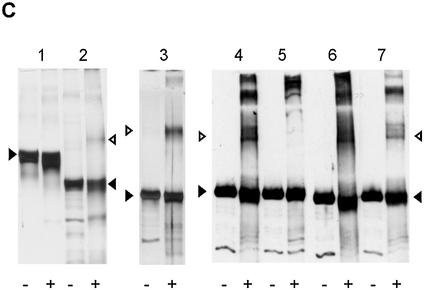
Cooperative pTP binding is most likely mediated by protein/protein contacts. (A) Increasing amounts of pTP were incubated with different DNA duplexes consisting of TD(1–20) with the following 10 bp long insertions between bp 10 and 11. DIM-wt, Ad ori bp 11–20; DIM-AT, AT-rich; DIM-CG, CG-rich; DIM-Oct, octamer site. Amount of pTP2 complex as fraction of the total amount of shifted pTP/DNA complex is plotted against pTP concentration. Values are averaged over two experiments; error margins were <10% fraction pTP2. Indicated schematically at the bottom of the figure are two pTP molecules (large circles) binding to the DIM-wt probe with the variable insertion sequence indicated by parallel lines. (B) Binding of pTP to DIM-Oct was studied using EMSA. 0, (lanes 1–3), 80 (4–6) or 160 (7–9) fmol pTP was incubated with DNA in the presence of 5 (lanes 2, 5 and 8) or 10 (lanes 3, 6 and 9) ng Oct-1 POU domain. Indicated schematically at the bottom of the figure are two pTP molecules bound to DIM-Oct in the absence (left) and presence (right) of Oct-1 POU (black oval) binding the insertion sequence in the middle of the DIM-Oct probe. (C) EDC crosslinking analysis of protein/protein interaction. An aliquot of 5 µM of each protein was incubated without (– lanes) or with (+ lanes) 10 mM EDC, separated by SDS–PAGE (lanes 1 and 2, 10%; lanes 3–7, 7.5%) and detected by silver-staining (lanes 1–3) or western blotting using an α-pTP polyclonal antibody (lanes 4–7). Lane 1, ovalbumin; lane 2, NFI DNA binding domain; lane 3, Ad5 pTP; lane 4, Ad5 pTP; lane 5, Ad5 pTP + pol; lane 6, Ad4 pTP; lane 7, Ad5 pTP + 7 µM TD(1–20). Filled triangles indicate monomers, open triangles indicate dimers formed after EDC treatment.
We then tested the presence of a potential pTP/pTP interaction. pTP was incubated with the DIM-Oct probe containing an octamer site between the two termini (Fig. 3B). We tested whether the addition of the Oct-1 POU domain, responsible for binding to the Oct-1 site, might interfere with pTP2 formation (see Fig. 3B). Addition of 5.0 ng Oct-1 POU inhibited the formation of the pTP2 complex on DIM-Oct 10-fold (lanes 4–6 and 7–9), while binding to DIM-wt was inhibited 2-fold (not shown). We did not observe pTP monomers on DIM-Oct in the presence of Oct-1 POU, so it cannot be concluded that pTP dimerization is specifically inhibited: Oct-1 might also block DNA binding of pTP to DIM-Oct. The effect of Oct-1 addition, however, was similar to that of the addition of α-His, which also inhibited all His-pTP binding to TD(30–50) (Fig. 2B). Our interpretation of these results is that dimerization is a prerequisite to obtain stable pTP binding on small DNA duplexes.
The potential presence of a direct pTP/pTP interaction was therefore analyzed in solution using the zero length crosslinking agent EDC (Fig. 3C). We incubated different proteins without (–) and with (+) EDC to test if crosslinking of putative protein dimers resulted in the formation of a denaturation-resistant higher molecular weight product. EDC treatment of ovalbumin did not result in the formation of dimers, but some products with slightly higher mobility were observed, presumably caused by intramolecular crosslinking (lanes 1– and 1+). In contrast, EDC was able to crosslink NFI monomers (dark triangle) into a denaturation-resistant dimer (open triangle) as published previously (24) (lanes 2– and 2+). pTP was crosslinked into a dimer even more efficiently (lane 3+), suggesting the formation of pTP dimers (open triangles) in solution. In the stacking gel, higher order aggregates were also observed (lane 3+). To verify that the higher molecular weight protein products observed after EDC treatment contained pTP, a similar experiment was analyzed by western blotting using an α-pTP antibody. Both Ad5 pTP and Ad4 pTP could be crosslinked into homodimers (open triangles) efficiently, while higher order multimers were detected as well (lanes 4+ and 6+).
During DNA replication, pTP is present as a heterodimer with the 140 kDa Ad DNA pol. As expected, this pTP/pol heterodimer crosslinked into a high molecular weight product in the stacking gel after EDC treament (lane 5+), while the pTP dimer caused by crosslinking of free pTP was now absent. Also in the presence of TD(1–20), pTP could be crosslinked into a homodimer (lane 7+), albeit less efficiently. These data suggest that pTP molecules can interact in solution and might be able to do so on DNA as well.
pTP binds dsDNA and ssDNA with overlapping protein surfaces
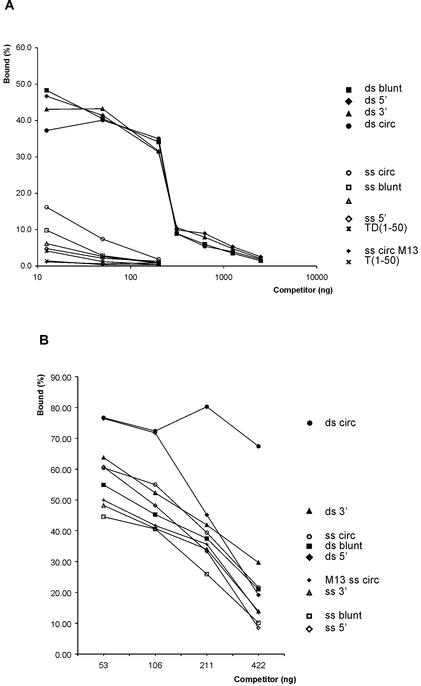
The binding of pTP to TD(30–50) suggested only limited sequence specificity (Fig. 2A), but pTP binding to TD(1–18) did not show sensitivity up to 100-fold excess competitor DNA unrelated to the origin in previous work (3). We tested unlabeled TD(1–50) and different restriction digests of plasmid dsDNA for their efficiency to compete for pTP bound to TD(1–50) (Fig. 4A). Approximately 3.1 ng of any competitor reduced pTP binding to 50% of wild-type; we assume that this represents non-specific DNA binding by pTP. TD(1–50) competed much more efficiently than plasmid DNA, suggesting at least limited sequence specificity. The loss of cooperative pTP binding on longer DNA probes suggested that pTP might bind DNA termini preferentially (Fig. 1A). We did not observe a preference for DNA termini, however, circular DNA and linear DNA competed with similar efficiency, regardless of the overhang created by the restriction enzyme.
Figure 4.
pTP binds dsDNA and ssDNA with overlapping protein surfaces. DNA bound by 160 fmol pTP as a fraction of the total amount of DNA was used as the index (100%); the signals of pTP1 and pTP2 were summed. Error margins for individual points are less than 5% of binding. (A) Binding of 160 fmol pTP to TD(1–50) was challenged with increasing amounts of competitor DNA. Circular plasmid DNA (ds circ) was digested using AluI, Sau3A or CfoI to create blunt (ds blunt), 5′ overhang (ds 5′) or 3′ overhang (ds 3′) fragments. Single-stranded competitors were generated by heat denaturation followed by rapid cooling for 1 min to keep the single strands separated, after which pTP was immediately added. As a control on heat denaturation, single-stranded circular M13 DNA was used as a competitor. Controls for specific binding were unlabeled TD(1–50) probe and single-stranded T(1–50). (B) pTP binding to T(1–50) was challenged with increasing amounts of dsDNA and ssDNA competitors as described in (A).
While fragmentation of competitor DNA contributed little to its binding efficiency, heat denaturation increased its inhibiting effect considerably: approximately 25-fold more dsDNA than ssDNA was needed to reduce TD(1–50) binding by pTP to 10%. M13 control circular ssDNA competed with even higher efficiency than ssDNA vector fragments (Fig. 4A), showing that the single-stranded character and not the fragmentation of competitor DNA is essential for its binding effciency. The pTP binding preference for ssDNA was not clear from Figure 1, which might be explained by the low amounts of DNA used in that experiment. In contrast, the addition of competitor DNA (Fig. 4A) selects for high affinity binding sites within a large pool of DNA.
When we assayed the binding of pTP to the single-stranded T(1–50) in the presence of DNA competitors, dsDNA competed for pTP binding to a ssDNA probe as well (Fig. 4B). This suggests that pTP binds both ssDNA and dsDNA with at least partly overlapping protein surfaces. Again, denaturing the dsDNA competitors increased their ability to compete for pTP binding, but the difference in efficiency was much less pronounced on the single-stranded T(1–50) probe. This might be explained by more stable pTP binding to T(1–50), requiring higher affinity sequences to displace pTP from T(1–50) (see Fig. 6B).
pTP binds ssDNA with a minimal length of 35 nt
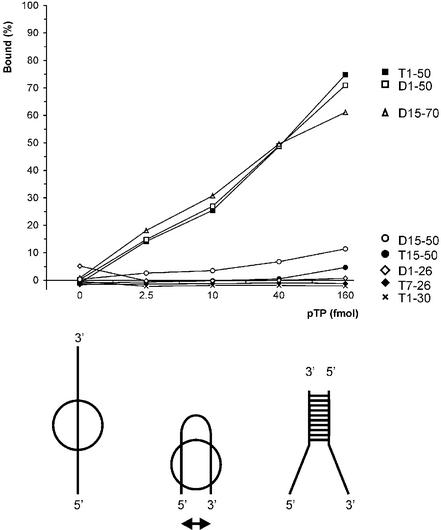
To be able to base pair the incoming nucleotide, the pTP/pol complex has to bind the genome terminus in an unwound state. The affinity of pTP for ssDNA could contribute to pTP/pol complex stability on such an unwound origin. We examined the ssDNA binding behavior of pTP using oligonucleotides containing Ad origin sequences (Fig. 5). Both T(1–50) and D(1–50) were bound with similar efficiency. Since replication starts specifically at the fourth nucleotide from the genome terminus, we tested whether the single strands contain sequences recognized by pTP. pTP did not bind D(1–26) or T(1–30) as a single strand, although T(1–30) is a template capable of supporting efficient DNA replication initiation (Fig. 5). In contrast, D(15–70) was bound with an efficiency similar to that of T(1–50) and D(1–50). The only common theme in pTP binding behavior was the minimal size of ssDNA needed to bind pTP. pTP bound to oligonucleotides with a minimal length of 35 nt, but failed to interact with the ssDNA fragments of shorter lengths that were tested.
Figure 5.
pTP binds ssDNA with a minimal length of 35 nt without sequence dependence. pTP binding to different origin sequences was analyzed in EMSA. The DNA bound by pTP as a fraction of the total amount of DNA was determined (%). Results are averaged over two experiments with individually labeled probes with error margins <10% binding. At the bottom of the figure, a model is indicated that might explain the size of ssDNA needed for stable pTP binding. The ssDNA strand (left) could loop back to allow the simultaneous interaction of two DNA binding regions within pTP (middle), a situation similar to a partially unwound replication origin DNA structure (right).
We were surprised by the size of the DNA needed to observe stable pTP binding, since we can make an estimate of the pTP binding site based on dsDNA binding. Assuming that the pTP2 complexes formed by pTP on TD(1–20) and TD(30–50) contain two pTP molecules, a third pTP could be loaded on TD(1–26) (Fig. 1C) and the DIM probes (30 bp) at high pTP concentrations (Fig. 3B, lane 7). This indicates a double-stranded binding site size of ∼8–10 bp. The large size of ssDNA needed to observe pTP binding might be explained if the ssDNA has to loop back to support stable pTP binding (Fig. 5, lower panel). This could allow simultaneous binding of the two opposing single strands by two pTP contact surfaces that are spaced by the dimensions of the 77 kDa protein (indicated in Fig. 5 by a double-headed arrow), a structure similar to an unwound origin (Fig. 5, bottom right). Such a binding mode is observed for the transcriptional coactivator PC4, which binds to two juxtaposing single strands as a dimer using the antiparallel binding grooves of each monomer (21,25).
To test this hypothesis, we assayed the binding of pTP to a DNA structure consisting of an 18 bp CG-rich stem and one or two 8 nt T overhangs that face each other (Fig. 6A). pTP formed a pTP2 complex on the CG-stem (YG:YC) and the stems with only a 1 nt T overhang on the 3′ side (YGT:YC) and 5′ side (YG:YTC), respectively. Since the CG-stem supported pTP binding as well, one cannot conclude that opposing single strands are a prerequisite for pTP binding. We do observe a qualitative difference, however, only when two opposing single strands were present in a ‘forked’ structure was pTP able to bind as a monomer (YGT:YTC). In fact, this was the only DNA duplex of size 20–30 giving rise to significant monomeric binding.
We compared the binding stability of pTP on YGT:YTC with that on other DNA probes after the addition of competitor vector dsDNA in a time course experiment (Fig. 6B). The fraction probe bound by 16 nM pTP during preincubation (time point 0) was indexed at 100% to facilitate comparison. On all DNA probes, the new association/dissociation equilibrium was reached within 1 min. pTP bound to T(1–50) was most resistant to competition with vector DNA. We observed a relative binding stability of T(1–50) > TD(1–50) > YGT:YTC > TD(1–20). When we inhibited pTP2 formation using α-His, Oct-1 and a tryptic digest of pTP, the formation of pTP1 was not observed on short DNA duplexes (Figs 2 and 3). The observation that pTP binds more stably to YGT:YTC than to TD(1–20) suggests that opposing single strands facilitate pTP binding to short DNA oligomers. The optimal binding site for pTP is, however, completely single stranded.
pTP inhibits DNA unwinding by DBP
The observed affinity of pTP for ssDNA immediately raises the question whether pTP might be able to assist in origin unwinding. Both the large size of the single strand needed to observe stable ssDNA binding by pTP (Fig. 5) and the binding behavior of pTP on YGT:YTC (Fig. 6A) suggest that pTP might contact two opposing single strands. However, Ad already encodes a ssDNA binding and unwinding protein, DBP. DBP can displace single strands by multimerizing on the strand that is not bound by pol (4,26). Ad DNA replication is strictly dependent on DBP during the elongation phase, but replication initiation is also observed in the absence of DBP. DNA unwinding by DBP can be assayed by the displacement of single strands that are prevented from reannealing by stopping the unwinding reaction with 1.0% SDS. We tested whether pTP can displace single strands or assist in strand displacement by DBP (Fig. 7).
In an unwinding assay, 2.5 pmol DBP displaced the single strands of TD(1–30) and a double-stranded probe unrelated to the origin (Fig. 7, lanes 2 and 12). In contrast, even 6.3 pmol pTP was unable to displace single strands (lanes 9 and 19). When we tested whether pTP might assist DBP in unwinding, pTP did not stimulate, but inhibited, DNA unwinding by DBP. On TD(1–30), 0.4 pmol pTP reduced DNA unwinding by DBP by 50% (lane 3). Unwinding of a 19mer dsDNA probe unrelated to the origin was inhibited by 50% by 1.6–3.2 pmol pTP (lanes 16 and 17). This suggests that DBP might be incapable of unwinding the replication origin in the presence of pTP, for example during the initiation reaction. Never theless, the strand displacement activity of DBP is essential during the elongation phase of Ad DNA replication (4).
pTP inhibited DNA unwinding by DBP more efficiently on TD(1–30), indicating that DNA binding by pTP might prevent DBP binding. We tested whether pTP and DBP could bind simultaneously to TD(1–30) under conditions that inhibit unwinding (5 mM Mg2+). Both proteins could bind to TD(1–30), but we did not observe ternary DBP/pTP/DNA complexes (not shown). Since DBP needs to multimerize on DNA in order to separate the two strands (4), DNA binding by pTP most likely inhibits unwinding by DBP.
DISCUSSION
pTP dimerizes on short DNA duplexes
During the DNA replication of Ad, pTP presents its priming Ser580 to the DNA pol at the origin by interactions with multiple replication components, including DNA, pol and Oct-1. In this study, we focused on the DNA binding properties of pTP in the absence of pol and observed cooperative binding of two pTP molecules to short DNA duplexes, most likely mediated by a protein/protein interaction (Figs 2 and 3). We interpret the pTP1 and pTP2 complexes as monomers and dimers of pTP on DNA based on the following observations. First, increasing the pTP concentration on probes supporting pTP1 binding results in the formation of pTP2 on DNA. Second, the addition of an α-His antibody blocked pTP2 formation of pTP-His on TD(30–50), but not pTP1 formation on TD(1–50). Third, Oct-1, recognizing an internal binding site, blocked pTP2 formation completely on DIM-Oct, but not on DIM-wt. Fourth, pTP could be crosslinked into homodimers both in the absence and presence of DNA by treatment with the zero length crosslinking agent EDC. Although this evidence is indirect, we consider dimerization of pTP the most likely mechanism to explain these combined observations. The different DNA binding behavior of pTP on 20mer and 50mer DNA duplexes was not dependent on the protein preparation, since it was also observed with purified bacterially expressed Ad5pTP-His (Fig. 2B) and purified baculovirally expressed Ad4 pTP (not shown).
Previously, two pTP complexes on DNA were observed on a 23mer Ad origin probe with 2 nt single-stranded overhangs, while only one complex was observed on a 19mer NF-κB probe (3). Since only the extra (low mobility) complex on the origin probe was sensitive to competition with origin DNA, this complex was interpreted as specific origin DNA binding by pTP. Our data now suggest an alternative explanation. Based on the protein concentrations used, the complex observed on both the origin and NF-κB probes could be a pTP dimer, while only the 4 bp longer origin probe could permit loading of a third pTP molecule, which is relatively sensitive to competition. Webster et al. observed that the faster migrating pTP complex on this probe displayed a mobility similar to that of pTP/pol, which would not be expected because of the 140 kDa mass difference (10). As suggested by Webster et al., this might be explained by dimeric binding of pTP, in agreement with our current observations.
We noted that monomeric pTP complexes were hardly detectable on short DNA duplexes other than TD(1–20) and YGT:YTC, even when dimerization was inhibited. This might be explained, when pTP binding to short duplexes is so unstable (Fig. 6B), by the need for an additional contact with a second pTP to remain bound under EMSA conditions. Stabilization of pTP binding to TD(1–18) was observed by the addition of monoclonal antibodies recognizing the C-terminus of pTP (10). This was interpreted as regulation of pTP DNA binding by the C-terminus of pTP, but in the light of our observations, a different explanation is possible: the binding of a dimer of pTP molecules on TD(1–18) might be stabilized by an antibody using both epitope recognizing sites to bridge the two dimeric components. An antibody recognizing a C-terminal His tag on pTP inhibited the binding of two adjacent pTP molecules (Fig. 2B); these two observations indicate that the pTP C-terminus might contribute to the dimeric interface.
Does the parental TP contact the pTP primer during replication initiation?
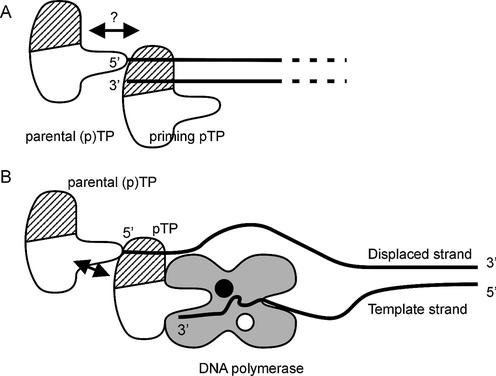
Since the replication origins are the termini of a 36 kb long dsDNA molecule, cooperative binding to both ends simultaneously is unlikely, assuming that the termini do not interact during initiation. After priming the replication reaction, however, pTP remains covalently linked to the 5′ end of the genome on either terminus. This covalently linked (p)TP is called parental TP, to distinguish it from the incoming pTP that primes the next round of replication. The observed cooperative binding of pTP to short DNA duplexes could mimic a priming pTP/parental (p)TP interaction as indicated in Figure 8A. Direct evidence for such an interaction is lacking, but one way to demonstrate such an interaction could come from terminal protein mutants that fail to stimulate DNA replication as a parental protein. Such mutants have been identified in the protein primed DNA replication system of bacteriophage φ29 (27,28), which bears strong functional similarity to that of Ad (9). Mutations of phage φ29 TP amino acids Asn80 and Tyr82 did not affect the priming capability, but interfered only with consecutive replication rounds. Likewise, mutations between φ29 TP amino acids 84 and 118 disrupted the second round of replication. Replication of mutant templates could only be restored by the addition of wild-type, not mutant, TP, showing that the parental TP/priming TP interaction is important for φ29 replication initiation.
Figure 8.
Model of pTP interactions during replication initiation. (A) A parental (p)TP molecule stays covalently bound to the 5′ end of the Ad genome (lines) after having primed the preceding replication round. The observed cooperative binding to short DNA duplexes could mimic the interaction of an incoming priming pTP with the parental (p)TP (arrow). The ds/ssDNA binding region of pTP is indicated by hatching. (B) The ssDNA binding affinity of pTP could contribute to pTP/pol binding stability on an unwound origin structure, while the pTP region providing the priming Ser580 should present the Ser hydroxyl group to the catalytic center (black circle) in the Ad DNA pol. Potential stabilizing contacts can be made with the parental (p)TP (arrow) and two ssDNA contact points spaced by the dimensions of pTP: the displaced strand (bound in the DNA binding region) and the three 3′ terminal nucleotides of the template strand (3′).
Previous work on Ad DNA replication had already suggested a role for the parental terminal protein. Indeed, TP-DNA and pTP-DNA are >30 times more efficient replication templates than naked DNA (15,29,30). DNase I footprints of TP-DNA suggested that the parental TP induces changes in the DNA structure and pTP/pol bound more efficiently to TP/DNA than to naked duplex DNA (30). When the parental TP is removed, exonucleolytic degradation of the displaced strand stimulates DNA replication, suggesting that the parental TP might assist in origin unwinding (29,31). We report two observations consistent with such a mechanism: the cooperative binding of adjacent pTP molecules and the affinity of pTP for ssDNA.
pTP binds ssDNA
We observed efficient pTP binding to single-stranded oligonucleotides with a minimal length of 35 nt and without any apparent sequence specificity (Fig. 5). In Figure 8B, we have indicated how both a priming pTP/parental (p)TP and a pTP/ssDNA interaction could contribute to initiation complex stability. In this model, the parental TP at the 5′ end of the genome contacts a pTP bound to the displaced DNA strand. In this way, pTP could participate in partial unwinding or stabilize an unwound origin caused by DNA end ‘breathing’. This enables the polymerase to base pair the incoming nucleotide. Apart from potential contacts with the displaced strand, pTP has to present its Ser580 to the polymerase active site of Ad polymerase (indicated in Fig. 8B with a black circle), most likely via contacts in the primer binding cleft (32). During replication initiation, the three 3′ terminal template nucleotides are present in this binding cleft, providing a second potential ssDNA contact point for pTP.
If indeed both the displaced and template strands can be contacted by pTP simultaneously, the striking length of the single-stranded oligonucleotide needed to observe stable pTP binding could be explained. Based on the size of the dsDNA needed to bind a third pTP molecule (Figs 1C and 3B), we estimate the size of the pTP binding site at ∼8–10 bp. If the 35 bases of ssDNA loop back to contact two pTP regions as indicated in the model at the bottom of Figure 5, the steric dimensions of the 77 kDa pTP (Fig. 5, arrowheads) determine the spacing between the two contact points. Evidence suggesting pTP binding to such an unwound structure is presented in Figure 6. While a CG-stem (YG:YC) or a stem with one single-stranded extension (YG:YTC) were bound by pTP dimers, a stem with two opposing single strands (YGT:YTC) supported monomeric pTP binding. Monomeric pTP binding correlated with relatively stable pTP binding (Fig. 6B), suggesting that opposing single strands help pTP to bind DNA.
Origin unwinding is a common theme in many, if not all, dsDNA replication systems (33). When we tested whether pTP could actively unwind short DNA stretches in the absence of the Ad unwinding protein DBP, we did not detect displacement of DNA strands (Fig. 7). If pTP binds and holds two opposing single strands simultaneously (Fig. 8), this would not necessarily lead to strand displacement as detected in our unwinding assays, but we did not observe any direct evidence that pTP partially unwinds DNA either, for example using KMnO4 footprinting (data not shown). pTP binding to DNA might be more reminiscent of the binding mechanism of the papillomavirus E1 protein, which causes subtle distortions of its origin recognition site. In crystal structures of a dimer and tetramer of the papillomavirus E1 protein, which binds both ssDNA and dsDNA and serves as the replication helicase in its hexameric form, two distinct DNA binding amino acid stretches contact either strand, causing increasing duplex destabilization on loading of multiple E1 proteins (34).
Whereas the pTP/pol complex displays moderately specific binding to the terminal 18 bp of the viral genome in a DNase I footprinting assay, pTP showed broad protection patterns on the Ad origin, but not on control DNA (3). Binding of pTP/pol to a partially single-stranded origin derived probe was sensitive to competition with both linear and circular ssDNA and dsDNA, but not to competition with 31 nt long oligonucleotides (22). The binding specificity of pTP/pol is enhanced during replication by the association with the highly specific DNA binding proteins NFI and Oct-1. pTP showed a broad binding spectrum, but was most efficiently competed with ssDNA (Fig. 4). There are probably no strict structural requirements on the side of the DNA for pTP binding, since dsDNA and ssDNA are bound by overlapping protein surfaces (Fig. 4). More likely, pTP contacts backbone charges enabling it to bind both ssDNA and dsDNA. When the positively charged arginines at positions 384–389 in pTP were changed to alanines, a strong decrease in DNA affinity was observed (18).
There is some evidence that the non-conserved N-terminus of Ad DNA polymerase might be involved in the recognition of origin base pairs 9–18 during initiation (35). The contribution of pTP to pTP/pol origin binding on the other hand is most likely not sequence specific. Rather, the affinity for ssDNA and potentially for the parental terminal protein could enable it to stabilize an initiation competent pTP/pol complex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank B. van Breukelen for purified baculovirally expressed Ad5 DBP. We thank A.B. Brenkman, M. Klejman and M. Mysiak for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.de Jong R.N. and van der Vliet,P.C. (1999) Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene, 236, 1–12. [DOI] [PubMed] [Google Scholar]
- 2.Hay R.T., Freeman,A., Leith,I., Monaghan,A. and Webster,A. (1995) Molecular interactions during adenovirus DNA replication. Curr. Top. Microbiol. Immunol., 199, 31–48. [DOI] [PubMed] [Google Scholar]
- 3.Temperley S.M. and Hay,R.T. (1992) Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J., 11, 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J., Kanellopoulos,P.N., Loonstra,A.K., van Oosterhout,J.A., Leonard,K., Tucker,P.A. and van der Vliet,P.C. (1997) Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J., 16, 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagata K., Guggenheimer,R.A. and Hurwitz,J. (1983) Adenovirus DNA replication in vitro: synthesis of full-length DNA with purified proteins. Proc. Natl Acad. Sci. USA, 80, 4266–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King A.J. and van der Vliet,P.C. (1994) A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J., 13, 5786–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King A.J., Teertstra,W.R. and van der Vliet,P.C. (1997) Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J. Biol. Chem., 272, 24617–24623. [DOI] [PubMed] [Google Scholar]
- 8.Mendez J., Blanco,L., Esteban,J.A., Bernad,A. and Salas,M. (1992) Initiation of phi 29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl Acad. Sci. USA, 89, 9579–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salas M., Miller,J.T., Leis,J. and DePamphilis,M.L. (1996) Mechanisms of priming DNA synthesis. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 131–176.
- 10.Webster A., Leith,I.R. and Hay,R.T. (1997) Domain organization of the adenovirus preterminal protein. J. Virol., 71, 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botting C.H. and Hay,R.T. (1999) Characterisation of the adenovirus preterminal protein and its interaction with the POU homeodomain of NFIII (Oct-1). Nucleic Acids Res., 27, 2799–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webster A., Leith,I.R. and Hay,R.T. (1994) Activation of adenovirus-coded protease and processing of preterminal protein. J. Virol., 68, 7292–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webster A., Hay,R.T. and Kemp,G. (1993) The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell, 72, 97–104. [DOI] [PubMed] [Google Scholar]
- 14.Mangel W.F., Toledo,D.L., Ding,J., Sweet,R.M. and McGrath,W.J. (1997) Temporal and spatial control of the adenovirus proteinase by both a peptide and the viral DNA. Trends Biochem. Sci., 22, 393–398. [DOI] [PubMed] [Google Scholar]
- 15.Webster A., Leith,I.R., Nicholson,J., Hounsell,J. and Hay,R.T. (1997) Role of preterminal protein processing in adenovirus replication. J. Virol., 71, 6381–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L.J. and Padmanabhan,R. (1988) Nuclear transport of adenovirus DNA polymerase is facilitated by interaction with preterminal protein. Cell, 55, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 17.Angeletti P.C. and Engler,J.A. (1998) Adenovirus preterminal protein binds to the CAD enzyme at active sites of viral DNA replication on the nuclear matrix. J. Virol., 72, 2896–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botting C.H. and Hay,R.T. (2001) Role of conserved residues in the activity of adenovirus preterminal protein. J. Gen. Virol., 82, 1917–1927. [DOI] [PubMed] [Google Scholar]
- 19.de Jong R.N., Mysiak,M.E., Meijer,L.A., van der,L.M. and van der Vliet,P.C. (2002) Recruitment of the priming protein pTP and DNA binding occur by overlapping Oct-1 POU homeodomain surfaces. EMBO J., 21, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coenjaerts F.E., van Oosterhout,J.A. and van der Vliet,P.C. (1994) The Oct-1 POU domain stimulates adenovirus DNA replication by a direct interaction between the viral precursor terminal protein–DNA polymerase complex and the POU homeodomain. EMBO J., 13, 5401–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werten S., Langen,F.W., van Schaik,R., Timmers,H.T., Meisterernst,M. and van der Vliet,P.C. (1998) High-affinity DNA binding by the C-terminal domain of the transcriptional coactivator PC4 requires simultaneous interaction with two opposing unpaired strands and results in helix destabilization. J. Mol. Biol., 276, 367–377. [DOI] [PubMed] [Google Scholar]
- 22.Kenny M.K. and Hurwitz,J. (1988) Initiation of adenovirus DNA replication. II. Structural requirements using synthetic oligonucleotide adenovirus templates. J. Biol. Chem., 263, 9809–9817. [PubMed] [Google Scholar]
- 23.Dekker J., Kanellopoulos,P.N., van Oosterhout,J.A., Stier,G., Tucker,P.A. and van der Vliet,P.C. (1998) ATP-independent DNA unwinding by the adenovirus single-stranded DNA binding protein requires a flexible DNA binding loop. J. Mol. Biol., 277, 825–838. [DOI] [PubMed] [Google Scholar]
- 24.Gounari F., De Francesco,R., Schmitt,J., van der Vliet,P.C., Cortese,R. and Stunnenberg,H. (1990) Amino-terminal domain of NF1 binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J., 9, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandsen J., Werten,S., van der Vliet,P.C., Meisterernst,M., Kroon,J. and Gros,P. (1997) C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nature Struct. Biol., 4, 900–903. [DOI] [PubMed] [Google Scholar]
- 26.Zijderveld D.C. and van der Vliet,P.C. (1994) Helix-destabilizing properties of the adenovirus DNA-binding protein. J. Virol., 68, 1158–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serna-Rico A., Illana,B., Salas,M. and Meijer,W.J. (2000) The putative coiled coil domain of the phi 29 terminal protein is a major determinant involved in recognition of the origin of replication. J. Biol. Chem., 275, 40529–40538. [DOI] [PubMed] [Google Scholar]
- 28.Illana B., Lazaro,J.M., Gutierrez,C., Meijer,W.J., Blanco,L. and Salas,M. (1999) Phage phi29 terminal protein residues Asn80 and Tyr82 are recognition elements of the replication origins. J. Biol. Chem., 274, 15073–15079. [DOI] [PubMed] [Google Scholar]
- 29.Guggenheimer R.A., Nagata,K., Kenny,M. and Hurwitz,J. (1984) Protein-primed replication of plasmids containing the terminus of the adenovirus genome. II. Purification and characterization of a host protein required for the replication of DNA templates devoid of the terminal protein. J. Biol. Chem., 259, 7815–7825. [PubMed] [Google Scholar]
- 30.Pronk R. and van der Vliet,P.C. (1993) The adenovirus terminal protein influences binding of replication proteins and changes the origin structure. Nucleic Acids Res., 21, 2293–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenny M.K., Balogh,L.A. and Hurwitz,J. (1988) Initiation of adenovirus DNA replication. I. Mechanism of action of a host protein required for replication of adenovirus DNA templates devoid of the terminal protein. J. Biol. Chem., 263, 9801–9808. [PubMed] [Google Scholar]
- 32.Liu H., Naismith,J.H. and Hay,R.T. (2000) Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J. Virol., 74, 11681–11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labib K. and Diffley,J.F. (2001) Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- 34.Enemark E.J., Stenlund,A. and Joshua-Tor,L. (2002) Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. EMBO J., 21, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joung I. and Engler,J.A. (1992) Mutations in two cysteine-histidine-rich clusters in adenovirus type 2 DNA polymerase affect DNA binding. J. Virol., 66, 5788–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]