Abstract
The chromatin environment and the sites of integration in the host genome are critical determinants of human immunodeficiency virus (HIV) transcription and replication. Depending on the chromosomal location of provirus integration within the genome, HIV-1 long terminal repeat (LTR)-mediated transcription may vary from 0- to 70-fold. Cis-elements such as topoisomerase II cleavage sites, Alu repeats and matrix attachment regions (MARs) are thought to be targets for retroviral integration. Here we show that a novel MAR sequence from the T-cell receptor β locus (MARβ) and the IgH MAR mediate transcriptional augmentation when placed upstream of the HIV-1 LTR promoter. The effect of transcriptional augmentation is seen in both transient and stable transfection, indicating its effect even upon integration in the genome. MAR-mediated transcriptional elevation is independent of Tat, and occurs synergistically in the presence of Tat. Further, we show that MAR-mediated transcriptional elevation is specific to the HIV-1 LTR and the Moloney murine leukemia virus LTR promoter. In a transient transfection assay using over-expressed IκB, the inhibitor of NF-κB, we show that MAR-induced processive transcription is NF-κB dependent, signifying the role of local enhancers within the LTR promoter. Furthermore, by RNase protection experiments using proximal and distal probes, we show that MAR-mediated transcriptional upregulation is more prominent at the distal rather than the proximal end, thus indicating the potential role of MARs in promoting elongation.
INTRODUCTION
The long terminal repeats (LTRs) of human immunodeficiency virus type-1 (HIV-1) act as an inducible promoter that can be selectively stimulated by the potent transactivator protein Tat (1). HIV encodes Tat which increases the processivity of transcription due to the cooperative binding of Tat to the upper stem and bulge region of the Tat-responsive element TAR, a structured element in the nascent viral RNA, resulting in hyperphosphorylation of RNA polymerase II (RNAP II) (2). Recent studies indicate that HIV-1 Tat associates tightly with the CDK9-containing positive transcription elongation complex P-TEFb (3,4) and with its component cyclin T1, inducing the loop-specific binding of the P-TEFb complex to TAR RNA (5). Upon phosphorylation by this complex, RNAP transcribes downstream genes by overcoming terminators. Although HIV transcription is dependent on transactivator Tat protein, in the initial phases at least, a few transcripts seem to be made independently of Tat.
Integration of retroviral DNA into the host genome is an obligatory step for viral replication (6). During the process of HIV infection, a linear cDNA molecule is integrated into the host genome as a provirus (7). However, due to the heterogeneity of the chromatin, the site of the integration of HIV in the genome could have dramatic effects on its transcriptional activation. Thus, the activity of the de novo promoter is determined by the site of integration and local enhancers of the integrated promoter. In an in vitro study on the integration reaction, the specificity for integration is shown to be favored in target DNAs containing sequence-directed bends or DNA distortions caused by bound proteins (8). In many cases, the backbone DNA conformation, such as a bend or curvature, may influence transcription in the absence of trans-factors (9). In another in vitro study on the integration reaction and flanking sequence analysis at the nucleotide level, a preference for integration at AT-rich regions was observed (10,11). A similar study in which analysis of 24 distinct human T-cell leukemia virus type-1 (HTLV-1) integration sites derived from asymptomatic carriers was carried out, the mean AT content of the hexameric repeat was found to be 59% (12).
In eukaryotes, chromatin is heterogeneous as it comprises euchromatin and heterochromatin; this heterogeneity is seen in both structural and functional aspects. In eukaryotic cells, the primary level of genome organization consists of nucleosomes that are formed by wrapping of double-stranded DNA onto the core histones. The spatial arrangement of the nucleosomes is affected both by trans-factors and by structural features of DNA (13). Upon integration, the HIV LTR is bound by a nucleosome (nuc-1) immediately downstream of the transcription start site. Remodeling of nuc-1 plays a significant role in transcriptional activation of the HIV LTR promoter (14). Thus, the results of in vivo integration studies can potentially address the influence of chromatin on target sites. In the case of retroviruses, it has been shown that integration preferentially occurs near DNase I-hypersensitive sites, Alu elements or topoisomerase (Topo) cleavage sites (15,16). While searching for global accessibility regions, it has been found that the integration complex could be specifically directed towards replication sites that might be preferred due to their association with the nuclear matrix (17). A recent report on integration target selection by HIV-1 in the genome and transcriptional profiling revealed a strong correlation between gene activity and integration targeting. Specifically, active genes were shown to be preferred sites for the integration (18).
Matrix attachment regions (MARs) are a class of cis-regulatory elements that are typically ∼200 bp long AT-rich DNA sequences, occurring an average of one in every 50 kb of eukaryotic DNA (19–21), and possess high affinity for the isolated nuclear matrix (22–24). MARs are often closely associated with transcriptional promoters and enhancers of several genes (25,26), e.g. the IgH enhancer (27,28). These are proposed to organize the genomic DNA into topologically independent loop domains and are implicated in chromatin accessibility, replication and transcription (29–32).
We therefore studied the effect of MARs on HIV LTR-mediated transcription. This report highlights the role of a novel MARβ and the well-characterized IgH MAR in the context of HIV LTR-mediated transcription. Our studies indicate that a short MAR sequence plays an essential role in HIV LTR-mediated transcription over a distance. For the first time, we report that MAR sequences upregulate HIV LTR-mediated transcription even in the absence of transactivator protein Tat by increasing the basal level of transcription both before and after integration. By RNase protection analysis, we further show that MARs enhance basal level transcription by increasing the processivity of transcription from the HIV-1 LTR promoter.
MATERIALS AND METHODS
PCR amplification and cloning of HIV-1 LTR
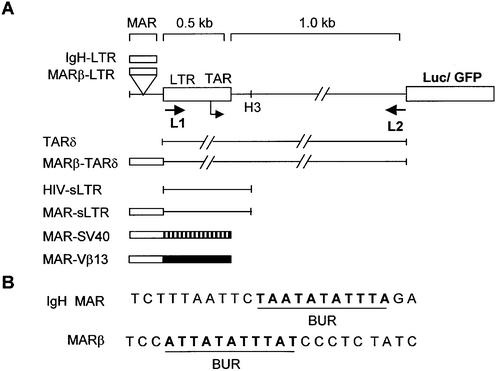
L1 (5′-CAAGATATCCTTGATCTGTGG-3′) and L2 (5′-AAGGGGAACCAAGAGA-3′) primers were used for amplification of the 1.5 kb region comprising the 5′ LTR and its downstream sequence (1.5 kb LTR) from the HXB3 genomic clone of HIV-1 (NIH AIDS reagent program) (33). The amplified product was purified and cloned into pGEM-T easy which was then cut out using EcoRI, Klenowed and further subcloned into the SmaI site of SK+ (Stratagene), producing NC-1. The EcoRI- and BamHI-digested 1.5 kb LTR fragment from NC-1 was then subcloned into promoterless pEGFP1 (Clontech), and named LTR-green fluorescent protein (GFP). Similarly, NC-1 was digested with XhoI and BamHI, and the LTR fragment was cloned into pGL3-Basic vector (Promega), and named LTR-Luc (Fig. 1). MARβ and a dimer of IgH sequences in pBluescript-SK+ were digested with XhoI and SalI and cloned into the XhoI sites upstream of either LTR-GFP or LTR-Luc vector plasmids, creating IgH-LTR-Luc or MARβ-LTR-Luc constructs. For construction of the 0.5 kb core LTR constructs (small LTR; sLTR), both MAR-LTR and LTR-only constructs were digested with HindIII and religated. XhoI-digested MARβ was cloned upstream of the SV40 promoter using the SalI site in the PGL3 promoter. pcDNA-Tat, the expression vector for Tat, was constructed by cloning HXB3 Tat in pcDNA 3.1 using EcoRI and NotI sites. For deletion of the TAR sequence from the LTR-Luc construct, the plasmid DNA was digested with BglII and SacI and religated to generate TARδ; for cloning of MARβ upstream of TARδ, the KpnI site was used. The construct obtained upon subcloning was named MARβ TARδ. Subcloning of the Moloney murine leukemia virus (MoMuLV) LTR was carried out from the MSCV 2.2 vector using KpnI and SalI into the KpnI and XhoI sites of pGL3-Basic. For cloning of MARβ upstream of the MoMuLV LTR, the KpnI site was used. All the constructs were confirmed by nucleotide sequencing.
Figure 1.
Schematic representation of the 1.5 kb HIV-1 LTR together with GFP-Luc as the reporter system. (A) L1 and L2 primers used for amplification are indicated by arrows. To make the short LTR (sLTR), the HindIII site was used as depicted. Hatched and solid boxes show SV40 and Vβ13 promoters, respectively. (B) Sequences of IgH and MARβ used in the studies. Underlined bold letters show the base unpairing region within the MAR sequences.
Cell culture and transfection experiments
293 and CHO (Chinese hamster ovary) cell lines were used for most of the transfections. Cells were grown on Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) in the presence of 5% CO2 at 37°C. Cells were seeded in a 35-mm diameter 6-well plate at 1 × 106 cells per well. Transfections were done using Lipofectamine 2000 (Gibco-BRL) in plain DMEM. A 2 µg aliquot of plasmid DNA was used for each transfection. In all Tat-dependent experiments, 1 µg of pcDNA-Tat was used for co-transfection. For raising stable cell lines, CHO cells were transfected with 2 µg of LTR-GFP and MARβ-LTR-GFP plasmid. After 3 days of transfection, cells were selected for neomycin resistance using G418 in DMEM. After 18 days of selection at various concentrations of G418, GFP-positive cells with integrated plasmids were obtained. Finally, stable reporter-expressing cells were maintained at 800 µg/ml G418 in DMEM + 10% FCS.
Luciferase assay
Luciferase assays were performed using a single Luciferase Assay Reporter system (Promega), and luciferase activity was calculated using a Fluroskan Ascent Luminometer (Labsystems). Forty hours after transfection, the culture medium was removed; cells were washed with phosphate-buffered saline (PBS) and processed further. Cells were resuspended into 200 µl of reporter lysis buffer and kept at –70°C. After freeze–thawing twice, lysed cells were spun at 9.5 g for 15 min. For accurate quantitation of luciferase activity, an equal amount (50 µg) of the protein was assayed. Protein concentrations of the lysates were measured using Bradford reagent (Bio-Rad).
GFP constructs and microscopy
The cells were transfected with the plasmid constructs containing LTR-GFP or MARβ-LTR-GFP into 293 cells. After 40 h of transfection, GFP-positive cells were viewed under an inverted microscope (Olympus) using a GFP filter. For fluorescence-activated cell sorting (FACS) analysis, cells were washed and resuspended in PBS, and finally GFP fluorescence was measured using FACS Vantage (Becton Dickinson).
Isolation of genomic DNA and Southern blot analysis
Genomic DNA was extracted from cell lines, which were stably transfected with LTR-GFP and MARβ-LTR-GFP, using the DNA-Zol method (Gibco-BRL). The optical density (OD) of each genomic DNA sample was calculated using a Smartspec spectrophotometer (Bio-Rad).
Genomic DNAs isolated from LTR-GFP and MARβ-LTR-GFP stable cell lines were subjected to restriction digestion using HindIII. Digested DNA was electrophoretically separated on a 0.9% agarose gel at 80 V and transferred onto Zetaprobe (Bio-Rad). Southern blot analysis was performed using a 1 kb HindIII fragment having a 0.5 kb LTR core sequence and 0.5 kb of additional downstream sequence. The LTR fragment was labeled with [α-32P]dATP using the protocol supplied by Gibco-BRL.
RNase protection assay
293 cells were transfected with LTR, MARβ-LTR and IgH-LTR in either the presence or absence of pcDNA-Tat. Cells were harvested at 48 h post-transfection, washed in PBS and pelleted at 9.5 g. Total RNA was isolated using Tri-Reagent (Sigma) and treated with 10 U of RNase-free DNase I (Boerhinger Mannheim) and 80 U of RNasein for 30 min at 37°C. RNA samples were then extracted with phenol/ chloroform and isoamyl alcohol, and finally precipitated with 2.5 vols of ethanol. Extracted RNA was resuspended in 30 µl of sterile diethylpyrocarbonate-treated water. A 10 µg aliquot of total RNA was used for each protection assay. An antisense probe for the distal and proximal sequences of the LTR was made by in vitro transcription in the presence of T3 and T7 RNAP, respectively. In a 25 µl transcription reaction, 1 µg of linearized DNA fragment, 1× transcription buffer (Stratagene), 0.4 mM ATP, 0.4 mM CTP, 0.4 mM GTP, 0.25 mM UTP, 50 µCi of [α-32P]UTP, 40 U of RNasin and 20 U of T3 RNAP (Stratagene) were used. Labeled RNA transcripts were purified using probequant G50 columns (Amersham, Pharmacia). Samples were heated at 90°C for 3 min before incubation for hybridization. For the hybridization reaction, 10 µg of total RNA was incubated with 105 c.p.m. of probe in hybridization buffer at 37°C for 18 h. After 18 h, the reaction mixture was diluted using 300 µl of digestion buffer. Single-stranded RNA was then digested using RNase T1 (15 µg/ml) and RNase A (1 µg/ml) for 2 h at 30°C. After phenol/chloroform extraction and ethanol precipitation, samples containing protected transcripts were analyzed on a 6% urea–polyacrylamide gel. Band intensity was quantified using a phosphoimager (Bio-Rad).
RESULTS
Activation of transcription from the HIV-1 LTR by TCR MARβ and IgH MAR sequences
MARs have been shown to be present near or within various enhancers, playing a pivotal role in transcriptional regulation. We made the HIV LTR promoter constructs with or without MARs to check the transcriptional profile by expression of reporter genes. As discussed in Materials and Methods, we have PCR amplified a 1.5 kb LTR containing 0.5 kb of core promoter along with 1 kb of downstream sequence up to the beginning of the Gag gene from the HXB3 clone of HIV-1 (Fig. 1A).
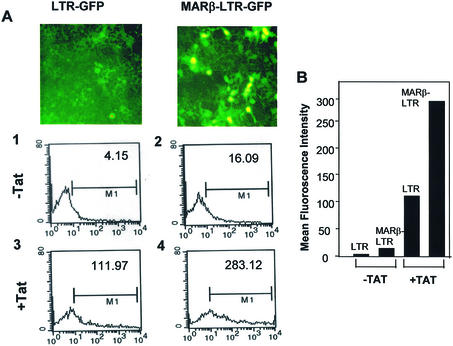
Since most of the strong terminators are located downstream of the core of the LTR promoter up to the Gag open reading frame, making the transcription poor, we analyzed the role of MARs using 1.5 kb LTR. Transcriptional activation was visualized by GFP expression upon transient transfection of either LTR-GFP or MARβ-LTR-GFP plasmid expressing GFP as a reporter. Visualizing the reaction by UV fluorescence showed very weak expression of GFP from the LTR promoter, while GFP expression was enhanced when MARβ was present upstream of the LTR. With the co-transfection of pcDNA-Tat, a significant increase in GFP expression was observed in the MARβ-LTR compared with the LTR alone (Fig. 2A). Quantitation of the percentage of GFP expression was performed by FACS and displayed an ∼4-fold increase in the GFP fluorescence in the presence of MARβ compared with LTR-GFP alone. Under similar conditions, co-transfection of Tat induced transcription which was 14-fold higher in LTR-GFP and 38-fold higher in MARβ-LTR-GFP (Fig. 2B).
Figure 2.
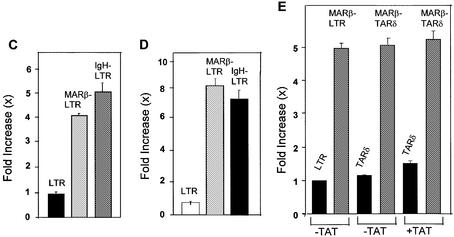
MAR-mediated enhancement of transcription through the LTR promoter. (A) FACS analysis of the GFP-positive 293 cells. One million 293 cells were seeded on a 30 mm plate and transiently transfected with the plasmid constructs expressing GFP under the influence of the LTR and MARβ-LTR promoters. A 2 µg aliquot of LTR-GFP and MARβ-LTR-GFP plasmid DNAs was transfected. In the case of the presence of Tat protein, 1 µg of pcDNA-Tat was co-transfected. The photographs were taken after 40 h using an inverted microscope (Olympus) under UV fluorescence. (B) Similarly, FACS analysis was performed from the same cells and the percentage of GFP-positive cells was plotted using a Sigma plot. (C and D) 293 cells were seeded as described above and transiently transfected with LTR-Luc, MARβ-LTR-Luc and IgH-LTR-Luc at 2 µg/well. In the case of Tat, 1 µg of pcDNA-Tat was co-transfected. Relative luciferase activities were calculated 40 h post-transcription by loading an equal amount of protein (50 µg). (E) 293 cells were seeded as described above and transiently transfected with TARδ and MARβ-TARδ at 2 µg/well. In the case of pcDNA-Tat, 1 µg of the plasmid was co-transfected. Relative lucierase activity was calculated 40 h after transfection. A 50 µg aliquot of protein was assayed for luciferase assay.
Further, in order to check if MAR-mediated enhancement of transcription is specific to MARβ or is also the case for other MARs, we placed the well-characterized IgH MAR upstream of the LTR. Upon transient expression of these plasmids, a 4-fold increase in luciferase activity was observed in the MARβ-LTR and IgH-LTR compared with the LTR only (Fig. 2C). Upon co-transfection with pcDNA-Tat in an experiment similar to that mentioned above, an ∼7- to 9-fold upregulation of the MARβ-LTR and IgH-LTR was observed when compared with the LTR with Tat (Fig. 2D). Thus, like the novel MARβ, the well-studied IgH MAR also significantly upregulated HIV LTR-mediated transcription. Since transcriptional activation of the HIV LTR is strongly dependent on transactivator protein Tat, our results demonstrate that MAR-mediated transcriptional enhancement is independent of Tat and increases proportionally with the induction of Tat protein.
In addition to Tat, TAR is known to be bound by cyclin T1 and CDK9, which influence the transcriptional activity of the HIV LTR. Thus, TAR was deleted from either the LTR or the MARβ-LTR construct (Fig. 1A) and a reporter gene expression assay was carried out as described earlier. Upon transient transfection, the MARβ-mediated 5-fold upregulation was similar in both the presence and absence of the TAR sequence. A similar experiment was performed using the above constructs in the presence of Tat (Fig. 2E). The level of transcriptional activation did not increase with induction of Tat when TAR was deleted; rather the result was similar to that observed in the absence of Tat. Thus, MAR-mediated upregulation in transcription is independent of Tat–TAR interaction.
Effect of MAR sequences on a truncated LTR
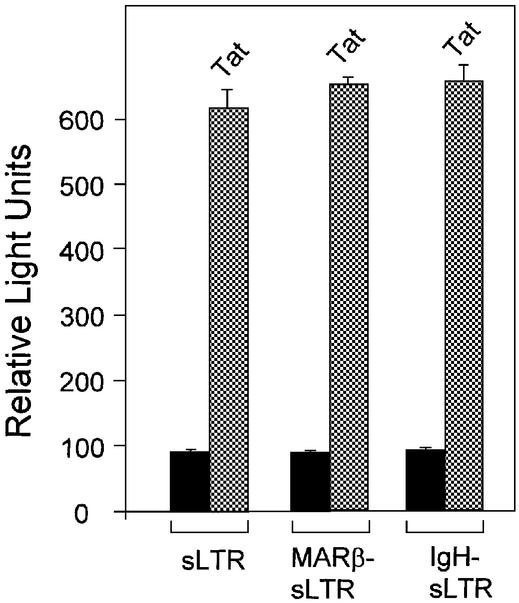
Since MARs are shown to enhance transcription in the presence of a 1.5 kb long LTR, we tested if MARs have any influence on the kinetics of transcription initiation. sLTR constructs were made using a HindIII site at around position +78 downstream of the transcription start site (Fig. 1A). Both MARβ and IgH MAR were inserted adjacent to the sLTR. By transient expression of the sLTR, MARβ-sLTR and IgH-sLTR, there is no significant MAR-mediated enhancement of transcription from the sLTR promoter; similar results were observed by co-transfection of pcDNA-Tat (Fig. 3). These results indicate that LTR-mediated transcription in either the presence or absence of MAR remained unaltered.
Figure 3.
MAR-mediated enhancement of transcription occurs at a distance. 293 cells were seeded as described above. Briefly, cells were transiently transfected with sLTR-Luc, MARβ-sLTR-Luc and IgH-sLTR-Luc. The experiment was performed in the presence and absence of HIV Tat. Relative light units were calculated 40 h post-transfection by loading an equal amount of protein (50 µg).
MAR-mediated transcriptional augmentation is specific for the LTR promoter
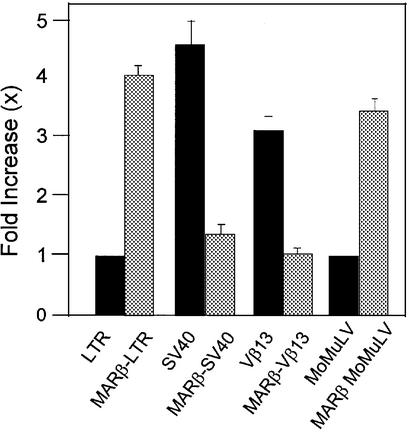
To examine if MAR-mediated transcriptional enhancement is retroviral promoter specific, MARβ was cloned in proximity to either the SV40 viral promoter or the mouse Vβ13 promoter and MoMuLV LTR promoters. The promoters associated with the respective MAR sequences were tested for transcriptional activity using luciferase as the reporter system as described earlier. Quantitative analysis shows a 2- to 3-fold downregulation in transcriptional activity in the context of either the Vβ13 or SV40 promoter when MARβ was placed upstream of the respective promoters (Fig. 4). In contrast, under identical conditions, MARβ-driven transcription was 4- and 3.4-fold higher in MARβ-LTR and MARβ-MoMuLV LTR, respectively, compared with LTR and MoMuLV LTR controls. Thus, MARβ-mediated enhancement of transcription is LTR promoter specific and shows a reverse pattern for both a DNA viral promoter and a eukaryotic promoter.
Figure 4.
MAR-mediated transcriptional enhancement is specific for the HIV LTR promoter. 293 cells were seeded as described previously and transiently transfected with LTR-Luc, MARβ-LTR-Luc, SV40-Luc, MARβ-SV40-Luc, Vβ13-Luc, MARβ-Vβ13-Luc, MoMuLV LTR and MARβ-MoMuLV LTR-Luc plasmids at 2 µg/well. Luciferase activity was calculated 40 h after transfection by loading an equal amount of protein (50 µg).
Influence of MARs in LTR-mediated transcription during elongation
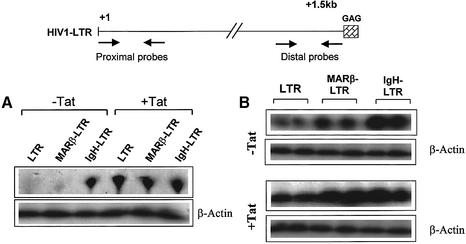
Cellular enhancers are shown to promote recruitment of elongation-competent transcription complexes to the HIV LTR promoter. Since we did not see any significant difference in the promoter activity using sLTR and MAR-sLTR, quantitative analysis at the RNA level becomes important. To determine if MAR-mediated transcriptional augmentation from the HIV LTR is by enhancement of the elongation process, RNase protection assays were performed using proximal and distal probes. The difference in the number of RNA transcripts corresponding to the proximal region with the LTR and MARβ-LTR was not very significant, but there was a notable increase in IgH-LTR transcripts (Fig. 5A). A probe of 213 bases which is at the extreme end of the 1.5 kb LTR was used for the protection of the distal region. The number of transcripts elongated from LTR, MARβ-LTR and IgH-LTR protected using the distal probe was quantified using a phosphoimager. Compared with the LTR only, MAR- mediated transcription resulted in the protection of 3-fold more transcripts (Fig. 5B), consistent with the higher reporter gene expression as shown in Figure 2. Since we observed a fold increase of the luciferase reporter expression by co-transfection of Tat proportionate to the above-mentioned constructs, a similar type of assay was performed to check the protection of transcripts in the presence of Tat. The number of transcripts protected supported the earlier results of the luciferase assay. As shown in Figure 5B, there is an ∼3-fold difference in the level of protected RNA when the MAR was located upstream of the LTR promoter compared with only the LTR. A β-actin probe was used for normalization of the assay in 10 µg of total RNA used for each reaction. Thus, addition of a stretch of MAR sequences upsteam of the LTR significantly increases the level of transcript that corresponds to the distal region. Interestingly, in the presence of IgH MAR, we see an increase in the protected RNA for both the proximal and distal region, indicating that unlike MARβ, IgH MAR influences initiation, at least in part.
Figure 5.
MAR-mediated enhancement of transcription by elongation. 293 cells were seeded as described previously and transiently transfected with LTR-Luc, MARβ LTR-Luc and IgH LTR-Luc plasmids at 2 µg/well. Transfections in the presence of Tat were performed by co-transfecting 1 µg of pcDNA-Tat. Total RNA was isolated using TRIZOL (Sigma), 40 h after transfection. (A) The proximal probe overlapping 300 bases from the +1 start site of the LTR was in vitro transcribed from a linearized template (NC-1 digested by XmnI) by T7 RNA polymerase. (B) For the distal probe, overlapping 213 bp from +1267 to +1480, in vitro transcription was carried out using a linearized template (XmnI-digested NC1) and T3 polymerase. A 10 µg aliquot of total RNA was analyzed for hybridization reaction by using 10 000 c.p.m. of probe RNA. Protected transcripts after digestion with RNase A and RNase T1 were analyzed on a 6% urea–polyacrylamide gel.
Role of MARs in LTR transcription upon stable integration
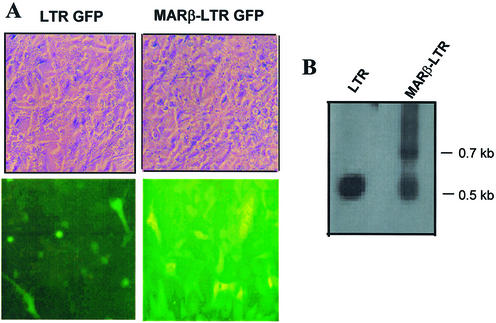
Previously, it has been shown that HIV-1 LTR-mediated transcription depends on the site of integration and cis-regulatory elements next to it (34). To address the question of whether a short stretch of MAR enhances HIV LTR-mediated transcription upon integration, plasmid constructs containing MARβ-LTR-GFP and LTR-GFP were stably transfected into CHO cells. The cells were selected for neomycin resistance using G418 (800 µg/ml). After 3 weeks, the cells were visualized under a microscope using a GFP filter. The GFP fluorescence was distinct and comparably higher from the cells that have a short stretch of MARβ-LTR sequence, indicating the processive transcription from the MARβ-LTR promoter at a distance (Fig. 6A). Interestingly, in an LTR-GFP stable line, very few cells (<1%) showed bright fluorescence indicating higher transcription activity. It is possible that in these cells, the plasmid molecules may have integrated near open and active chromatin. Thus, the role of MAR in LTR-mediated transcription at a distance is also true even after integration.
Figure 6.
Effect of MAR on LTR-mediated transcription upon stable integration. CHO cells were seeded at a rate of 1 × 106 cells per 6-well plate and transfected by LTR-GFP and MARβ-LTR-GFP at 2 µg/well. Transfected cells were stably selected using 800 µg/ml neomycin. A single cell population was obtained and grown. (A) Photographs of stably transfected LTR-GFP and MARβ-LTR-GFP cells were taken by an inverted microscope (Olympus) under UV fluorescence together with the bright field images. (B) Genomic DNA was prepared from LTR-GFP and MARβ-LTR-GFP stable cell lines using DNA-Zol reagent. A 20 µg aliquot of genomic DNA was digested with HindIII and BamHI. A 1 kb LTR probe core was used for hybridization with 10 000 c.p.m.
To further show the integration of the plasmid DNA into the genome, Southern blot analysis was performed using a 1 kb LTR probe. The results obtained gave a pattern of a 0.5 kb doublet band for LTR-GFP. MARβ-LTR-GFP gave rise to two fragments corresponding to 0.5 and 0.7 kb, as predicted. Doublet bands of 0.5 kb contain 540 and 555 bp fragments corresponding to the core LTR promoter and downstream sequence, respectively. The 0.5 kb and 0.7 kb fragments for MARβ-LTR-GFP correspond to the 540 bp LTR core promoter along with MARβ (Fig. 6B). Thus, genomic Southern blot confirms the presence of respective insertions in the chromosome.
NF-κB is required for MAR-LTR-mediated transcription
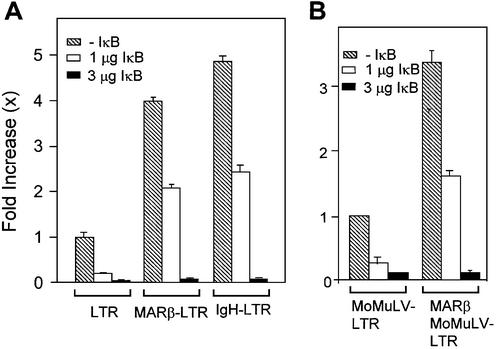
The HIV LTR promoter contains at least two indispensable NF-κB sites (35). Depletion of NF-κB by its inhibitor IκB-α results in drastic downregulation of LTR-mediated transcription. To determine whether MAR-mediated enhancement of transcription is influenced by NF-κB, co-transfection of 1 and 3 µg of IκB-α-expressing plasmid together with 2 µg of LTR, MARβ-LTR or IgH-LTR was carried out. After 40 h of transfection, luciferase assays were performed by loading an equal amount of protein. As shown in an earlier experiment, MARβ or IgH fusion constructs display transcriptional enhancement of ∼4- and 5-fold, respectively, compared with LTR only. In the presence of the lower dose (1 µg) of IκB, the inhibition of transcription was ∼2- to 2.5-fold (Fig. 2C). This inhibition of transcription from the LTR and MAR-LTR was not significantly different. With the higher dose of IκB (3 µg of plasmid DNA), there is negligible reporter gene expression, indicating complete suppression of transcription (Fig. 7A). In order to determine the role of NF-κB in MoMuLV LTR-mediated transcription in the presence and absence of MARβ, IκB was co-transfected in a dose-dependent manner (1 and 3 µg) together with either MoMuLV LTR or MARβ-MoMuLV LTR. The reporter assay was performed by loading an equal amount of protein (50 µg). As for the HIV LTR, dose-dependent inhibition of transcription was observed from the MoMuLV LTR in the presence and absence of MARβ, indicating that MAR-mediated transcription is NF-κB dependent.
Figure 7.
Transcriptional inhibition from LTR and MAR-LTR promoters by IκB in a dose-dependent manner. (A and B) 293 cells were seeded as described previously and transiently transfected with LTR-Luc, MARβ-LTR-Luc, IgH-LTR-Luc, MoMuLV LTR and MARβ-MoMuLV LTR plasmids at 2 µg/well. IκB plasmid was co-transfected at concentrations of 1 and 3 µg. Decreases in the relative light units were calculated 40 h after transfection by loading an equal amount of protein (50 µg).
DISCUSSION
The retroviral genome integrates into multiple sites in chromatin and, depending on the chromatin environment of sequences flanking the sites of the integration, the transcriptional rate at a distance changes dramatically (34). Integration at hotspots has been shown to increase the accessibility of the region of integration that promotes transcription. Earlier studies on MARs showed that because of their structural properties, they could augment the transcription from various promoters by anchoring them to the nuclear matrix (36,37). Phi Van and Stratling showed that MAR of chicken lysozyme stimulated transgene expression and mitigated position effect variegation (38). Similarly, a concept of use of MARs for efficient expression and transfer of the transgene by retroviral delivery has been established (39). Thus, a probable hotspot may increase the basal transcription and protect the transcription domain from the surrounding chromatin environment. In this regard, we present evidence suggesting the role of MARs in LTR-mediated transcription, the probable mechanism by which MARs act.
An AT-rich MARβ sequence was identified in the context of V(D)J recombination of the T-cell receptor β (TCRβ) locus (40). The sequence comparison analysis of this novel MARβ shows strong homology between human and mouse (41). The IgH MAR used in this study is located at the 3′ end of the 1 kb IgH enhancer and is bound by SATB1, a T-cell-specific MAR-binding protein (42).
Upon transient and stable transfection of MAR-LTR constructs, MARs are shown to elevate the transcription to significantly high levels in the absence of Tat protein. Since Tat is known to increase LTR-mediated transcription, the presence of Tat in MAR-LTR-mediated transfection would be expected to increase transcriptional augmentation synergistically. TAR deletion experiments suggest TAR–Tat-independent transcription from the LTR in the presence of MARβ. Transcription complexes that initiated from the HIV LTR promoter are poorly processive and are subsequently converted into a more processive form after induction with Tat and CDK9 kinases. However, in vivo, it has been seen that in certain circumstances, Tat-independent HIV LTR transcription rises to a significantly high level.
Recently, it has been shown that cellular enhancers promote recruitment of elongation-competent transcription complexes to the HIV LTR promoter. After studying the role of MARs in LTR-mediated transcription, it is important to understand the mechanism by which MARs work. RNase protection assays using distal and proximal probes showed that initiation levels were the same in LTR and MARβ-LTR constructs, but there was a notable increase in initiation with the IgH MAR-LTR. Indeed, both the MARs increased the elongation of the transcript by increasing the basal level of transcription in the absence and presence of Tat. It has been discussed in earlier reports that the recruitment of CDK9 and cyclin T1 to the HIV TAR RNA and subsequent induction with the Tat protein make this interaction fruitful by modifying the C-terminal domain of RNAP II. Phosphorylation of RNAP II increases the processivity of transcription by overcoming the terminator signals within the LTR. Since MAR-mediated transcriptional elongation is independent of Tat, RNAP might be modified by the induction of unknown cellular proteins recruited through MARs so as to overcome the downstream terminators. Earlier it was reported that the IgH enhancer increases HIV LTR transcription by elongation, and the process is independent of NF-κB (43). According to these studies, transcription complexes initiated in the presence of enhancer were highly processive. With respect to the above studies, we narrowed the area down to a small core region of MARs that potentially can regulate HIV transcription elongation and can function in a manner similar to enhancers. Since we observed a notable increase in initiation with the use of IgH MAR, we hypothesize that the MARs act upon initiation possibly through a strong base unpairing region. Despite a considerable difference in the initiation levels, both the MARs do seem to positively influence the elongation.
Integration is a crucial event in retroviral replication (44). Retroviruses have evolved with different strategies for exploiting the host machinery by using either host proteins or hotspots in the genome. The retrovirus LTR contains many cis-elements apart from the core promoter that distinguishes them from other viral promoters. Bode et al. have proposed that all the integration events by retroviruses occurred in MARs (45). To examine whether the influence of MARs on transcription is specific to HIV or is also the case for other viruses, we conducted experiments with the SV40 and MoMuLV viral promoters. MARβ is bound by MAR-binding protein SMAR1 and represses MARβ-Vβ13 promoter- mediated transcription (46). Our data demonstrate that the SV40 promoter behaves similarly to the Vβ13 promoter in the presence of MARβ; in contrast, a positive influence of MARβ is seen in the HIV and MoMuLV LTR. Thus, it is possible that retroviruses may use MARs for upregulation of basal transcription even in the presence of many endogenous repressor proteins.
One of the important features regarding the HIV-1 life cycle is that its genome remains quiescent in the form of a provirus within the host genome for a considerable period of time. In latently infected cells, the LTR promoter of the proviral genome is turned on upon sudden induction of NF-κB. Thus, two of the NF-κB-binding sites residing within the HIV LTR are known as ‘enhancer’ elements. (35). These sites act as promoter-proximal activation elements within the LTR. To understand the influence of MARs on the recruitment of NF-κB and in allowing processive transcription, we used its inhibitor IκB. A decrease in the transcriptional profile in a manner proportional to the dose of IκB indicated that MAR-mediated transcriptional enhancement is dependent on NF-κB, suggesting that the integrity of the LTR is essential for this phenomenon. NF-κB-independent upregulation of transcription has been seen with the use of heterologous enhancers in HIV LTR-mediated transcription, which indicates that promoter integrity is not essential. Enhancers often consist of long stretches of DNA and contain binding sites for different transcription factors, which can replace NF-κB functions. In contrast, MARs are very short sequences to which not many transcription factors bind, other than the specific MAR-binding proteins. Thus, this independent system has a requirement for NF-κB that binds to intrinsic enhancer NF-κB sites in the HIV LTR.
During the early phase of HIV infection, it transcribes short aberrant transcripts of 60–80 nt together with 2 and 4 kb, and sometimes full-length transcripts (47). For production of proteins, the transcript needs to be transported into the cytoplasm. The 2 kb transcripts localize to the cytoplasm without a requirement for regulatory proteins, which then leads to synthesis of Tat and Rev proteins. These proteins are responsible for transactivation of the LTR, favoring the kinetics of transcription and transport of larger transcripts of HIV. As MARs help in increasing the basal transcription by an increase in the processivity from the LTR promoter, we hypothesize that this may augment LTR-mediated transcription in the early phase of the HIV life cycle when regulatory proteins such as Tat are absent, and help in obtaining threshold levels of regulatory proteins. Accumulated new data suggest that retroviral integration is not random in the genome (11,16,18). Since MARs are AT-rich sequences found near active genes and often possess the peculiar characteristics of a Topo II cleavage site, DNase I hypersensitivity, it is plausible that they may serve as hotspots for retroviral integration.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr G. C. Mishra, the Director of the NCCS, for his generous and moral support in carrying out the experiments, Dr Terumi Kohwi-Shigematsu for IgH-MAR, Ajith Mathew of our laboratory for the pcDNA-Tat construct, Professor Jianzhu Chen, MIT, Boston for providing MSCV 2.2 vector as a kind gift, and the NIH AIDS Research and Reference Reagent Program catalog for providing us with the HXB3 clone. This work was mostly supported by the Jai Vigyan Mission project from the Department of Biotechnology, New Delhi, India.
REFERENCES
- 1.Sodroski J.G., Rosen,C.A., Wong-Stall,F., Salahuddinn,S.Z., Popovic,M., Arya,S., Gallo,R.C. and Hasteline,W.A. (1985) Trans-acting transcriptional regulation of human T-cell leukemia virus type III LTR. Science, 227, 171–173. [DOI] [PubMed] [Google Scholar]
- 2.Mancebo H.S.Y., Lee,G., Fygare,J., Tomassine,J., Luu,P., Zhu,Y., Peng,J., Blau,C., Hazuda,D., Price,D. and Flores,O. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev., 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emerman M. and Malim,M. (1998) HIV regulatory/accessory genes; keys to unraveling viral and host cell biology. Science, 280, 1880–1884. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y., Pe’ery,T., Peng, Ramanathan,Y., Marshall,N., Marshall,T., Amendt,B., Mathews,M.B. and Price,D.H. (1997) Transcription elongation factor p-TEFb is required for HIV-1 Tat transcription in vitro. Genes Dev., 11, 2622–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei P., Garber,M.E., Fang,S.M., Fischer,W.H. and Jones,K.A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop specific binding to TAR RNA. Cell, 92, 451–462. [DOI] [PubMed] [Google Scholar]
- 6.Bushman F.D., Fujiwara,T. and Craigie,R. (1990) Retroviral DNA integration directed by HIV integration protein in vitro. Science, 249, 1555–1558. [DOI] [PubMed] [Google Scholar]
- 7.Wiskerchen M. and Musing,M.A. (1995) Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates and sustain viral propagation in primary cells. J. Virol., 69, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller H.P. and Varmus,H.E. (1994) DNA bending creates favoured sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J., 13, 4740–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Withers-Ward E.S., Kitamura,Y., Barnes,J.P. and Coffin,J.M. (1994) Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev., 8, 1473–1487. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald M.L. and Grandgenett.D.P. (1994) Retroviral integration: in vitro host site selection by avian integrase. J. Virol., 68, 4314–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih C.C., Stoye,J.P. and Coffin,J.M. (1988) Highly preferred targets for retrovirus integration. Cell, 53, 531–537. [DOI] [PubMed] [Google Scholar]
- 12.Wattel E., Vartanian,J.P., Pannetier,C. and Wain-Hobson,S. (1995) Clonal expansion of human T-cell leukemia virus type-1 infected cells in asymptomatic and symptomatic carriers without malignancy. J. Virol., 69, 2863–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolffe A.P. and Guschin,D. (2000) Chromatin structural features and targets that regulate transcription. J. Struct. Biol., 129, 102–122. [DOI] [PubMed] [Google Scholar]
- 14.VanLint C., Emiliani,S., Ott,M. and Verdin,E. (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J., 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 15.Scherdin U., Rhodes,C. and Briendl,M. (1990) Transcriptionally active genome regions are preferred targets for retrovirus integration. J. Virol., 64, 907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard M.T. and Griffith,J.D. (1993) A cluster of strong topoisomerase II cleavage sites located near an integrated human immunodeficiency virus. J. Mol. Biol., 232, 1060–1068. [DOI] [PubMed] [Google Scholar]
- 17.Berezney R. (1991) The nuclear matrix: a heuristic model for investigating genomic organization and function in the cell nucleus. J. Cell. Biochem. 47, 109–123. [DOI] [PubMed] [Google Scholar]
- 18.Schroder A.R.W., Shinn,P., Chen,H., Berry,C., Ecker,J.R. and Bushman,F. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell, 110, 521–529. [DOI] [PubMed] [Google Scholar]
- 19.Garrard W.T. (1990) Chromosomal loop organization in eukaryotic genomes. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Heidelberg, Germany. Vol. 4, pp. 163–175.
- 20.Adachi Y., Kas,E. and Laemmli,U.K. (1989) Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J., 8, 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pommier Y., Cockerill,P.N., Kohn,K.W. and Garrard,W.T. (1990) Identification within the simian virus 40 genome of a chromosomal loop attachment site that contains topoisomerase II cleavage sites. J. Virol., 64, 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperry A.O., Blasquez,V.C. and Garrard,W.T. (1989) Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II. Proc. Natl Acad. Sci. USA, 86, 5497–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webb C.F., Das,C., Eneff,K.L. and Tucker,P.W. (1991) Identification of a matrix-associated region 5′ of an immunoglobulin heavy chain variable region gene. Mol. Cell. Biol., 11, 5206–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirkovitch J., Mirault,M.E. and Laemmli,U.K. (1984) Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell, 9, 223–232. [DOI] [PubMed] [Google Scholar]
- 25.Gasser S.M. and Laemmli,U.K. (1986) Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D.melanogaster. Cell, 46, 521–530. [DOI] [PubMed] [Google Scholar]
- 26.Cockerill P.N. and Garrard,W.T. (1986) Chromosomal loop anchorage of the κ immunoglobulin gene occurs next to the enhancer in a region containing topoismerases II sites. Cell, 44, 273–282. [DOI] [PubMed] [Google Scholar]
- 27.Cockerill P.N., Yuen,M.H. and Garrard,W.T. (1987) The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J. Biol. Chem., 262, 5394–5397. [PubMed] [Google Scholar]
- 28.Kohwi-Shigematsu T. and Kohwi,Y. (1990) Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry, 29, 9551–9560. [DOI] [PubMed] [Google Scholar]
- 29.Forrester W.C., vanGenderen,C., Jenuwein,T. and Grosschedl,R. (1994) Dependence of enhancer-mediated transcription of the immunoglobulin µ gene on nuclear matrix attachment regions. Science, 265, 1221–1225. [DOI] [PubMed] [Google Scholar]
- 30.Kirillov A., Kistler,B., Mostsolasky,R., Cedar,H., Wirth,T. and Bergman,Y. (1996) A role for nuclear NF-κB-cell-specific demethylation of the Igκ locus. Nature Genet., 13, 435–441. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T., Forrester,W.C., Fernandez-Herrero,L.A., Laible,G., Dull,M. and Grosschedl,R. (1997) Extension of chromatin accessibility by nuclear matrix attachment regions. Nature, 385, 269–272. [DOI] [PubMed] [Google Scholar]
- 32.Hart C.M. and Laemmli,U.K. (1998) Facilitation of chromatin dynamics by SARs. Curr. Opin. Genet. Dev., 8, 519–525. [DOI] [PubMed] [Google Scholar]
- 33.Shaw G.M., Hahn,B.H., Arya.S.K., Groopman,J.E., Gallo,R.C. and Wong-Staal,F. (1984) Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in acquired immunodeficiency syndrome. Science, 226, 1165–1170. [DOI] [PubMed] [Google Scholar]
- 34.Joradan A., Defechereux,P. and Verdin,E. (2001) The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J., 20, 1726–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nable G. and Baltimore,D.A. (1987) An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature, 326, 711–713. [DOI] [PubMed] [Google Scholar]
- 36.Klehr D., Maass,K. and Bode,J. (1991) SAR elements from the human IFN-β domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry, 30, 1264–1270. [DOI] [PubMed] [Google Scholar]
- 37.Bode J., Schlake,T., Rios-Ramfrez,M., Mielke,C., Stengert.M., Kay,V. and Klehr-Wirth,D. (1995) Scaffold/ matrix attached regions (S/MARs): structural properties creating transcriptionally active loci. Int. Rev. Cytol., 162A, 389–453. [DOI] [PubMed] [Google Scholar]
- 38.Phi-Van L. and Stratling,W.H. (1996) Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry, 35, 10735–10742. [DOI] [PubMed] [Google Scholar]
- 39.Schubeler D., Mielke,C., Maass,K. and Bode,J. (1996) Scaffold/ matrix-attached regions act upon transcription in a context-dependent manner. Biochemistry, 35, 11160–11169. [DOI] [PubMed] [Google Scholar]
- 40.Chattopadhyay S., Whitehurst,C., Schwenk,F. and Chen,J. (1998) Biochemical and functional analyses of chromatin changes at the TCRβ gene locus during CD4– CD8– to CD4+ CD8+ thymocyte differentiation. J. Immunol., 160, 1256–1267. [PubMed] [Google Scholar]
- 41.Chattopadhyay S., Whitehurst,C., Schwenk,F. and Chen,J. (1998) A nuclear matrix attachment region upstream of the T cell receptor β gene enhancer binds Cux/CDP and SATB1 and modulates enhancer dependent reporter gene expression but not endogeneous gene expression. J. Biol. Chem., 273, 29838–29846. [DOI] [PubMed] [Google Scholar]
- 42.Dickinson L.A., Joh,T., Kohiwi,Y. and Kohwi-Shigimatsu,T.A. (1992) A tissue specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell, 70, 631–645. [DOI] [PubMed] [Google Scholar]
- 43.West M.J. and Karn,J. (1999) Stimulation of Tat associated kinase-independent transcriptional elongation from the human immunodeficiency virus type-1 long terminal repeat by cellular enhancer. EMBO J., 18, 1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandmeyer S.B., Hansen,L.J. and Chalker,D.L. (1990) Integration specificity of retrotransposons and retroviruses. Annu. Rev. Genet., 24, 491–515. [DOI] [PubMed] [Google Scholar]
- 45.Bode J., Bartsch,J., Boulikas,T., Iber,M., Mielke,C., Schubeler,D., Seibler,J. and Benham,C. (1998) Transcription-promoting genomic sites in mammalia: their elucidation and architectural principles. Gene Ther. Mol. Biol., 1, 551–580. [Google Scholar]
- 46.Chattopadhyay S., Kaul,R., Charest,.A., Housman,D. and Chen,J. (2000) SMAR1, a novel alternatively spliced gene product, binds the scaffold/matrix-associated region at the T cell receptor β locus. Genomics, 68, 93–96. [DOI] [PubMed] [Google Scholar]
- 47.Pollard V.W. and Malim,M.H. (1998) The HIV Rev protein. Annu. Rev. Microbiol., 52, 491–532. [DOI] [PubMed] [Google Scholar]