Abstract
p53 function is modulated by several covalent and non-covalent modifiers. The architectural DNA- binding protein, High Mobility Group protein B-1 is a unique activator of p53. HMGB-1 protein is structured into two HMG-box domains, namely A-box and B-box, connected to a long highly acidic C-terminal domain. Here we report that both the C-terminal domain and A-box of HMGB-1 are critical for stimulation of p53-mediated DNA binding to its cognate site. Though deletion of these domains showed minimal effect in activation of p53-mediated transcription from the DNA template as compared to full-length HMGB-1, truncation of both the domains indeed showed significant reduction of transcriptional activation from the chromatin template as observed in DNA binding. Using transient transfection assays we showed that the C-terminal acidic domain and A-box of HMGB-1 are critical for the enhancement of the p53-mediated transactivation in vivo. Furthermore, the C-terminal domain and A-box deleted HMGB-1 could not activate p53-dependent apoptosis above the basal level. In conclusion, these results elucidate the role of acidic C-terminal domain and A-box of HMGB-1 in p53-mediated transcriptional activation and its further downstream effect.
INTRODUCTION
p53, a transcription factor and tumor suppressor, regulates expression of several genes that are critical for cell cycle arrest and apoptosis. Genes that are directly regulated by p53 include GADD45, Bax, p21/WAF1, Cyclin G, Mdm2, IGFBP3, ACTA, MMP2, PCNA and TGF-α (1). Each of these downstream effectors, which are activated by p53 during genotoxic insult contain, one or more p53-responsive sites in their promoter region. p53 contains three functionally important domains: transactivation domain, sequence-specific DNA binding domain and C-terminal oligomerization domain. The sequence-specific DNA binding domain is frequently mutated in several malignant tumors and most of the mutant forms have impaired DNA binding activities.
Post-translational modification (i.e. phosphorylation and acetylation), antibody binding and deletion of highly basic C-terminal 30 amino acid (363–393) activates p53-mediated transactivation (2–4). The C-terminal basic 30 amino acid (363–393) stretch in the oligomerization domain was found to be a negative regulator of sequence-specific DNA binding of p53 (2). The majority of the p53 activators stimulate p53 function by relieving the repression of this domain (5). The notable exception is the High Mobility Group protein B-1 (HMGB-1). It has been shown that HMGB-1 can stimulate DNA binding as well as transactivation of both full-length p53 and C-terminal deleted p53 (p53Δ30), suggesting a unique mechanism of activation (5). HMGB-1 is a nonhistone chromosomal protein having architectural DNA binding properties. Apart from p53, it also activates sequence-specific DNA binding of several other transcription factors, which includes the estrogen receptor (6), HOX proteins (7), MTLF (8) and the POU domain containing proteins Oct1, Oct2 and Oct6 (9). In most of these instances, it was reported that HMGB-1 enhances transcriptional activation of these proteins in transient transfection assays (10). Induction of transcription by MTLF was shown in an in vitro transcription assay using adenovirus major late promoter (8). However, the mechanism of HMGB-1-mediated stimulation of DNA binding as well as transcriptional activation is poorly understood.
HMGB-1 protein contains three domains: two highly basic N-terminal domains, namely, A-box and B-box, and a long acidic C-terminal domain. The A-box and B-box functionally interact with Hox proteins, steroid hormone receptors, Oct1 and Oct2 (6,7,9,11) and the C-terminal domain regulates the DNA binding of HMGB-1 (12). Significantly, p53 directly interacts with the A-box and results in an enhancement of its DNA binding to cisplatin-modified DNA (13). This provides a molecular link between DNA damage and p53-mediated DNA repair. Though HMGB-1 is not a member of the nucleosome binding family proteins, like HMGN-1/-2, it directly interacts with the chromatin through the linker DNA and core histones (14). The C-terminal domain of HMGB-1 interacts with the core histone dimer H2A–H2B (15,16) and the A-box interacts with core histones H3 and H4 (16).
The present study examined the role of the C-terminal acidic domain and the A-box of HMGB-1 in the regulation of p53 function. The C-terminal domain and the A-box have been found to be essential for the stimulation of p53-mediated DNA binding to its cognate site. Transcription from chromatin template showed significant decrease in transcriptional activation upon truncation of the C-terminal domain and the A-box of HMGB-1. The absolute requirements of these domains are specific for the stimulation of p53-dependent chromatin transcription. Transient transfection experiments using the H1299 (p53 null) cell line also showed that the C-terminal domain and A-box are critical for the reporter gene expression specifically in the p53 context. Furthermore, deletion of the C-terminal domain and A-box of HMGB-1 did not activate p53-induced apoptosis, strengthening our hypothesis that the C-terminal domain and A-box are critical for HMGB-1-mediated p53 activation.
MATERIALS AND METHODS
Plasmid constructions
Recombinant N-terminal His6-tagged truncated forms of HMGB-1, HMGB-1ΔC and HMGB-1ΔA were cloned by PCR-based subcloning using primers complementary to the respective 5′ and 3′ ends of the full-length human HMGB-1 (HMGB-1FL) expression clone (17). The amplicons were inserted between NheI and HindIII sites of pET28b (Novagen) vector. Similarly, recombinant N-terminal His6-tagged HMGB-2 was also cloned into pET28b using the HMGB-2 cDNA clone (18). In order to construct the template plasmid for in vitro transcription experiments, p(p53)5ML plasmid containing five p53 binding sites was constructed by annealing the synthetic oligonucleotides, based on the GADD45 promoter sequence, followed by digestion of PstI and XbaI and inserted into the PstI and XbaI site of p5GalML plasmid (19). The mammalian expression clone of HMGB-1FL and its truncated forms (HMGB-1ΔC and HMGB-1ΔA) were constructed by PCR-based subcloning into the NheI and HindIII site of pCDNA 3.1 (+) vector (Invitrogen). By employing the same strategy, full-length human p53 was cloned into the EcoRI and BamHI site of pIRESpuro mammalian expression vector (Clontech), from a FLAG-tagged p53 expression clone (3). The sequences of all the constructs were confirmed by DNA sequencing using an ABI prism automated DNA sequencer.
Raising antibody
Rabbit polyclonal anti-HMGB-2 antibody was generated against His6-tagged recombinant HMGB-2 protein and affinity purified using Protein A–agarose (Invitrogen)
Purification of recombinant proteins
HMGB-1FL and HMGB-1ΔA were purified by Ni-nitrilotriacetic acid agarose (Ni-NTA) (Qiagen) followed by P11 (Whatman) ion-exchange chromatography. The C-terminal truncated form of HMGB-1, HMGB-1ΔC, was purified by Ni-NTA beads followed by Q-Sepharose ion-exchange chromatography. The recombinant proteins were analyzed by 18% SDS–PAGE and the authenticity of the protein was confirmed by western blot using HMGB-2 antibody and anti-His15 antibody (Santa Cruz) (data not shown). The FLAG-tagged full-length p53 was expressed in Escherichia coli and purified by immunoaffinity chromatography using M2 agarose beads (Sigma). Recombinant His6-tagged NAP1 was purified by Ni-NTA agarose beads as described (20). The His6-tagged p300 was purified by Ni-NTA agarose as described (21) (data not shown). The ACF complex (containing Acf and ISWI) was immunopurified by M2-agarose from recombinant Acf and ISWI baculovirus (22) infected Sf21 cells as described (22). FLAG-tagged human topoisomerase I was purified by M2-agarose from Sf21 cells according to a standard procedure (23) (data not shown).
Purification of HeLa core histones and preparation of HeLa nuclear extract
Human core histones were purified from a HeLa nuclear pellet as described (20). HeLa nuclear extract was prepared as described previously (24).
Electrophoretic mobility shift assay (EMSA)
A 40mer oligonucleotide containing p53 binding sites (5′-AATTCTCGAGCAGAACATGTCTA AGCATGCTGGGC TCGAG-3′) derived from GADD45 promoter was labeled using T4 polynucleotide kinase (New England Biolabs). The radiolabeled strand was annealed with the complementary strand and EMSA was performed as described (5). Reaction mixtures (40 µl) contained 8 µl of 5× EMSA buffer (100 mM HEPES at pH 7.9, 125 mM KCl, 0.5 mM EDTA, 50% glycerol, 10 mM MgCl2), 2 µl of 60 µg/ml double-stranded poly[d(I–C)], 4 µl of BSA (1 mg/ml), 32P-labeled probe DNA (3 ng), proteins as indicated and water in a total volume of 40 µl.
Reaction mixtures were incubated at 30°C for 30 min and the total volume of each reaction mixture was then loaded on a native 4% polyacrylamide gel containing 0.5× Tris-borate/EDTA buffer and 0.05% NP-40, and electrophoresed at 4°C for 2 h (250–260 V). The gel was then dried and subjected to autoradiography. The DNA–protein complex was quantified by phosphorimaging (Fuji Film) using Image Guage software. Quantitation of DNA binding data represents the average of three independent experiments.
Chromatin assembly
Chromatin assembly was performed by using covalently closed circular p(p53)5ML plasmid that had been relaxed previously by recombinant human topoisomerase I. The standard reaction mixture (70 µl) contained relaxed covalently closed circular DNA (1.75 µg), core histone (2 µg), NAP1 (15 µg), BSA (2 mg/ml), KCl (50 mM), HEPES at pH 7.6 (10 mM), EDTA (2.5 mM), glycerol (10%), ACF complex (10 ng of ACF, ISWI), 5 mM MgCl2, 3 mM ATP and an ATP regeneration system (30 mM phosphocreatine and 1 mg/ml creatine phosphokinase). All components except DNA, the ACF complex and the ATP regeneration system containing MgCl2 and ATP were incubated at 27°C for 30 min. After this preincubation, the ACF complex, ATP regeneration system and DNA were added in succession and then the reaction mixture was further incubated at 27°C for 4 h. In vitro assembled chromatin was analyzed by supercoiling assay as described (20) and partial micrococcal nuclease (MNase) (Sigma) digestion with different concentrations of MNase as well as varying times of digestion as described (20). Freshly assembled chromatin template was used directly for chromatin transcription.
In vitro transcription assay
Transcription assays were performed by incubating reconstituted chromatin template (containing 100 ng of DNA) or an equimolar amount of histone-free DNA [incubated in assembly buffer at 30°C for 30 min with p300 (25 ng) and acetyl CoA (40 µM)] in a total volume of 10 µl containing histone acetyl transferase (HAT) reaction buffer (10 mM HEPES pH 7.8, 50 mM KCl, 5 mM DTT, 0.5 mM PMSF, 10 mM sodium butyrate, 0.25 mg/ml BSA and 5% glycerol) followed by further incubation with Lys-CoA, a specific inhibitor of p300 HAT activity (10 µM) (25) for 15 min at 30°C. After the remodeling reaction and inhibition of p300 HAT activity, activator binding was done by the addition of p53 (50 ng) or Gal4VP16 (50 ng) and different concentrations (50, 100 and 200 ng) of HMGB-1, HMGB-1ΔC and HMGB-1ΔA at 30°C for 30 min for DNA transcription and 50 ng of each protein in chromatin transcription. Upon completion of the factor binding, 10 µl of nuclear extract and 2.5 µl of 20× transcription buffer (0.4 mM HEPES pH 8.4, 100 mM MgCl2) and 40 U of ribonuclease (RNase) inhibitor (RNase OUT; Invitrogen) was added to each reaction and incubated for 20 min, after which 3.74 µl of reaction mix (12 mM ATP, 12 mM CTP, 2 mM 3′-O-methyl GTP and 15 µCi UTP) was added in a total volume of 50 µl and then incubated for 50 min at 30°C. Ten units of T1 RNase (Invitrogen) was added and the incubation continued for another 15 min. Transcription was terminated by the addition of 250 µl of stop buffer (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 100 mM NaCl, 1% SDS and 0.025 ng/ml tRNA). The radiolabeled transcript was extracted with phenol–chloroform and ethanol precipitated, the dried pellet dissolved in loading dye (8 M urea, 0.005% bromophenol blue and xyelencynol) and analyzed on 5% urea– polyacrylamide gel. Gels were then dried and subjected to autoradiography at –70°C. Quantitation of transcription was done as described for EMSA. Quantitation of DNA and chromatin transcription data represents the average of three independent experiments.
Transfection assay
H1299 cells (p53 null) were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. p53, HMGB-1FL and the truncated forms of HMGB-1 (HMGB-1ΔC and HMGB-1ΔA) used in this study were constructed under the control of cytomegalovirus (CMV) promoter. PG13Luc and G10Luc were used as a reporter construct. PG13Luc contains 13 consensus p53-binding sites in tandem followed by polyoma virus promoter, which drives the luciferase gene (26). G10Luc contains 10 Gal binding sites followed by SV40 promoter, which drives the luciferase gene. The pCMV-βgal (Clontech) was used as an internal control. Cells were seeded at 0.6 × 106 cells per 60 mm diameter dish and transfected using the indicated amount of DNA for respective clones using the calcium phosphate method. The precipitate was then left on the plate for 12 h, after which time cells were supplemented with fresh DMEM and 10% FBS. Empty vector was included in the transfection mixture to keep the total amount of transfected DNA constant in each sample. Luciferase activity and β-galactosidase activity were measured 24 h later using a luciferase assay and β-galactosidase assay system, according to the procedures provided by the manufacturer (Promega). The relative luciferase activity was the average of three experiments.
Apoptosis assay
H1299 cells were seeded at 0.6 × 106 cells per 60 mm diameter dishes 14 h prior to transfection. Cells were transfected with the indicated amount of p53, HMGB-1FL and its truncated forms either alone or in combination using Lipofectamine 2000 plus (Invitrogen). Transfection was performed according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were fixed with 2.5% paraformaldehyde and nucleus stained with Hoechst (33258) stain and observed under a fluorescence microscope using a blind approach.
RESULTS
C-terminal acidic domain and A-box of HMGB-1 are crucial for the stimulation of p53-mediated DNA binding
HMGB-1 has been shown to enhance DNA binding of p53 to its cognate site (5). Since the HMGB-1 activates the C-terminal 30 residues deleted p53, the mechanism of activation is somewhat distinct and unique (5). In order to elucidate this mechanism it is essential to find out the role of each domain of HMGB-1 in the stimulation of p53 DNA binding. We have constructed two deletion mutants for this purpose: (i) p53 interacting A-box deleted, HMGB-1ΔA and (ii) C-terminal deleted HMGB-1ΔC (Fig. 1A). These proteins were expressed and purified from E.coli (Fig. 1B) to homogeneity. The authenticity of these proteins was confirmed by western blot analysis (Fig. 1C) using HMGB-2 antibody. The effect of these truncated forms of HMGB-1 on p53 DNA binding was studied in an EMSA using radiolabeled 40mer double-stranded oligonucleotide containing a p53 cognate site and recombinant p53 (Fig. 2). Our EMSA analysis showed that HMGB-1FL activates p53 DNA binding by 6-fold as compared to p53 alone (lane 2 versus lanes 9–11), whereas acidic C-terminal deleted HMGB-1ΔC could not activate the DNA binding above the basal level (Fig. 2, compare lane 2 with lanes 6–8 and with lanes 9–11). The A-box of HMGB-1 directly interacts with p53. Thus, in order to find out the role of A-box in the stimulation of p53-mediated DNA binding, the A-box truncated form of HMGB-1, HMGB-1ΔA, was used in the DNA binding experiment. We found that the A-box deleted HMGB-1 was also unable to stimulate the p53 binding to its cognate site as compared to HMGB-1FL (Fig. 2, compare lane 2 with lanes 3–5 and with lanes 9–11). However, the HMGB-1FL, HMGB-1ΔC and HMGB-1ΔA did not bind to the DNA sequence nonspecifically (Fig. 2, lanes 12–20). These results strongly suggest that the C-terminal acidic domain and A-box of HMGB-1 are crucial for the stimulation of p53-mediated DNA binding.
Figure 1.
Recombinant and native proteins used in the different experiments. (A) Diagrammatic representation of HMGB-1 and its truncated forms, HMGB-1ΔC and HMGB-1ΔA. (B) 200 ng of His6-tagged HMGB-1FL (lane 1), 100 ng of HMGB-1ΔC (lane 2) and 200 ng of HMGB-1ΔA (lane 3) were analyzed by SDS–PAGE (18%). (C) The authenticity of these proteins was analyzed by western blot using HMGB-2 rabbit polyclonal antibody. HMG proteins were separated by 12% SDS–PAGE followed by western blot: HMGB-1FL (lane 1), HMGB-1ΔC (lane 2) and HMGB-1ΔA (lane 3). (D) FLAG-tagged full-length human p53 purified from E.coli and analyzed by SDS–PAGE (10%). The authenticity of the p53 protein was confirmed by western blot using p53 monoclonal antibody, DO1 (Oncogene). (E) Analysis of the recombinant Drosophila ACF complex by SDS–PAGE (8%) which was purified from FLAG-tagged ACF and ISWI baculovirus infected Sf21 cells. (F) Recombinant His6-tagged mouse NAP1 purified from E.coli. (G) Native core histone purified from HeLa nuclear pellet and analyzed by SDS–PAGE (15%).
Figure 2.
The C-terminal acidic domain and A-box of HMGB-1 is essential to enhance sequence-specific DNA binding of p53. The effect of the C-terminal domain of HMGB-1 on p53-mediated DNA binding is shown. Human p53 (50 ng) was incubated with 3 ng of a 32P-labeled p53 binding oligonucleotide in the absence (lane 1) or presence (lane 2) of 100, 200 and 300 ng of HMGB-1ΔA (lanes 3–5), HMGB-1ΔC (lanes 6–8) and HMGB-1FL (lanes 9–11). The 32P-labeled probe was also incubated with only 100, 200 and 300 ng of HMGB-1ΔA (lanes 12–14), HMGB-1ΔC (lanes 15–17) and HMGB-1FL (lanes 18–20). Lane 1 contains only probe without any protein.
C-terminal acidic domain and A-box of HMGB-1 regulate p53-mediated transcription
Since HMGB-1 is known to function as a transcriptional coactivator and regulate the RNA polymerase II-mediated transcription (17), we were interested in investigating the effect of HMGB-1 and its different domains on p53-mediated transcription in vitro. To address this, we have constructed the DNA template with five p53-binding sites (based on GADD45 promoter) by replacing Gal4 sites into a plasmid (p5GalML) (19) comprised of adenovirus major late core promoter and a G-less cassette (Fig. 3A). The sequence and the position of the p53 binding sites were confirmed by DNA sequencing. An in vitro DNA transcription assay using HeLa nuclear extract and increasing concentrations of bacterially expressed FLAG-tagged p53 (30–200 ng) showed corresponding transcriptional activation from the template (Fig. 3B), indicating that the template is appropriate for in vitro transcription studies.
Figure 3.
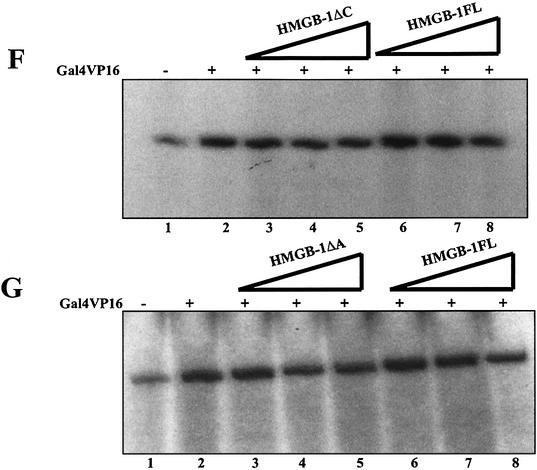
In vitro transcription from naked DNA template by p53 in the presence of HMGB-1FL and its truncated forms. (A) Schematic representation of the plasmid template (used for transcription) containing five p53 binding sites upstream of adenovirus major late core promoter (MLP) and G-less cassette. The transcription start site is represented as +1. (B) The newly constructed template P(p53)5ML subjected to an in vitro transcription experiment using HeLa nuclear extract and without or with increasing concentrations of p53: lane 1, without p53; lane 2, 30 ng of p53; lane 3, 50 ng of p53; lane 4, 100 ng of p53; lane 5, 200 ng of p53. (C) Schematic representation of transcription protocol. (D) Effect of C-terminal domain on p53-mediated transcription. The DNA template (100 ng) was subjected to transcription according to the protocol in (C) with (lanes 2–8) or without (lane 1), 50 ng of p53 (lane 2) and the indicated amount of HMGB-1 and its truncated forms. Fifty, 100 and 200 ng of HMGB-1ΔC (lanes 3–5), and 50, 100 and 200 ng of HMGB-1FL (lanes 6–8) were added along with p53 (50 ng). (E) The effect of A-box of HMGB-1 on p53-mediated transcription from p(p53)5ML naked DNA template. Transcription reactions were performed according to the protocol in (C), without p53 (lane 1) or with 50 ng of p53 (lane 2) and 50, 100 and 200 ng of HMGB-1ΔA (lanes 3–5), and 50, 100 and 200 ng of HMGB-1FL (lanes 6–8) in the presence of p53 (50 ng). (F) Effect of C-terminal domain on Gal4VP16-mediated transcription. The DNA template (100 ng) was subjected to transcription according to protocol (C). The transcription reaction was performed with Gal4VP16 (lanes 2–8) or without Gal4VP16 (lane 1). Fifty nanograms of Gal4VP16 (lane 2) were incubated with 50, 100 and 200 ng of HMGB-1ΔC (lanes 3–5), and 50, 100 and 200 ng of HMGB-1FL (lanes 6–8). (G) The effect of A-box of Gal4VP16-mediated transcription. The DNA template (100 ng) was subjected to transcription according to the protocol in (C). The transcription reaction was performed with Gal4VP16 (lanes 2–8) or without Gal4VP16 (lane 1). Fifty nanograms of Gal4VP16 (lane 2) were incubated with 50, 100 and 200 ng of HMGB-1ΔA (lanes 3–5), and 50, 100 and 200 ng of HMGB-1FL (lanes 6–8).
Transcription from DNA template was carried out according to the protocol shown in Figure 3C. The transcription from DNA template showed that the HMGB-1FL activates p53-mediated transcription by ∼2.1- and ∼1.9-fold (for 50 and 100 ng, respectively) (Fig. 3D, compare lane 2 with lanes 6 and 7). Interestingly, the C-terminal deleted HMGB-1, HMGB-1ΔC, could minimally activate the p53-mediated transcription at 50 and 100 ng concentration, which is considerably lower than the activation brought about by HMGB-1FL (Fig. 3D, compare lane 2 with lanes 3 and 4). As observed in the case of HMGB-1FL, a higher concentration (200 ng) of HMGB-1ΔC also repressed the p53 driven transcription from DNA template (Fig. 3D, lanes 8 and 4, respectively). Similarly, the A-box truncated HMGB-1, HMGB-1ΔA, could minimally activate the transcription at 50 ng concentration (Fig. 3E, compare lane 2 with lane 3 and lane 2 with lane 6), whereas 100 and 200 ng of HMGB-1ΔA repressed the transcription as compared to p53 alone (Fig. 3E, compare lane 2 with lanes 4 and 5).
In order to find out the specificity of HMGB-1 towards p53 in the stimulation of transcriptional activation, we have further performed in vitro transcription experiments using p5GalML template containing five gal binding sites and a subsaturation amount of Gal4VP16 as an activator. The result showed that HMGB-1ΔC, HMGB-1ΔA and HMGB-1FL could not activate Gal4VP16-mediated transcription from the DNA template (Fig. 3F, compare lane 2 with lanes 3–5, lane 2 with lanes 6–8; Fig. 3G, compare lane 2 with lanes 3–5 and compare lanes 6–8). In agreement with the results from the DNA binding experiment these data suggest that the C-terminal domain and A-box of HMGB-1 are required to activate p53-mediated transcription from the DNA template and the stimulation of transcriptional activation by HMGB-1 is specific for p53.
The HMGB-1 is a chromatin protein and has been implicated in the maintenance and establishment of chromatin structure. These observations lead us to investigate the phenomenon in the chromatin context. The chromatin template was assembled on p(p53)5ML plasmid using recombinant Drosophila ACF complex (Fig. 1E), His6-tagged mouse NAP1 (Fig. 1F), recombinant topoisomerase I and highly purified human (HeLa) core histones (Fig. 1G) as described in Materials and Methods. The assembled chromatin was characterized by a supercoiling assay (Fig. 4A) and partial micrococcal digestion (Fig. 4B). The assembly reaction without ATP showed several intermediate DNA forms, whereas a complete absence of these forms was observed in the ATP-containing assembly reaction as revealed by the supercoiling assay (Fig. 4A compare lanes 3 and 4). This suggests that the assembly is ATP dependent as expected (22). The MNase digestion pattern of the reconstituted chromatin showed well resolved regularly spaced nucleosomes (Fig. 4B). These results indicate that the reconstituted chromatin is appropriate for in vitro transcription experiments. The assembled chromatin template was used for the transcription experiment as shown in the protocol of Figure 4C. In this protocol, prior to the activator binding step, chromatin was subjected to acetylation in the presence of p300 and acetyl CoA. It was necessary to overcome the chromatin-mediated repression, and without this step, chromatin template was completely inert to transcription. After chromatin remodeling, the HAT activity of p300 was inhibited by a specific inhibitor, Lys-CoA. The inhibition was confirmed by incubating p53, HMGB-1 and core histone with 3H acetyl CoA and the inhibited enzyme in a separate reaction which showed that p53, HMGB-1 and core histones could not be acetylated by Lys-CoA treated p300 (data not shown). The ectopic addition of full-length recombinant HMGB-1 enhances the p53-dependent transcription ∼4-fold as compared to p53 alone (Fig. 4D, compare lane 2 with lane 5), whereas addition of HMGB-1 truncated forms, HMGB-1ΔC and HMGB-1ΔA, could not stimulate p53-mediated transcriptional activation (Fig. 4D, compare lane 2 with lanes 3 and 4). Similar to transcription from histone-free DNA template, HMGB-1ΔC, HMGB-1ΔA and HMGB-1FL could not activate Gal4VP16-mediated transcription also from chromatin template (Fig. 4E, compare lane 2 with lanes 3–5). These results lead to a very significant indication that the C-terminal domain and A-box of HMGB-1 play a critical role in the stimulation of p53-mediated transcription in the chromatin context (see Discussion) and the HMB-1-mediated transcriptional activation is specific for p53.
Figure 4.

The C-terminal acidic domain and A-box of HMGB-1 is essential for p53-mediated transcriptional activation from chromatin template. (A) DNA supercoiling assays for in vitro assembled chromatin. Chromatin was assembled using the ATP regeneration system without ATP (lane 3) or with ATP (lane 4). Lane 1, supercoiled DNA used for chromatin assembly; lane 2, relaxed DNA after recombinant topoisomerase I treatment of the DNA of lane 1. (B) MNase digestion analysis of assembled chromatin. The chromatin was treated with varying concentrations of MNase for 6 min (lanes 2–5) and fixed concentrations of MNase with varying times of 5 (lane 6) and 10 min (lane 7) at room temperature. After deproteinization, the resulting DNA was resolved on a 1.25% agarose gel and stained with ethidium bromide. (C) Schematic diagram of in vitro transcription from chromatin template. (D) In vitro transcription from chromatin template. Freshly assembled chromatin template subjected to in vitro transcription as depicted in the protocol (C). Transcription reactions were performed without p53 (lane 1) or with 50 ng of p53 alone (lane 2) or in combination with 50 ng of HMGB-1ΔC (lane 3), 50 ng of HMGB-1ΔA (lane 4), 50 ng of HMGB-1FL (lane 5) and 100 ng of histone-free DNA treated in an assembly reaction was preincubated with 50 ng of p53 and 50 ng of HMGB-1FL and subjected to a transcription reaction (lane 6). (E) HMGB-1 and its truncated form cannot activate transcription of Gal4VP16 from the chromatin template. Freshly assembled chromatin template was subjected to in vitro transcription as depicted in the protocol (C). Transcription reactions were performed without Gal4VP16 (lane 1) or with 50 ng of Gal4VP16 alone (lane 2) or in combination with 50 ng of HMGB-1ΔC (lane 3), 50 ng of HMGB-1ΔA (lane 4), 50 ng of HMGB-1FL (lane 5) and 100 ng of histone-free DNA treated in an assembly reaction was preincubated with 50 ng of Gal4VP16 and 50 ng of HMGB-1FL and subjected to a transcription reaction (lane 6).
Stimulation of transactivation of p53 by C-terminal acidic domain and A-box of HMGB-1
The significant difference in p53-mediated transcriptional activation from chromatin template upon truncation of the C-terminal domain and A-box of HMGB-1 prompted us to examine the role of these domains in an another physiological condition, which is activation of the p53-responsive promoter in transient transfection. For this purpose, we have cloned HMGB-1 and its truncated forms in pCDNA 3.1(+) vector under the regulation of the CMV promoter. In order to test the ability of these constructs to activate p53-mediated (placed under the CMV promoter) transcriptional activation of a promoter containing a p53-responsive site (PG13Luc) (26), transient transfection assays were performed using the H1299 (p53 null) cell line. The H1299 cells were transfected with plasmid expressing p53 and HMGB-1FL and its truncated forms either separately or together along with the luciferase reporter construct. The pCMVβgal was used as an internal control. Our results showed that there was significant activation (∼9-fold) of the p53-responsive promoter upon cotransfection of HMGB-1FL and p53 as compared to p53 alone (Fig. 5A). The cotransfection of HMGB-1ΔC and HMGB-1ΔA with p53 yielded minimal transactivation (∼2-fold) in relation to only p53 that could be contributed by some unknown cellular factor. However, HMGB-1ΔC, HMGB-1ΔA and HMGB-1FL could not stimulate Gal4VP16-mediated reporter gene expression in vivo (Fig. 5B). These observations suggest that the truncation of both the domains of HMGB-1 lead to a significant decrease in the HMGB-1-mediated activation of p53 function and this activation is specific for p53. These results further strengthen our hypothesis that the C-terminal domain and A-box of HMGB-1 are crucial for the activation of p53 function.
Figure 5.
Acidic C-terminal domain and A-box of HMGB-1 specifically stimulates transactivation by p53. (A) H1299 cells were transiently transfected with p53 (250 ng) and HMGB-1FL, HMGB-1ΔC and HMGB-1ΔA (2.5 µg) either alone or in combination. The amount of transfected DNA was normalized with equivalent amounts of control parental vectors. The reporter constructs used was PG13Luc (1 µg). CMV-βgal (1 µg) was used as internal control for all the transfection experiments. Relative luciferase activity is plotted (y-axis) after normalization of luciferase activity with β-galactosidase activity. The relative luciferase activity is an average of three experiments. (B) H1299 cells were transiently transfected with Gal4VP16 (100 ng) and HMGB-1FL, HMGB-1ΔC and HMGB-1ΔA (1 µg) either alone or in combination. The amount of transfected DNA was normalized with the equivalent amount of control parental vectors. The reporter construct used was G10Luc (500 ng). CMV-βgal (500 ng) was used as an internal control for all the transfection experiments. Relative luciferase activity is plotted (y-axis) after normalization of luciferase activity with β-galactosidase activity. The relative luciferase activity is an average of three experiments. VP16, Gal4 Vp16.
C-terminal domain and A-box of HMGB-1 induces p53-dependent apoptosis
In order to maintain the normal cellular function, p53 induces apoptosis in cells undergoing irreparable genotoxic damage by triggering the apoptotic machinery for its silent death. The factors that activate p53 function finally result in apoptosis and/or cell cycle arrest. Since HMGB-1 has been shown to activate p53 function from different promoters, we are interested in investigating the ultimate fate of the cell when HMGB-1 and its truncated forms are over-expressed along with p53 in the H1299 cell line. Interestingly, we found that the HMGB-1FL indeed activated p53-dependent apoptosis by 81% as compared to p53 alone (compare Fig. 6D with A and I). The truncation of the C-terminal domain and A-box of HMGB-1 could not activate apoptosis above the basal (p53) level (compare Fig. 6D with B and C; Fig. 6I). These data validate the role of the C-terminal domain and A-box of HMGB-1 in the activation of p53 function.
Figure 6.
Effect of C-terminal domain and A-box of HMGB-1 in p53-mediated apoptosis. H1299 cells were transfected with 2.5 µg of p53 or 5 µg of HMGB-1FL, HMGB-1ΔC and HMGB-1ΔA either alone or in combination. Hoechst stain cells were observed under a fluorescence microscope using a blind approach. Apoptotic nuclei are indicated (arrows). The percentage of apoptotic nuclei refers to the average number of apoptotic cells present in 300 nuclei over three independent experiments (I).
DISCUSSION
The highly abundant chromosomal protein HMGB-1 plays a significant role in the regulation of chromatin structure as well as transcriptional activation (12,27,28). It enhances the binding of several transcription factors to their cognate site resulting in the stimulation of transcriptional activation. One of these factors is p53, the tumor suppressor. Apparently, the mechanism of p53 activation by HMGB-1 is a unique one and remains less understood (5). This study has addressed the role of different domains of HMGB-1 in the stimulation of p53-mediated DNA binding and transcriptional activation. Here we report that (i) both the C-terminal domain and A-box of HMGB-1 are essential for the activation of p53-mediated DNA binding, (ii) these domains of HMGB-1 are critical for p53-mediated transcriptional activation specifically from chromatin template assembled by the ACF complex, (iii) the stimulation of transactivation from a p53-responsive promoter also requires the C-terminal acidic domain and A-box of HMGB-1 and (iv) truncation of the C-terminal domain and A-box failed to induce p53-mediated apoptosis, strengthening our observation in physiological conditions.
The biochemical activity of p53 which is required for tumor suppression and the cellular response to DNA damage involves the ability of the protein to bind to the cognate sites and to function as a transcription factor (29–33). By activating p53-mediated transcription, p53 rescues the cells from genotoxic damage. Thus, the proper regulation of p53 is essential to maintain normal cell growth and development. Most of the p53 modulator proteins stimulate the p53 function by inhibiting the self-repression (exerted by the C-terminal domain of p53) and thereby cannot activate the C-terminal domain deleted p53. HMGB-1 differs from the other p53 modulators because it can stimulate C-terminal domain deleted p53 suggesting a unique mechanism of p53 activation (5). In an another important study, it has been reported that the C-terminal domain of p53 interacts with the A-box of HMGB-1 and this interaction is required for the sequence non-specific binding of HMGB-1 to cisplatin-modified DNA (13). Thus, these independent studies clearly showed that the HMGB-1 enhances the activation of p53 function without the interaction between the C-terminal domain of p53 and the A-box of HMGB-1. Interestingly, we have found that in the absence of A-box, HMGB-1 cannot stimulate the p53 DNA binding above the basal level. These data suggest a novel and unidentified mechanism of HMGB-1-mediated activation of p53 wherein A-box of HMGB-1 plays a critical role. Our result also showed that HMGB-1ΔC, which lacks a C-terminal acidic domain, did not stimulate p53 DNA binding. Since the C-terminal acidic domain of HMGB-1 regulates its DNA binding properties (28,34,35), these data suggest a specific role of the C-terminal acidic domain in the activation of p53 DNA binding.
The results from the DNA binding experiments lead us to address the effect of DNA binding in transcriptional activation. Though deletion of the C-terminal domain and A-box of HMGB-1 minimally effect p53-mediated transcription from DNA template, chromatinization of the same template showed the absolute necessity of these domains. At this juncture it was important to address the specificity of HMGB-1-mediated enhancement of p53 driven transcription. We have performed in vitro transcription experiments using a potent chimeric transcriptional activator, Gal4VP16. The result showed that HMGB-1 and its derivatives could not stimulate transcription from DNA template containing five Gal4 binding sites. Chromatinization of this template resulted in only ∼2-fold repression as compared to transcription from the DNA template, whereas in p53-dependent transcription, this repression is much higher (∼6-fold) (compare Fig. 4D, lane 6 with Fig. 4E, lane 6). This could be attributed to more efficient Gal4VP16-mediated transcriptional activation. However, transcription from the chromatin template driven by Gal4VP16 also could not be stimulated further by the ectopic addition of HMGB-1 and its derivatives. This observation suggests a specific role of the C-terminal domain and A-box of HMGB-1 in the activation process wherein HMGB-1 cannot stimulate transcription globally by modulating the activity of general polymerase II factors including RNA polymerase II itself. We observed that higher concentrations of HMGB-1FL as well as the deletion constructs of HMGB-1 inhibit both DNA (Fig. 3D, lanes 8 and 5 and 3E, lanes 8 and 4, 5) and chromatin (data not shown) transcription mediated by both p53 and Gal4VP16. Presumably, higher concentrations of these proteins sequestrates general transcription factors (e.g. TBP and TFIIA) from the transcription reaction and thereby causes the repression (17). Furthermore, this chromatin-specific requirement of the C-terminal domain and A-box are in proper agreement with the observation that the acidic C-terminal domain and A-box of HMGB-1 are required for the stimulation of reporter gene expression in transient transfection assays (Figs 4D and 5) (36). The transient transfection assay using p53, HMGB-1 and its truncated forms showed that the deletion of both the C-terminal domain and A-box of HMGB-1 reduces the p53-responsive promoter activity by ∼4-fold (Fig. 5A). However, HMGB-1 and its truncated forms could not stimulate Gal4VP16-mediated transactivation from the Gal-responsive promoter, G10Luc (Fig. 5B). This observation further supports the in vitro transcription data regarding HMGB-1-mediated specific activation of p53 function. We have observed a very high activity of the Gal-responsive promoter in our transient transfection assay with Gal4VP16. This could be because of the fact that the Gal4VP16 is a highly efficient chimeric transcriptional activator, whereas p53 is a natural and comparatively less potent activator (compare Fig. 5A and B). Though it is not unambiguous, there is evidence suggesting that transiently transfected DNAs assemble into a physiological chromatin template. Thus, taken together, the chromatin transcription and transient transfection data argue for a chromatin-specific role of the C-terminal domain and A-box of HMGB-1 in transcriptional activation of p53. Furthermore, truncation of both the C-terminal acidic domain and A-box of HMGB-1 could not stimulate p53-mediated apoptosis suggesting the role of both the domains in activation of p53 function. Our apoptosis assay data strongly correlate HMGB-1-mediated activation of p53 DNA binding and transcription with functional consequences in vivo. This is the first example showing the role of different domains of HMGB-1 in the activation of p53 DNA binding and transcription from chromatin template linking p53-dependent apoptosis.
It is important to note that the C-terminal domain of HMGB-1 interacts with the H2A–H2B dimer of core histone and the A-box interacts with H3–H4 (15,16). In this present context, based on our observation, we are tempted to speculate that this histone-specific interaction of HMGB-1 domains may play an important role for the regulation of several transcription factors including p53. Significantly, both HMGB-1 and p53 are post-translationally modified with several functional consequences (37–39). It would be interesting to examine the intriguing effect of these post-translational modifications in the context of HMGB-1-mediated activation of p53 function.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Robert G. Roeder, M. M. Seidman, Bert Vogelstein, Philip A. Cole, James T. Kadonaga, Lee W. Kraus, K. Vousden, Masami Horikoshi and K. Somasundaram for providing valuable reagents used in this study; V. Swaminathan for helpful discussion and A. Hari Kishore for manuscript preparation. T.K.K. acknowledges Dr Alain Verreault, Cancer Research, London for hosting the UICC fellowship. The work is supported by Department of Atomic Energy, Government Of India and Jawaharlal Nehru Center for Advanced Scientific Research.
REFERENCES
- 1.Wang L., Wu,Q., Qiu,P., Mirza,A., McGuirk., Kirschmeier,P., Greenes,J.R., Wang,Y., Pickett,C.B. and Liu,S. (2001) Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J. Biol. Chem., 276, 43604–43610. [DOI] [PubMed] [Google Scholar]
- 2.Hupp T.R., Meek,D.W., Midglex,C.A. and Lane,D.P. (1992) Regulation of the specific DNA binding function of p53. Cell, 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 3.Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 4.Jayaraman L., Murthy,K.G.K., Zhu,C., Curran,T., Xanthoudakis,S. and Prives,C. (1997) Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev., 11, 558–570. [DOI] [PubMed] [Google Scholar]
- 5.Jayaraman L., Moorthy,N.C., Murthy,K.G.K., Manley,J.L., Bustin,M. and Prives,C. (1998) High mobility group protein-1 (HMGB-1) is a unique activator of p53. Genes Dev., 12, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verrier C.S., Roodi,N., Yee,C.J., Bailey,L.R., Jensen,R.A., Bustin,M. and Parl,F.F. (1997) High Mobility Group (HMG) protein and TATA-binding protein-associated factor TAF(II)30 affect estrogen receptor-mediated transcriptional activation. Mol. Endocrinol., 11, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 7.Zappavigna V., Falcida,L., Citterich,M., Mavilio,F. and Bianchi,M.E. (1996) HMGB-1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J., 15, 4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 8.Watt F. and Molloy,P.L. (1988) High mobility group proteins 1 and 2 stimulate binding of a specific transcription factor to the adenovirus major late promoter. Nucleic Acids Res., 16, 1471–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwilling S., Konig,H. and Wirth,T. (1995) High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J., 14, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogawa Y., Aizawa,S., Shirakawa,H. and Yoshida,M. (1995) Stimulation of transcription accompanying relaxation of chromatin structure in cells overexpressing high mobility group 1 protein. J. Biol. Chem., 270, 9272–9280. [DOI] [PubMed] [Google Scholar]
- 11.Onate S.A., Prendergast,J.P., Wagner,M., Nissen,R., Reeves,R., Pettijohn,D.E. and Edwards,D.P. (1994) The DNA bending protein HMGB-1 enhances progesterone receptor binding to its target DNA sequences. Mol. Cell. Biol., 14, 3376–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin M. and Reeves,R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acids Res., 54, 35–99. [DOI] [PubMed] [Google Scholar]
- 13.Immamera T., Izumi H., Nagatani,G., Ise,T., Nomoto,M., Iwamoto,Y. and Kohono,K. (2001) Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein. J. Biol. Chem., 276, 7534–7540. [DOI] [PubMed] [Google Scholar]
- 14.Carbalo M., Puigdomenech,P. and Palau,J. (1983) DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J., 2, 1759–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernues J., Espel,E. and Querol,E. (1986) Identification of the core-histone-binding domains of HMGB-1 and HMG2. Biochim. Biophys. Acta, 866, 242–251. [DOI] [PubMed] [Google Scholar]
- 16.Stros M. and Kolibalova,A. (1987) Interaction of non-histone proteins HMGB-1 and HMG2 with core histones in nucleosomes and core particles revealed by chemical cross-linking. Eur. J. Biochem., 162, 111–118. [DOI] [PubMed] [Google Scholar]
- 17.Ge H. and Roeder,R.G. (1994) The high mobility group protein HMGB-1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem., 269, 17136–17140. [PubMed] [Google Scholar]
- 18.Majumder A., Brown,D., Kerby,S., Rudzinski,I., Po He,T., Randhawa,Z. and Seidman,M.M. (1991) Sequence of human HMG2 cDNA. Nucleic Acids Res., 19, 6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge H., Martinez,E., Chiang,C.M. and Roeder,R.G. (1996) Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol., 274, 57–71. [DOI] [PubMed] [Google Scholar]
- 20.Kundu T.K., Wang,Z. and Roeder,R.G. (1999) Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol., 19, 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus W.L. and Kadonaga,J.T. (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev., 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.J. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundu T.K., Palhan,V.B., Wang,Z., Woojin,A., Cole,P.A. and Roeder,R.G. (2000) Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell, 6, 551–561. [DOI] [PubMed] [Google Scholar]
- 24.Dignam J.P., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau O.D., Kundu,T.K., Soccio,R.E., Ait-Si-Ali,S., Khalil,E.M., Vassilev,A., Wolffe,A.P., Nakatani,Y., Roeder,R.G. and Cole,P.A. (2000) HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell, 5, 589–595. [DOI] [PubMed] [Google Scholar]
- 26.Kern S.E., Pietenpol,J.A., Thiagalingam,S., Seymour,A., Kinzler,K.W. and Vogelstein,B. (1992) Oncogenic forms of p53 inhibit p53-regulated gene expression. Science, 256, 827–830. [DOI] [PubMed] [Google Scholar]
- 27.Thomas J.O. and Travers,A.A. (2001) HMGB-1 and 2 and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci., 26, 167–174. [DOI] [PubMed] [Google Scholar]
- 28.Landsman D. and Bustin,M. (1991) Assessment of the transcriptional activation potential of the HMG chromosomal proteins. Mol. Cell. Biol., 11, 4483–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funk W.D., Pak,D.T., Karas,R.H., Wright,W.E. and Shay,J.W. (1992) A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell Biol., 12, 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietenpol J.A., Tokino,T., Thiagalingam,S., El-Deiry,W.S., Kinzler,K.W. and Vogelstein,B. (1994) Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc. Natl Acad. Sci. USA, 91, 1998–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Deiry W.S., Kern,S.E., Pietenpol,J.A., Kinzler,K.W. and Vogelstein,B. (1992) Definition of a consensus binding site for p53. Nature Genet., 1, 45–49. [DOI] [PubMed] [Google Scholar]
- 32.Ko L.J. and Prives,C. (1996) p53: puzzle and paradigm. Genes Dev., 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- 33.Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 34.Lee K.B. and Thomas,J.O. (2000) The effect of the acidic tail on the DNA-binding properties of the HMGB-1,2 class of proteins: insights from tail switching and tail removal. J. Mol. Biol., 304, 135–149. [DOI] [PubMed] [Google Scholar]
- 35.Sheflin L.G., Fucile,N.W. and Spaulding,S.W. (1993) The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry, 32, 3238–3248. [DOI] [PubMed] [Google Scholar]
- 36.Aizawa S., Nishino,H., Saito,K., Kimura,K., Shirakawa,H. and Yoshida,M. (1994) Stimulation of transcription in cultured cells by high mobility group protein 1: essential role of the acidic carboxyl-terminal region. Biochemistry, 33, 14690–14695. [DOI] [PubMed] [Google Scholar]
- 37.Sakaguchi K., Herrera,J.E., Saito,S., Miki,T., Bustin,M., Vassilev,A., Anderson,C.W. and Appella,E. (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade Genes Dev., 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterner R., Vidali,G. and Alfrey,V.G. (1979) Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMGB-1. J. Biol. Chem., 254, 11577–11583. [PubMed] [Google Scholar]
- 39.Watanabe F., Shirakawa,H., Yoshida,M., Tsukada,K. and Teraoka,H. (1994) Stimulation of DNA-dependent protein kinase activity by high mobility group Proteins1 and 2. Biochem. Biophys. Res. Commun., 202, 736–742. [DOI] [PubMed] [Google Scholar]