Abstract
Locked nucleic acids (LNAs) and double-stranded small interfering RNAs (siRNAs) are rather new promising antisense molecules for cell culture and in vivo applications. Here, we compare LNA–DNA–LNA gapmer oligonucleotides and siRNAs with a phosphorothioate and a chimeric 2′-O-methyl RNA–DNA gapmer with respect to their capacities to knock down the expression of the vanilloid receptor subtype 1 (VR1). LNA–DNA–LNA gapmers with four or five LNAs on either side and a central stretch of 10 or 8 DNA monomers in the center were found to be active gapmers that inhibit gene expression. A comparative co-transfection study showed that siRNA is the most potent inhibitor of VR1–green fluorescent protein (GFP) expression. A specific inhibition was observed with an estimated IC50 of 0.06 nM. An LNA gapmer was found to be the most efficient single-stranded antisense oligonucleotide, with an IC50 of 0.4 nM being 175-fold lower than that of commonly used phosphorothioates (IC50 ∼70 nM). In contrast, the efficiency of a 2′-O-methyl-modified oligonucleotide (IC50 ∼220 nM) was 3-fold lower compared with the phosphorothioate. The high potency of siRNAs and chimeric LNA–DNA oligonucleotides make them valuable candidates for cell culture and in vivo applications targeting the VR1 mRNA.
INTRODUCTION
Antisense approaches that specifically suppress the expression of therapeutically relevant genes have been receiving increased attention over the last years (1–4). Several oligonucleotides (ONs) have reached the stage of clinical trials, and one antisense drug, Vitravene (Formivirsen), was approved by the Food and Drug Administration (5). Important progress in this field was made with the design of serum-stable analogs with an increased resistance to nucleases. The most commonly used antisense (AS) ONs to date are phosphorothioates [PS; for a review see Eckstein (6)], which combine many desired properties such as high biostability, RNase H activation and target specificity. The major disadvantages are, however, reduced binding affinities for complementary sequences and non-specific binding to several proteins (7–9), limiting many applications due to toxic side effects (10). Therefore, a second generation of AS ONs with modifications at the 2′-hydroxyl group was developed (11).
Recently, major advances for the use of AS ONs were achieved by the development of chemically modified nucleotides with improved properties such as enhanced serum stability, greater target affinity and reduced toxicity (12–14). One of the most promising modifications is locked nucleic acid (LNA), which contains a methylene bridge that connects the 2′-oxygen of ribose with the 4′-carbon (15–18). This 3′-endo conformation leads to improved binding to complementary DNA and RNA sequences and increases the melting temperature (Tm) by several degrees. In addition, chimeric LNA-containing ONs have significantly higher serum stability, with up to 10-fold increased half-lives in human serum compared with unmodified AS oligodeoxynucleotides (ODNs). Furthermore, it was shown that chimeric LNA–DNA ONs with a central DNA stretch of 7–8 monomers activate RNase H (19). Moreover, efficient cationic lipid-mediated uptake of LNA into different cell lines was demonstrated by the use of labeled LNA-containing ONs (20–23). In the first in vivo experiment, high antisense efficiency in the lack of toxicity was observed after intrathecal injection of ONs containing LNA (20). Subsequently, LNA ONs have been employed successfully to inhibit tumor growth in a xenograft mouse model (24).
Over the last few years, RNA interference (RNAi) has been found to be a highly efficient silencing mechanism based on double-stranded RNA (dsRNA) molecules that trigger the sequence-specific suppression of gene expression (25–27). Introduction of long dsRNA into mammalian cells, however, resulted in an interferon response by activating protein kinase R and RNase L (28,29). These unspecific effects could be bypassed by introducing 21–23mer small interfering RNAs (siRNAs), because non-specific responses are not triggered by dsRNA shorter than 30 bp (30). The potency of siRNA molecules to suppress gene expression specifically in vivo was demonstrated recently (31–33).
Taken together, a variety of new antisense approaches have emerged in the last years by the design of modified nucleotides with improved properties and by the development of highly efficient antisense strategies using the cellular RNAi pathway. In a recent review, Braasch and Corey called for comparative studies to demonstrate the relative strengths of different oligomer chemistries as being one of the major goals for improving ONs and their applications (13). We therefore compared the potency of LNA gapmers and siRNAs with widely applied AS ONs such as PS and 2′-O-methyl- (OMe-) modified RNA in mammalian cells. As a target to measure silencing efficacy, we used the vanilloid receptor subtype 1 (VR1) from rat, which was transiently expressed as a VR1–green fluorescent protein (GFP) fusion protein. VR1, which belongs to the TRP (transient receptor potential) channels (34), is of general therapeutic interest, because it is expressed predominantly in primary neurons and plays an important role in pain perception (35,36). Our comparative study revealed that VR1-specific siRNA and an LNA–DNA–LNA gapmer have significantly higher potencies as antisense molecules than the PS or OMe ONs investigated in the present study.
MATERIALS AND METHODS
Oligonucleotides
Unmodified DNA ONs and PS were obtained from MWG Biotech AG (Ebersberg, Germany), OMe-modified RNA was purchased from IBA GmbH (Göttingen, Germany). LNA–DNA gapmers were obtained from Proligo (Boulder, CO, and Paris, France).
LNA–DNA gapmers corresponding to the sequence of V29 were synthesized with 8 or 10 unmodified DNA monomers in the center and five or four LNA monomers at each end. All other LNA ONs as well as the OMe-modified V29 ON were designed as gapmers with five modified nucleotides at each end and eight deoxynucleotides in the center.
VR1-specific siRNAs were obtained from IBA GmbH or MWG Biotech AG as deprotected duplexes. Sequences of the ONs and siRNAs used in the present study are summarized in Table 1.
Table 1. Sequences of oligonucleotides used in this study.
| V15-PS | CAT GTC ATG ACG GTT AGG |
| V15inv-PS | GGA TTG GCA GTA CTG TAC |
| V16.1-PS | GCG CAT CTT CTA CTT CAA CTT |
| V16.1inv-PS | TTC AAC TTC ATC TTC TAC GCG |
| V29-PS | ATC TTG TTG ACG GTC TCA |
| V29inv-PS | ACT CTG GCA GTT GTT CTA |
| V4-L5 | CCG GGT ACG ACT CCT GGT |
| V15-L5 | CAT GTC ATG ACG GTT AGG |
| V16-L5 | ATG CGC TTG ACA AAT CTG |
| V25-L5 | GTG GTG TGG ACT CCA TAG |
| V27-L5 | CAG CTC CAG ACA TGT GGA |
| V29-L4 | ATC TTG TTG ACG GTC TCA |
| V29inv-L4 | ACT CTG GCA GTT GTT CTA |
| V29-L5 | ATC TTG TTG ACG GTC TCA |
| V29inv-L5 | ACT CTG GCA GTT GTT CTA |
| V31-L5 | CTC GGC CTG ACC TCA GGG |
| V29-OMe | atc ttG TTG ACG Gtc tca |
| V29inv-OMe | act ctG GCA GTT Gtt cta |
| VsiRNA1 | GCG CAU CUU CUA CUU CAA CTT |
| TT CGC GUA GAA GAU GAA GUU G | |
| VsiRNA2 | GUU CGU GAC AAG CAU GUA CTT |
| TT CAA GCA CUG UUC GUA CAU G | |
| VsiRNA3 | GCA UGU ACA ACG AGA UCU UTT |
| TT CGU ACA UGU UGC UCU AGA A | |
| VsiRNA4 | CCG UCA UGA CAU GCU UCU CTT |
| TT GGC AGU ACU GUA CGA AGA G | |
| VsiRNA5 | GAA UAA CUC UCU GCC UAU GTT |
| TT CUU AUU GAG AGA CGG AUA C | |
| VsiRNA6 | UGU GGG UAU CAU CAA CGA GTT |
| TT ACA CCC AUA GUA GUU GCU C | |
| K3a-L4 | TCC TTC TCT TTG CCC GGC |
| K3ainv-L4 | CGG CCC GTT TCT CTT CCT |
| K3a-L5 | TCC TTT TCT TTG CCC GGC |
| K3ainv-L5 | CGG CCC GTT TCT CTT CCT |
Upper case letters = DNA; lower case letters = 2′-O-methyl; underlined letters = phosphorothioates; bold letters = LNA; italic letters = RNA; inv = inverted sequence; V = vanilloid receptor 1; K = Pim-1 kinase.
Cell culture and transfection
Cos-7 cells (monkey African green kidney fibroblasts) were grown at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco’s modified Eagle’s medium (PAA laboratories, Germany), supplemented with 10% fetal calf serum (FCS; PAA laboratories), penicillin (100 µg/ml) and streptomycin (100 µg/ml) (Invitrogen, Germany). Cells were passaged by diluting them 1:10 before they reached confluency, to maintain exponential growth. The day before transfection, cells were trypsinized, resuspended in medium without antibiotics and transferred to 24-well plates at a density of 8 × 104 cells per well in a volume of 500 µl. Transfection and co-transfection experiments were carried out with Lipofectamine 2000 (Invitrogen, Germany). For each transfection, 1 µg of plasmid DNA [pcDNA3.1/CT-GFP-TOPO (Invitrogen, Germany) coding for the VR1–GFP fusion protein] and the respective amount of antisense molecules were mixed with 50 µl of OPTIMEM (Invitrogen, Germany). In a separate tube, 2.5 µl of Lipofectamine-2000 per reaction were added to 50 µl of OPTIMEM and incubated for 5 min at room temperature. Both solutions were mixed and incubated for an additional 20 min at room temperature to allow complex formation. The solutions were then added to the cells in the 24-well plate, giving an end volume of 600 µl. Cells were incubated at 37°C in the presence of the transfection solution for 24 h.
Fluorescence microscopy and immunoblotting
Transfection efficiency and antisense effects were analyzed by fluorescence microscopy and western blotting. Therefore, the medium was aspirated from the cells, 200 µl of phosphate-buffered saline were added and fluorescence images were taken directly from living cells using a Leica DM IRB fluorescence microscope.
For western blot experiments, cells were lysed directly in 24-well plates with lysis buffer (125 mM Tris–HCl, pH 6.8, 4% SDS, 1.4 M β-mercaptoethanol, 25% glycerol and 0.05% bromophenol blue). The complete lysate was boiled for 5 min at 95°C and equal amounts of protein were separated on 10% polyacrylamide gels. Transfer of separated proteins to PVDF membranes (Amersham, Germany) was performed with a semi-dry blotter (Bio-Rad, Munich, Germany). For immunostaining, membranes were incubated with a GFP antiserum (Invitrogen, Germany) (dilution 1:5000). Secondary antibodies were conjugated with alkaline phosphatase (AP) (Chemicon, Germany) and diluted 1:5000. As a chemiluminescence substrate, we used CDP-Star (Roche, Germany). To confirm equal loading of the samples, membranes were reprobed with a monoclonal mouse antibody against actin (Chemicon, Germany).
Northern blot
For northern blot experiments, a digoxigenin-labeled antisense RNA was used. Therefore, the vector pCRII TOPO that contained the VR1 cDNA was linearized with SmaI, and subsequently an in vitro transcription with labeled nucleotides was performed (Roche, Germany), resulting in a 1.7 kb RNA probe. Total RNA was prepared from transfected Cos-7 cells (RNeasy; Qiagen, Germany) and separated on 1.2% formaldehyde gels. Northern blotting was performed with the NorthernMax kit (Ambion, TX) according to the manufacturer’s recommendations. Hybridization and washing steps were performed at 68°C. Detection of the hybridized probe was done with the digoxigenin detection system (Roche, Germany) using a digoxigenin-specific antibody, which was conjugated with AP. CDP-Star was used as a substrate for AP, and chemiluminescence was detected with X-ray films.
Quantification of antisense effects on the protein level
Protein levels were quantified from western blots with Quantity One Software (Bio-Rad). The antisense effects were calculated by dividing the amount of VR1 protein in the presence of the AS and control ON, respectively. All signals were normalized to actin levels as internal standards. Values were fitted with a sigmoidal Boltzman equation using Origin (Microcal Software, Northampton, MA) to estimate IC50 values. Averages and standard deviations of three independent experiments are given.
Stability of oligonucleotides in cell culture medium
Single-stranded ONs and siRNAs were diluted in DMEM + 10% FCS to a final concentration of 10 pmol/µl and incubated at 37°C. Aliquots of 100 pmol of single-stranded ONs or 20 pmol of the siRNA were taken after 0.25, 0.5, 1, 2, 4, 6, 24 and 48 h. Reactions were terminated by addition of 10 µl of loading buffer [TBE (0.1 M Tris–HCl pH 8.4, 0.09 M boric acid, 2 mM EDTA) 7 M urea, 0.1% bromphenol blue, 0.01% xylene cyanol] and subsequent freezing in liquid nitrogen. The samples were separated on 20% polyacrylamide gels and stained with ethidium bromide (0.25 mg/l) for at least 20 min. Bands were evaluated quantitatively with the Quantity One program (Bio-Rad). All data were fitted with a single exponential decay function (Origin, Microcal Software, Northampton, MA) to obtain the half-lives of the ONs.
RESULTS
Antisense effects of 18mer phosphorothioates targeted against VR1
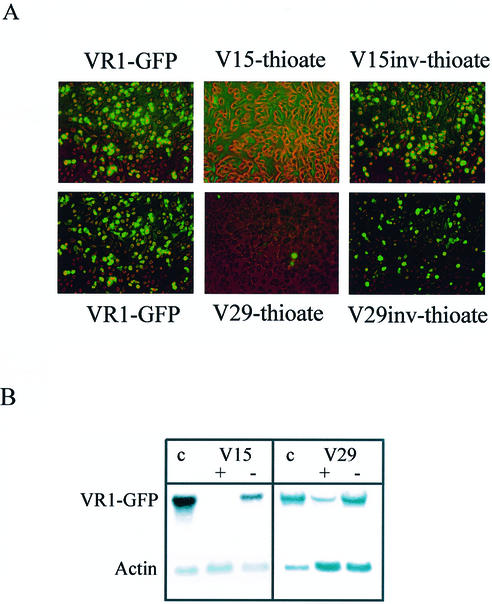
In a previous study, we characterized the VR1 mRNA in vitro by messenger walk screening with 32 different AS ONs and found highly accessible target sites for some of the ODNs (37). Here, we wanted to investigate their antisense effects in cell culture. Recently, the generation of fusion constructs of the gene of interest and the gene encoding GFP followed by co-transfection of the plasmid and AS ONs was described as a fast method to evaluate the potency of antisense molecules (38). Employing this approach, antisense effects can easily be determined by analyzing fluorescence or by western blotting using antibodies against the reporter protein, thereby avoiding the need to develop target-specific assays. Therefore, the VR1 cDNA was fused to the gene encoding GFP. Subsequently, the VR1–GFP plasmid was co-transfected with PS V15 and V29 at a concentration of 100 nM. These AS ONs previously were found to have the highest efficiency in vitro (37). For the V15-thioate, we observed a complete inhibition of the VR1–GFP green fluorescence (Fig. 1A), whereas the PS V29 reduces VR1–GFP gene expression to a level of ∼90% at this concentration. The observed antisense effects were sequence specific since control ONs with an inverted sequence (Fig. 1A) as well as an unrelated Pim-1 AS ON and an N18 randomer (data not shown) did not reduce VR1–GFP expression.
Figure 1.
(A) Transfection of Cos-7 cells with a VR1–GFP plasmid and PS V15 and its inverted sequence V15inv, as well as V29 and V29inv at a concentration of 100 nM. Cells with green fluorescence express the VR1–GFP fusion protein. (B) Antisense effect of PS V15 and V29 demonstrated by northern blotting: full-length VR1–GFP mRNA appeared in transfected cells and in the controls (V15inv, V29inv); no or only a low amount of full-length VR1 mRNA could be observed in cells incubated with PS V15 or V29. As a control for loading equal amounts of RNA, an actin-specific probe was used. c = control without AS ON; + = AS ON; – = control ON with an inverted sequence.
In order to prove that the observed antisense effects were due to degradation of the targeted VR1 mRNA, we performed northern blot analysis with a VR1-specific probe and found that VR1–GFP mRNA was degraded in the presence of V15 or V29 (Fig. 1B). Moreover, in the presence of the inverted controls, V15inv or V29inv, VR1–GFP mRNA was unaffected (Fig. 1B). From these results, we concluded that AS ONs against VR1 found to be efficient in vitro were also suitable for antisense approaches in cell culture.
Antisense effects of LNA gapmers against VR1
We previously have optimized the design of highly stable LNA–DNA ONs that activate RNase H in vitro (19). Gapmers with a stretch of seven to eight DNA monomers in the center initiate RNase H cleavage of the target RNA. Here, we used 18mer ONs with four or five LNA (L4 and L5) monomers at each end and 10 or 8 DNA nucleotides in between for co-transfection experiments against the VR1–GPF fusion mRNA. This design was chosen to fulfill the requirements for efficient RNase H activation. Control ONs with an inverted sequence and identical design were used to confirm sequence specificity of the AS ONs. A specific band with a molecular weight of ∼120 kDa could be detected with an anti-GFP antibody in our western blot experiments, demonstrating the correct expression of the VR1–GFP fusion protein. Subsequent incubation of the membranes with an anti-actin antibody demonstrated that equal amounts of proteins were loaded in each lane (Fig. 2A).
Figure 2.
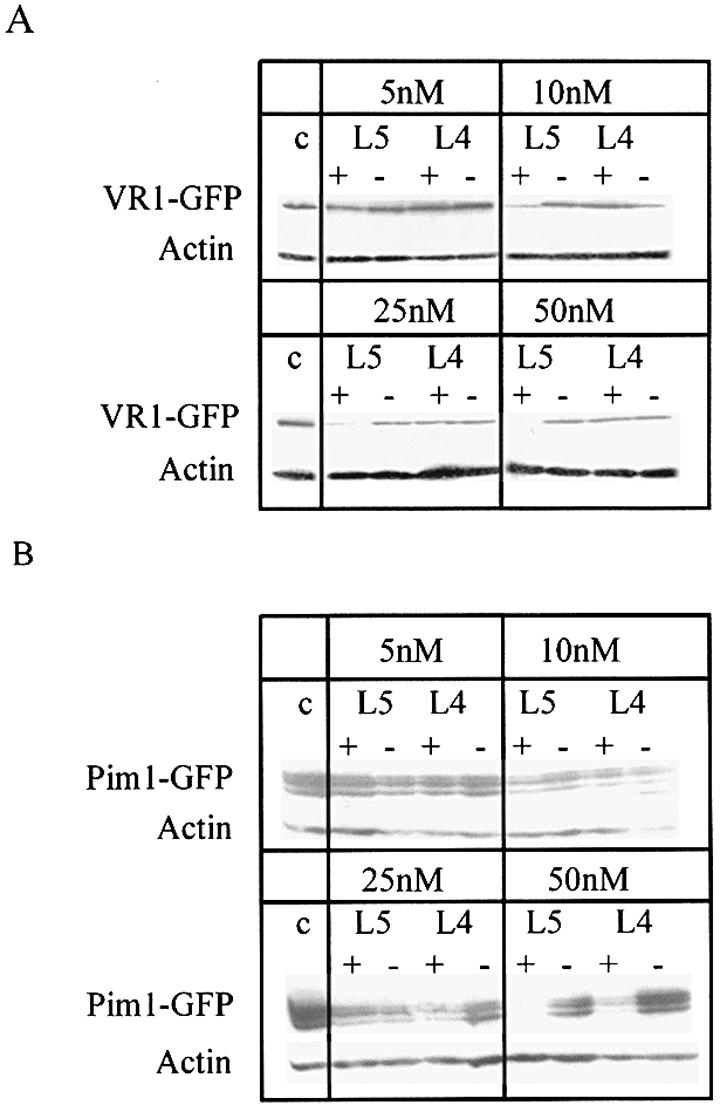
(A) Antisense effect of VR1-specific LNA gapmers with 4 or 5 nt end blocks (L4 and L5, respectively). Co-transfection experiments were performed with 1 µg of VR1–GFP-containing plasmid and the indicated concentrations of the L5 and L4 gapmers. VR1–GFP expression was detected by western blot analysis with an antibody directed against GFP. An anti-actin antibody serves as a control for equal loading of proteins in each lane. (B) Antisense effect of the Pim1-specific LNA gapmers L4 and L5. Co-transfection experiments were performed with 1 µg of Pim1–GFP- coding plasmid and the indicated concentrations of L5 and L4 gapmers. Detection of Pim1–GFP was performed as described above. + = AS ON; – = control ON with inverted sequence.
For the L4 gapmer, hardly any antisense effect was observed in the concentration range used (Fig. 2A). In contrast, the L5 gapmer was highly efficient at a concentration of 10 nM, and a complete inhibition of VR1–GPF expression was reached at a concentration of 50 nM.
The design of LNA gapmers was transferred to a second target, the serine/threonine kinase Pim-1 (39). As described previously for the VR1 mRNA (37), we optimized AS ODNs directed against the Pim-1 mRNA by messenger walk screening. One of the most efficient AS ODNs in vitro, K3a, also inhibits Pim1–GFP expression in mammalian cells at a concentration of 100 nM as a PS (data not shown). We designed the chimeric DNA–LNA gapmers L4 and L5 of K3a (see Table 1) and tested them together with the corresponding inverted controls for their potency to inhibit Pim1–GFP expression in co-transfection experiments. In contrast to the results observed with the VR1-specific L4 gapmer, we found an efficient antisense effect of the Pim1-specific L4 and L5 gapmers at a concentration of 50 nM (Fig. 2B), demonstrating the potency of these gapmers to act as AS ONs in cell culture.
Together with the results from our previous in vitro study (19), we conclude that the optimal design of a chimeric LNA–DNA AS ON is a stretch of 8 or 10 DNA monomers in the center and five or four LNA nucleotides at each end. The reason for the discrepancy in antisense potency between the VR1-specific L4 gapmer and the Pim1-specific L4 gapmer is not clear at the moment. Nevertheless, gapmers with 4 nt LNA end blocks seem to be active in principle, since inhibition of luciferase mRNA expression with gapmers containing four or five LNA monomers at each end in mammalian cells has recently been demonstrated (23).
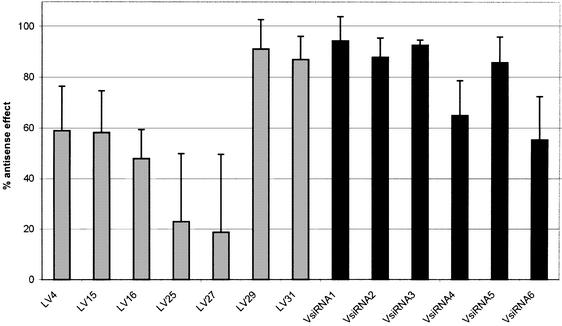
Hit rates for LNA oligonucleotides and siRNAs
To identify a highly efficient antisense molecule against a certain target, it is necessary to screen a number of candidates. We therefore investigated the rates of efficient LNA gapmers and siRNAs, respectively. A number of both types of antisense agents were screened, and the antisense efficiency was determined for each ON. For the LNA gapmers, we chose ONs with 5 nt end blocks against target sequences of the VR1 mRNA that were highly accessible in vitro (37). The siRNAs were designed according to the following criteria: start with AA; a GC content below 50%; and a unique sequence in the genome. The VsiRNA1 even obeyed the previously suggested sequence criterion AA(N19)TT (40). As can be seen in Figure 3, two out of seven LNA ONs (29%) and four out of six siRNAs (almost 67%) led to a >80% reduction of VR1–GFP expression. The most efficient antisense agents (V29-L5 and VsiRNA1) were used for the comparative experiments described below.
Figure 3.
Screening of LNA gapmers and siRNAs. The antisense potency of a number of LNA gapmers (gray bars) and siRNAs (black bars) has been determined based on the protein levels. ONs were used at a concentration of 10 nM and cells were harvested 24 h post-transfection. Averages and standard deviations of at least three independent experiments are shown.
Comparison of different antisense molecules in mammalian cell culture
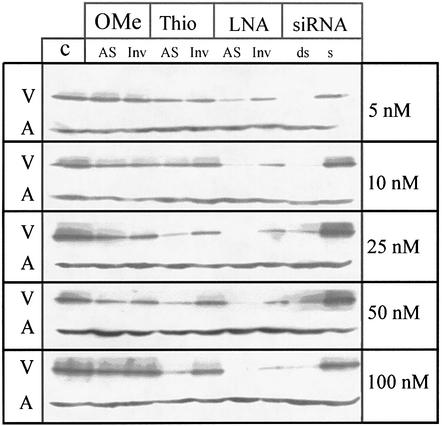
The main goal of our study was to compare the potencies of different antisense strategies against VR1. Therefore, we analyzed the antisense effects of VsiRNA1 and 18mer AS ONs against target site V29 that were modified as an all-PS, an OMe or an LNA gapmer. The VR1–GFP plasmid was co-transfected with the antisense molecules in a concentration range between 5 and 100 nM and analyzed by western blot (Fig. 4). Control ONs of identical design except for an inverted sequence are shown for comparison. We found that siRNA and LNA gapmers completely blocked the VR1–GFP expression at concentrations in the low nanomolar range. For the PS, a significant antisense effect was observed at concentrations higher than 25 nM, whereas no reduction of gene expression was found for the OMe-modified gapmer. Membranes subsequently were incubated with an anti-actin antibody to verify that equal amounts of protein were loaded in each lane.
Figure 4.
Western blot analysis from co-transfection experiments of the plasmid encoding the VR1–GFP fusion protein and modified AS ONs. OMe-RNA and LNA gapmers, PS and the siRNA were used at concentrations between 5 and 100 nM. Inverted control ONs for each modification and the sense strand of the siRNA were used as controls. Cells were harvested after 24 h and western blots were performed with an anti-GFP antibody. V = VR1–GFP band; A = actin band.
In order to compare silencing efficiencies of antisense molecules directed against the same target side, we also analyzed a PS, V16.1, that was isosequential to VsiRNA1. Interestingly, V16.1 shows hardly any inhibition of VR1–GFP expression at a concentration of 100 nM (data not shown).
Subsequently, the duration of gene silencing was investigated for LNA ONs and siRNAs. Up to 4 days after transfection, a constant level of VR1–GFP expression was observed by fluorescence microscopy for the control sample, but a complete suppression of gene expression was found for both types of antisense molecules (data not shown). At days 5 and 6, fluorescence reappeared in the samples co-transfected with LNA ONs or siRNA, accompanied by a decrease of fluorescence in the control without antisense agents. These results are indicative of a similar duration of gene silencing for both the LNA gapmer and the siRNA using a transient transfection system. This finding, however, will have to be confirmed with an endogenously expressed target gene.
Estimation of IC50 values
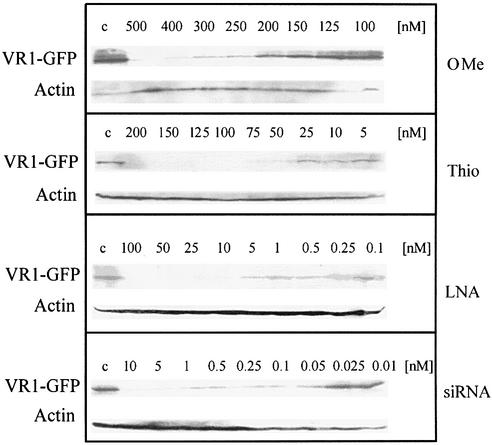
To further quantify potencies of the different strategies, we estimated IC50 values with experiments in a suitable concentration range for each antisense molecule (Fig. 5). Protein levels were determined from western blots with the Quantity One program (see Materials and Methods). Three independent experiments were performed for each type of ON in order to minimize errors due to typical variations of transfection efficiencies. Quantitative evaluation by sigmoid fitting of average values of the three independent experiments is shown in Figure 6. In order to indicate the variability between experiments, individual IC50 values of three independent experiments for the different types of antisense molecules to inhibit VR1–GFP expression are summarized in Table 2. Average IC50 values and standard deviations of the three experiments are given in column 5 of Table 2.
Figure 5.
Western blot analysis of the antisense efficiency of modified ONs (OMe-RNA gapmer, PS and LNA gapmer) and siRNA in an optimal concentration range for each antisense molecule.
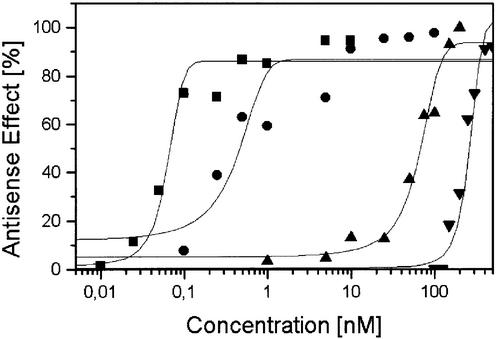
Figure 6.
Quantification of the antisense effects of OMe-RNA and LNA gapmers, a PS and the VR1-siRNA. The antisense effects were determined from the levels of the target protein in the western blots shown in Figure 5. Average values of three independent experiments are plotted as a function of AS ON concentrations (logarithmic scale) and fitted with a sigmoidal Boltzman function. Square = siRNA; circle = LNA; triangle = thioate; diamond = OMe.
Table 2. Estimated IC50 values for the PS, LNA and OMe-RNA gapmers and siRNA to inhibit expression of VR1–GFP and half-lives of the ONs in cell culture medium.
| IC50 Exp1 (nM) | IC50 Exp2 (nM) | IC50 Exp3 (nM) | IC50,average (nM) | t1/2 (h) | |
|---|---|---|---|---|---|
| DNA | ND | ND | ND | ND | 0.33 ± 0.03 |
| Thio | 48.9 | 61.0 | 85.7 | 70 ± 20 | >>48 |
| LNA | 0.47 | 0.34 | 0.37 | 0.4 ± 0.07 | 41 ± 16 |
| OMe | 211.4 | 197.5 | 264.8 | 220 ± 10 | 5.2 ± 1.2 |
| siRNA | 0.043 | 0.065 | 0.081 | 0.06 ± 0.02 | 2.1 ± 1.2 |
Individual results of three independent experiments are given to indicate variations of antisense effects between experiments. The fifth column shows the average and standard deviation of the three experiments. The last column shows estimated half-lives of the ONs in cell culture medium containing 10% FCS. Averages and standard deviation of three independent experiments are given. ND = not determined.
As could already be seen in the preceding experiments, siRNA revealed an extremely high antisense potency with a significant antisense effect at concentrations as low as 0.05 nM and an IC50 value of 0.06 nM. The efficiency of the siRNA approach was increased by a factor greater than 1000 compared with PS. The optimized LNA–DNA–LNA ON still had a 175-fold lower IC50 of 0.4 nM compared with the isosequential PS with an IC50 value of 70 nM, whereas the OMe-modified gapmer had a 3-fold lower antisense potency (IC50 ∼220 nM). These results demonstrated that siRNAs and LNA gapmers are promising antisense molecules because of their high efficacy, and, in the case of LNA, enhanced stability and high target affinity.
To further analyze possible reasons for the different potencies of the ONs investigated above, we estimated their half-lives in cell culture medium containing 10% FCS. Because LNAs are difficult to phosphorylate on the 5′ end and to avoid underestimation of stability due to phosphatases, we did not use the widely applied approach to end-label the ONs with radioactive 32P, but stained the full-length ONs with ethidium bromide. All ONs were incubated in cell culture medium under equal conditions, but a lower amount of the siRNA, as compared with the single-stranded ONs, was analyzed due to intensive staining of the dsRNA by ethidium bromide. In contrast, the LNA gapmer was weakly stained and its half-life could only be determined with a large error. Estimated half-lives of the ON of three independent experiments are summarized in Table 2. The PS and the LNA gapmer were highly stable under the conditions used. The OMe gapmer exhibited a much lower stability, which might explain its weak antisense effect. Interestingly, the siRNA had a short half-life of only ∼2 h. This might indicate that additional factors exist to protect short dsRNA molecules inside cells from nucleolytic degradation.
DISCUSSION
In recent years, a variety of nuclease-resistant AS ONs with high affinities towards their target sequences were developed. In addition, RNAi as a highly efficient mechanism for gene silencing was adapted for mammalian cells. As Braasch and Corey mentioned in a recent review (13), different antisense strategies should now be compared to optimize approaches for in vivo and therapeutic applications. In our present study, we therefore compare gapmers containing LNA or OMe-RNA with PS and target-specific siRNA.
We fused our gene of interest, VR1, to GFP which can easily be detected by fluorescence microscopy and western blotting with commercially available anti-GFP antibodies. After co-transfection of the plasmid coding the fusion protein with different AS ONs, a good correlation was observed between fluorescence intensity and signals in the northern and western blot, i.e. the use of GFP fusion proteins is an easy method to optimize AS ONs even if suitable target specific antibodies do not exist.
For initial cell culture experiments, PS against target sites were used which have previously been found to be highly accessible in vitro (37). Efficient transfection of >90% of the cells with AS ONs was achieved as determined by the use of Cy3-labeled ONs (data not shown). Both AS ONs (V15 and V29) could entirely inhibit VR1–GFP expression, the ON directed against target site V15 being slightly more efficient than the ON directed against target site V29. In our previous in vitro study, ON V29 had a somewhat higher efficiency than V15 (94% mRNA cleavage compared with 83% degradation). From our results, we conclude that suitable ONs can be identified by in vitro screening, but the efficiency of individual ONs in cell culture might differ slightly due to different folding of the mRNA and/or RNA-binding proteins.
One important goal of this study was to optimize the design of chimeric LNA–DNA ONs with high antisense potency in cells. Previously, we found that LNA gapmers with at least three LNA end blocks are sufficient to stabilize the ON in human serum at least 10-fold compared with the unmodified ODNs (19). Furthermore, we have shown that a stretch of seven to eight DNA monomers in the center of an 18mer ON is required to activate RNase H. In addition, it was shown that efficient antisense inhibition of the coding region of the firefly luciferase gene in cell culture requires activation of RNase H (23). In our study, we focused on LNA gapmers fulfilling the requirements to activate RNase H. Accordingly, we co-transfected LNA gapmers against target site V29 that contained five (L5) or four (L4) LNA end blocks with 8 or 10 DNA monomers in the center. The LNA ONs were efficient, the gapmer with five LNA monomers at either end being superior to the gapmer with only 4 nt end blocks. Moreover, a further target site in the Pim-1 mRNA was recognized by Pim1-specific L4 and L5 gapmers, leading to an efficient inhibition of Pim-1 expression. Together with the results from our previous in vitro study, these data show that chimeric ONs with five or four LNA monomers at the ends and a central stretch of 8 or 10 deoxynucleotides are potent antisense molecules. A further extension of the end block does not seem to be reasonable, since gapmers with a shorter stretch of consecutive DNA monomers in the center do not initiate efficient target RNA cleavage by RNase H. In line with our data, Corey and co-workers found an LNA gapmer with five LNA end-blocks directed against the coding region of luciferase to be a potent inhibitor of luciferase gene expression (23).
In the last years, AS ONs with novel modifications and siRNAs gained importance as molecules that specifically suppress the expression of a target gene. Here, we investigated the respective hit rates for LNA gapmers and siRNAs. The LNA ON sequences were chosen on the basis of our previous in vitro study (37), whereas the siRNAs were designed according to the following criteria: the sequences start with AA, the GC content was kept below 50%, regions close to the start and stop codon were avoided, and sequences with homologies to other coding sequences were eliminated. As can be seen in Figure 3, approximately one-third of the LNA ONs and two-thirds of the siRNAs efficiently suppressed target gene expression at a concentration of 10 nM. It should be noted that the hit rate varies from one target to the next. We found much lower portions of potent siRNAs for other targets (data not shown) and, as an example, Miyagishi and Taira reported that only one siRNA out of five was highly efficient in inhibiting luciferase expression, whereas one siRNA was completely inactive and the remaining three siRNAs had moderate activities (41). Nevertheless, there is a good chance of obtaining one efficient antisense agent when starting with three to five LNA gapmers or siRNAs according to the suggested criteria.
The major aim of our study was to evaluate the potency of different types of antisense strategies. We therefore compared the antisense effects of PS, LNA and OMe-RNA gapmers and siRNA. Interestingly, a PS isosequential to an siRNA, which was directed against the optimal AA(N)19TT target site for RNAi approaches, had hardly any specific antisense effect (data not shown). On the contrary, the target sites optimized for AS ONs do not start with two consecutive As, which are recommended for the design of siRNA. It is therefore likely that optimal target sites for AS ONs are not necessarily identical to optimal target sites for siRNA. This assumption, however, has to be proven in further experiments. Nevertheless, since the major goal of antisense approaches is to knock down target gene expression, we compared isosequential single-stranded molecules against the optimized target site for AS ONs and an siRNA directed against a target site, which fulfills optimal criteria for siRNA approaches.
We observed that VsiRNA1 almost completely suppresses VR1–GFP expression at the lowest concentration used for initial experiments. The LNA gapmer is also highly efficient in the low nanomolar range, whereas much higher concentrations are needed for PS and no effect is observed for the OMe-RNA gapmer in the concentration range used.
Furthermore, the time course for the suppression of VR1–GFP expression by the LNA ON and the siRNA was investigated. An almost complete silencing of the target gene for up to 4 days was observed for both types of antisense molecules (data not shown). At days 5 and 6, fluorescence reappeared, but gene expression of the untreated control decreased from day 4 due to the transient transfection procedure used. Nevertheless, our results obtained so far are in line with reports in the literature. Inhibition of human Bcl-X by siRNAs as well as 2′-O-methoxyethyl/deoxyPS gapmers was found to be maximal at 24 h post-transfection and the RNA level returned to normal by day 5 (42). In addition, the expression of the human tissue factor was efficiently suppressed by siRNA for up to 3 days, but silencing dropped off at day 5 (no data were shown for day 4) (43). It should be noted that inhibition of target gene expression was prolonged in the latter study by introduction of modified nucleotides into the siRNA.
Further experiments were performed to estimate IC50 values (Figs 5 and 6), which are summarized in Table 2. The LNA gapmer is the most efficient molecule of the single-stranded AS ONs. LNA-containing ONs have been shown to have potencies exceeding that of the isosequential unmodified DNA ON (20). From this study, however, it could not be concluded whether the LNA gapmer had improved antisense properties besides a higher stability against nucleolytic degradation. In another recent study, LNA and peptide nucleic acid (PNA) AS ONs against human telomerase were compared (21). The LNA ON had a significantly higher affinity towards the target RNA compared with the analogous PNA. IC50 values for LNA-mediated inhibition of human telomerase in cell lysates in the low nanomolar range were reported, whereas a PNA ON had a 200-fold lower potency. In our present study, we determined antisense efficiencies in living cells and found a comparable IC50 of 0.4 nM for the best LNA–DNA gapmer, which was 175- and 550-fold superior compared with IC50 values for the analogous PS and OMe-RNA/DNA gapmer, respectively. Two properties were reported in the literature to account for a good antisense efficiency of LNA ONs: high affinity toward complementary sequences (19,21) and good cellular uptake after delivery by lipofection (22,24).
The most efficient antisense molecule used in the present study was siRNA. Only recently it could be shown that RNAi can be induced in mammalian cells by introduction of 21 nt siRNA duplexes without activating unspecific responses, e.g. the interferon response (30). In our co-transfection experiments, we observed efficient and specific inhibition of VR1–GFP expression even in the subnanomolar range, which is in line with published data (40). Thus the potency of siRNA directed against VR1 is 7-fold higher compared with the LNA gapmer and >1000-fold higher compared with the PS. Details of the mechanism of RNAi in mammalian cells that account for the high efficiency remain to be clarified.
It should be noted that it is still being discussed controversially in the literature whether RNAi is superior compared with traditional antisense approaches. For example, Bertrand et al. (44) found siRNA to be more efficient in inhibiting GFP expression in cell culture and in vivo in xenografted mice compared with an AS ON stabilized by two PS linkages at both ends. In contrast, a detailed study with 80 siRNAs revealed that, in general, antisense efficiency correlated with the activity of RNase H-dependent 2′-O-methoxyethyl/PS ONs targeted to the same sites (42). In these experiments, both types of antisense agents behaved similarly in terms of potency, maximal effect, specificity and duration of action, and efficiency. However, for this study, siRNAs were not designed according to the optimized criteria described in the literature (see above). One might therefore speculate that good target sites for AS ONs are not necessarily suitable for siRNAs. Therefore, further studies are needed to decide whether RNAi is more efficient than conventional antisense approaches. In addition, single-stranded ONs might still have advantages over siRNA, such as lower production costs and easier handling. Furthermore, far more knowledge has been gained for AS ONs over the last two decades as compared with the poor range of information available for siRNAs. Pharmacokinetics for different types of AS ONs have been studied intensively in animal models (1), and more than a dozen AS ONs are currently being tested in clinical trials (14).
One factor that might influence antisense activity of an ON is its resistance to nucleolytic degradation. We found the PS as well as the LNA gapmer to be highly stable in cell culture medium containing 10% FCS. Unmodified DNA is degraded in <1 h and the OMe gapmer had a half-life of only 5 h, which might account for its low antisense activity. The siRNA exhibited a half-life of ∼2 h. In a recent study, double-stranded siRNA was found to be degraded in <1 h in HeLa cell lysate, but ∼75% of intact siRNA was recovered 24 h after lipofection into HeLa cells (44). It is therefore likely that additional factors, e.g. the transfection reagent or cellular components, further protect the ONs from degradation. In addition, nuclease resistance is known to be sequence dependent.
Taken together, we compared the efficiency of different antisense strategies in mammalian cell culture. Double-stranded siRNA molecules recruiting the intracellular RNAi mechanism were the most potent antisense molecules against the VR1–GFP mRNA (IC50 of 0.06 nM). Single-stranded ONs consisting of 5 nt LNA end blocks and a DNA stretch of eight monomers in the center have been found to be 175- and 550-fold superior in suppressing the expression of the gene targeted in the present study compared with isosequential PS and OMe gapmer ONs, respectively. We therefore conclude that siRNA as well as chimeric LNA–DNA ONs are attractive antisense agents that will now be tested in animal models.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank S. Schubert for initial help in cell culture experiments, and Dr C. Gillen, Dr E. Wade (Grünenthal GmbH, Germany) and Professor F. Hucho (Free University Berlin, Germany) for helpful discussions and support. In addition, we gratefully acknowledge Proligo, Boulder/Paris, for supplying LNA. This work was financed by the Bundesministerium für Bildung und Forschung (grant no. 01GG9818/0) and Fonds der Chemischen Industrie.
REFERENCES
- 1.Crooke S.T. (2000) Progress in antisense technology: the end of the beginning. Methods Enzymol., 313, 3–45. [DOI] [PubMed] [Google Scholar]
- 2.Phillips M.I. and Zhang,Y.C. (2000) Basic principles of using antisense oligonucleotides in vivo. Methods Enzymol., 313, 46–56. [DOI] [PubMed] [Google Scholar]
- 3.Stein C.A. (2001) The experimental use of antisense oligonucleotides: a guide for the perplexed. J. Clin. Invest., 108, 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dove A. (2002) Antisense and sensibility. Nat. Biotechnol., 20, 121–124. [DOI] [PubMed] [Google Scholar]
- 5.Marwick C. (1998) First ‘antisense’ drug will treat CMV retinitis. J. Am. Med. Assoc., 280, 871. [PubMed] [Google Scholar]
- 6.Eckstein F. (2000) Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev., 10, 117–121. [DOI] [PubMed] [Google Scholar]
- 7.Brown D.A., Kang,S.H., Gryaznov,S.M., DeDionisio,L., Heidenreich,O., Sullivan,S., Xu,X. and Nerenberg,M.I. (1994) Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem., 269, 26801–26805. [PubMed] [Google Scholar]
- 8.Guvakova M.A., Yakubov,L.A., Vlodavsky,I., Tonkinson,J.L. and Stein,C.A. (1995) Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors and remove it from low affinity binding sites on extracellular matrix. J. Biol. Chem., 270, 2620–2627. [DOI] [PubMed] [Google Scholar]
- 9.Rockwell P., O’Connor,W.J., King,K., Goldstein,N.I., Zhang,L.M. and Stein,C.A. (1997) Cell-surface perturbations of the epidermal growth factor and vascular endothelial growth factor receptors by phosphorothioate oligodeoxynucleotides. Proc. Natl Acad. Sci. USA, 94, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin A.A. (1999) A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim. Biophys Acta, 1489, 69–84. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal S. and Zhao,Q. (1998) Mixed backbone oligonucleotides: improvement in oligonucleotide-induced toxicity in vivo. Antisense Nucleic Acid Drug Dev., 8, 135–139. [DOI] [PubMed] [Google Scholar]
- 12.Toulme J.J. (2001) New candidates for true antisense. Nat. Biotechnol., 19, 17–18. [DOI] [PubMed] [Google Scholar]
- 13.Braasch D.A. and Corey,D.R. (2002) Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry, 41, 4503–4510. [DOI] [PubMed] [Google Scholar]
- 14.Kurreck J. (2003) Antisense technologies: improvement through novel chemical modifications. Eur. J. Biochem., 270, 1628–1644. [DOI] [PubMed] [Google Scholar]
- 15.Braasch D.A. and Corey,D.R. (2001) Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol., 8, 1–7. [DOI] [PubMed] [Google Scholar]
- 16.Elayadi A.N. and Corey,D.R. (2001) Application of PNA and LNA oligomers to chemotherapy. Curr. Opin. Invest. Drugs, 2, 558–561. [PubMed] [Google Scholar]
- 17.Ørum H. and Wengel,J. (2001) Locked nucleic acids: a promising molecular family for gene-function analysis and antisense drug development. Curr. Opin. Mol. Ther., 3, 239–243. [PubMed] [Google Scholar]
- 18.Petersen M. and Wengel,J. (2003) LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol., 21, 74–81. [DOI] [PubMed] [Google Scholar]
- 19.Kurreck J., Wyszko,E., Gillen,C. and Erdmann,V.A. (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res., 30, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahlestedt C., Salmi,P., Good,L., Kela,J., Johnsson,T., Hokfelt,T., Broberger,C., Porreca,F., Lai,J., Ren,K., Ossipov,M., Koshkin,A., Jakobsen,N., Skouv,J., Oerum,H., Jacobsen,M.H. and Wengel,J. (2000) Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA, 97, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elayadi A.N., Braasch,D.A. and Corey,D.R. (2002) Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry, 41, 9973–9981. [DOI] [PubMed] [Google Scholar]
- 22.Arzumanov A., Walsh,A.P., Rajwanshi,V.K., Kumar,R., Wengel,J. and Gait,M.J. (2001) Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry, 40, 14645–14654. [DOI] [PubMed] [Google Scholar]
- 23.Braasch D.A., Liu,Y. and Corey,D.R. (2002) Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res., 30, 5160–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fluiter K., ten Asbroek,L.M.A., de Wissel,M., Jakobs,M.E., Wissenbach,M., Olsson,H., Olsen,O., Oerum,H. and Baas,F. (2003) In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res., 31, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamore P.D. (2001) RNA interference: listening to the sound of silence. Nature Struct. Biol., 8, 746–750. [DOI] [PubMed] [Google Scholar]
- 26.Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- 27.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 28.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 29.He Y and Katze,M.G. (2002) To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol., 15, 95–119. [DOI] [PubMed] [Google Scholar]
- 30.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 31.McCaffrey A.P., Meuse,L., Pham,T.T., Conklin,D.S., Hannon,G.J. and Kay,M.A. (2002) RNA interference in adult mice. Nature, 418, 38–39. [DOI] [PubMed] [Google Scholar]
- 32.Xia H., Mao,Q., Paulson,H. and Davidson,B.L. (2002) siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol., 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 33.Brummelkamp T., Bernards,R. and Agami,R. (2002) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell, 2, 243–247. [DOI] [PubMed] [Google Scholar]
- 34.Montell C., Birnbaumer,L. and Flockerzi,V. (2002) The TRP channels, a remarkably functional family. Cell, 108, 595–598. [DOI] [PubMed] [Google Scholar]
- 35.Caterina M.J., Schumacher,M.A., Tominaga,M., Rosen,T.A., Levine,J.D. and Julius,D. (1997) The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature, 389, 816–824. [DOI] [PubMed] [Google Scholar]
- 36.Tominaga M., Caterina,M.J., Malmberg,A.B., Rosen,T.A., Gilbert,H., Skinner,K., Raumann,B.E., Basbaum,A.I. and Julius,D. (1998) The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron, 21, 531–543. [DOI] [PubMed] [Google Scholar]
- 37.Kurreck J., Bieber,B., Jahnel,R. and Erdmann,V.A. (2002) Comparative study of DNA enzymes and ribozymes against the same full-length messenger RNA of the vanilloid receptor subtype I. J. Biol. Chem., 277, 7099–7107. [DOI] [PubMed] [Google Scholar]
- 38.‘t Hoen P.A., Rosema,B.S., Commandeur,J.N., Vermeulen,N.P., Manoharan,M, van Berkel,T.J., Biessen,E.A. and Bijsterbosch,M.K. (2002) Selection of effective antisense oligodeoxynucleotides with a green fluorescent protein-based assay. Discovery of selective and potent inhibitors of glutathione S-transferase Mu expression. Eur. J. Biochem., 269, 2574–2583. [DOI] [PubMed] [Google Scholar]
- 39.Saris C.J., Domen,J. and Berns,A. (1991) The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J., 10, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elbashir S.M., Harborth,J., Weber,K. and Tuschl,T. (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Method, 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 41.Miyagishi M. and Taira,K. (2002) U6 promotor-driven siRNA with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 42.Vickers T.A., Koo,S., Bennett,C.F., Crook,S.T., Dean,N.M. and Baker,B.F. (2003) Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. J. Biol. Chem., 278, 7108–7118. [DOI] [PubMed] [Google Scholar]
- 43.Amarzguioui M., Holen,T., Babaie,E. and Prydz,H. (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res., 31, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand J.-R., Pottier,M., Vekris,A., Opolon,P., Maksimenko,A. and Malvy,C. (2002) Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun., 296, 1000–1004. [DOI] [PubMed] [Google Scholar]