Abstract
Bacterial staphylococcal enterotoxin B is involved in several severe disease patterns and it was therefore used as a target for the generation of biologically stable mirror-image oligonucleotide ligands, so called Spiegelmers. The toxin is a 28 kDa protein consisting of 239 amino acids. Since the full-length protein is not accessible to chemical peptide synthesis, a stable domain of 25 amino acids was identified as a suitable selection target. DNA in vitro selection experiments were carried out against the equivalent mirror-image d-peptide domain resulting in high affinity d-DNA aptamers. As expected, the corresponding enantiomeric l-DNA Spiegelmer showed comparable binding characteristics to the l-peptide domain. Moreover, the Spiegelmer bound the whole protein target with only slightly reduced affinity. Dissociation constants of both peptide–oligonucleotide complexes were measured in the range of 200 nM, whereas the Spiegelmer binding to the full-length protein was determined at ∼420 nM. These data demonstrate the possibility to identify Spiegelmers against large protein targets by a domain approach.
INTRODUCTION
Standard in vitro selection techniques revealed a significant number of nucleic acid based ligands (so called aptamers) binding to their respective target molecules with high affinity and specificity, rivalling the characteristics of antigen–antibody interactions (reviewed in 1). Aptamers were identified to recognise all kinds of target classes such as small molecules, peptides, proteins but also viral particles and whole cells (2). The mode of high affinity and specificity recognition is mediated by the three-dimensional structure of the oligonucleotide ligand. However, the application of aptamers as therapeutical substances is severely hampered as a result of their susceptibility to enzymatic degradation. The use of 2′-modified RNA libraries or the introduction of base, sugar or phosphate-backbone modifications improves the biological half-life significantly. Nevertheless, these modified oligonucleotides are still substrates of metabolising activities in vivo. An approach to identify nuclease resistant oligonucleotides requires the introduction of chiral principles into the in vitro selection process. First applications led to the identification of l-enantiomeric RNA or DNA ligands, termed Spiegelmers, that bind to arginine (3), adenosine (4), vasopressin (5) and gonadotropin-releasing hormone I (6,7). Spiegelmers recognise their targets in the same way as aptamers, but they are unsusceptible to enzymatic degradation by nucleases. To identify a Spiegelmer, the target of choice has to be prepared in its mirror-image configuration first of all. A standard in vitro selection scheme is performed to isolate an aptamer (d-nucleic acid) that binds to this mirror-image target. After synthesising the isolated aptamer sequence in its respective l-enantiomeric form, the resulting Spiegelmer binds the natural target (in the natural configuration) with comparable affinity. This mirror-image approach has also been shown for the generation of d-peptide ligands from phage display libraries (8). In all cases published so far, the targets were relatively small and the respective enantiomers could be chemically synthesised in total for the selection process. However, due to limitations in solid phase peptide synthesis technology, larger peptide or protein targets are hardly accessible as full-length molecules in the mirror-image configuration. In order to tackle more complex targets, substructures such as domains or epitopes would have to be defined in advance. This procedure is clearly described for the generation of antibodies (9,10). In principle, antibodies are able to recognise epitopes that are divorced from their original structural context. For aptamers, only two similar examples have been shown so far. In 1996, Xu and Ellington reported an RNA aptamer that was generated against a 16 amino acid long substructure of the HIV-rev protein, a so called arginine-rich motif (ARM) (11) that naturally interacts with an RNA molecule, the Rev-binding element (RBE). The RNA aptamers raised against the 16mer peptide showed cross-reactivity to the full-length rev protein that consists of 116 amino acids. An additional example is reported by Proske et al. (12) who identified 2′-amino-2′-deoxypyrimidine-modified aptamers to a 40 amino acid long epitope of the prion protein. Further investigations revealed that these aptamers were also able to bind to full-length prion protein.
In our study, we show the results of a mirror-image in vitro selection approach against the staphylococcal enterotoxin B (SEB) (13), a 28 kDa protein, employing a DNA library. SEB is synthesised by several Staphylococcus aureus strains. The toxin is involved in different disease patterns such as food poisoning (14), rheumatoid arthritis (15,16), atopic eczema (17–19), and most severely in the toxic shock syndrome with a mortality rate of more than 50% (20). SEB belongs to the protein family of so-called superantigens, which induce a polyclonal immune response by direct binding to class II major histocompatibility complex proteins (MHC II) and T-cell receptors on the surface of B and T cells without previously being internalised and processed like a normal antigen (21,22). So far, SEB has not been reported as a nucleic acid binding protein.
In order to generate a Spiegelmer with wide application potential for the diagnosis and/or treatment of SEB mediated diseases, we envisaged a domain approach to perform the mirror-image in vitro selection. Therefore, the structural motifs of the protein were analysed. In the context of the whole protein structure, a 25mer domain that forms a loop structure, which is closed by a disulphide bridge, was identified. In vitro selection was performed against the d-configured peptide. Here we demonstrate that this domain approach led to a Spiegelmer that recognises the l-configured peptide as well as the full-length SEB protein.
MATERIALS AND METHODS
Materials
A 25mer peptide sequence (YYYQCYFSKKTNDINSHQ TDKRKTC) corresponding to positions 89–113 of the SEB protein was synthesised in the d- and l-configuration, respectively, at Jerini AG (Berlin, Germany) using Fmoc chemistry. A biotin residue was then covalently coupled to the N-terminus via an aminohexane linker (LC-linker from Pierce Biotechnology, Rockford, IL). SEB protein was produced as a lyophylisate at Toxin Technology (Sarasota, FL) with more than 95% purity and was distributed by Alexis (Grünberg, Germany). For binding assays, SEB was dissolved in H2O to 1 µg/µl. d- and l-oligonucleotides were synthesised at NOXXON Pharma AG (Berlin, Germany) using standard phosphoramidite chemistry. l-DNA-phosphoramidites were obtained from ChemGenes Corporation (Boston, MA). NeutrAvidin agarose was supplied by Pierce Biotechnology. Taq DNA polymerase was purchased from Life Technologies (Karlsruhe, Germany). Nucleotides were obtained from Larova Biochemie (Teltow, Germany). Radioactive [α-32P]dCTP was supplied by Hartmann Analytic (Braunschweig, Germany). Other chemicals in analytical grade were obtained from AppliChem (Darmstadt, Germany). Cloning and sequencing was carried out at SEQLAB (Göttingen, Germany).
In vitro selection
In vitro selection was performed with a single-stranded DNA pool containing 60 internal, randomised nucleotides and two constant, flanking primer binding sites. At first, the synthetic DNA template 5′-TCAGCTGGACGTCTTCGAAT-N60-TGTCAGGAGCTCGAATTCCC-3′ was transformed into dsDNA using the forward primer A [5′-(T)20-XXACT ATAGGGAATTCGAGCTCCTGACA-3′] and the reverse primer B (5′-TCAGCTGGACGTCTTCGAAT-3′). Forward primer A contains a T20 tail that cannot be copied and an XX spacer (X is spacer 9 from Glen Research, Sterling, VA) that cannot be read through by Taq DNA polymerase (23). The resulting PCR product, radioactively labelled by the use of [α-32P]dCTP, yielded strands of unequal length (106 and 126 nt). The 106 nt long strand was separated and isolated by preparative denaturing gel electrophoresis [8% PAA (19:1); 8 M urea]. Approximately 3 × 1014 different molecules of the ssDNA strand were used in the initial selection library. Compared to the amount of the DNA library, a 2-fold excess of biotinylated d-SEB peptide was immobilised on NeutrAvidin agarose in selection buffer (20 mM Tris, pH 7.4; 250 mM NaCl; 5 mM KCl; 2 mM CaCl2; 1 mM MgCl2; 0.005% Triton X-100) using 1.5 ml of MoBiTec mini columns (Göttingen, Germany) that were constantly rotated end-over-end for 30 min. Before and after peptide immobilisation the NeutrAvidin agarose was washed and equilibrated with several volumes of selection buffer. Then, the immobilised d-peptide was brought into contact with the DNA pool that was previously denatured (5 min at 94°C) and renatured (20 min at room temperature). The mixture was constantly moved end-over-end in a defined volume of selection buffer at room temperature for 1 h (Supplementary Material, Table S1). Non-binding DNA molecules were removed up to the remaining quantity of ∼0.5% in the final washing fraction. Binding species were affinity eluted with two matrix volumes of free d-peptide in selection buffer under constant movement. Elution was started with a 10-fold molar excess of free over immobilised d-peptide for 10 min, followed by a second elution step employing a 20-fold excess for 1 h. Finally, the matrix was washed with selection buffer. The eluted amount of DNA was precipitated with ethanol and amplified by PCR for the next selection round. Beginning with the second selection round, the enriched DNA pool was preincubated with NeutrAvidin agarose for 30 min, before adding the flow-through and a first washing volume (two matrix volumes) to the immobilised d-peptide in order to avoid selection of DNA molecules that bind to NeutrAvidin agarose itself. During the course of the selection rounds, the selection pressure was increased by decreasing the d-peptide concentration from 11.8 to 0.7 µM (Table S1). After 12 selection rounds, the eluted DNA was amplified with forward primer A′ (5′-TTCTAATACGACTCACTATAGGGAATT CGAGCTCCTGACA-3′) and reverse primer B. The resulting PCR product was TA cloned and sequenced.
Binding analysis on the Biacore 2000
Binding characteristics of aptamers and Spiegelmers to the l- and d-enantiomeric SEB peptides were determined by real-time kinetic measurement on a Biacore 2000 (Biacore AB, Freiburg, Germany) using a 1:1 binding model. The respective N-terminally biotinylated peptide was immobilised on a Sensor SA Chip (Biacore AB) and binding of DNA ligands was measured in the peptide flow cell, which was corrected for the reaction in the reference flow cell. Analysis of the Spiegelmer binding to whole SEB protein was carried out with a competition assay (24,25). A constant amount of free SEB protein that binds to the immobilised Spiegelmer on the chip surface was competed with increasing amounts of free Spiegelmer. The experimental setting according to Friguet (25) enabled the determination of the binding constant for the SEB Spiegelmer interaction in solution.
Specificity assay
The binding specificity of the SEB Spiegelmer was tested in a different competition assay. 500 pmol of N-terminally biotinylated l-SEB peptide was immobilised on NeutrAvidin agarose that was washed and pre-equilibrated with selection buffer or with 10 µg/µl casein in selection buffer. Casein (blocking reagent; Roche Diagnostics, Mannheim, Germany) served as a model for an extrinsic protein. After de- and renaturation, the Spiegelmer, 32P-labelled with T4 polynucleotidkinase at the 5′ end, was pre-incubated for 1 h with either selection buffer, 10 µg/µl casein in selection buffer or with 250 and 500 pmol of SEB protein in the presence of 10 µg/µl casein in selection buffer, respectively. The pre-incubated probes were added to the immobilised l-peptide for 1 h. The Spiegelmer binding to the l-peptide matrix was analysed in a Beckman LS 6500 liquid scintillation counter (Beckman Coulter, Fullerton, CA). In order to verify the results, binding to non-peptide derivatised NeutrAvidin agarose was determined.
RESULTS AND DISCUSSION
Domain approach
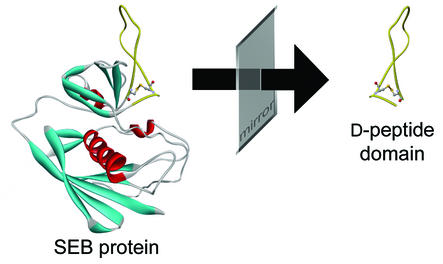
Since the full-length SEB protein is not accessible to chemical peptide synthesis, it was necessary to identify a synthesisable peptide domain. In this context, the availability of the crystal structure of the 28 kDa protein that was determined to 1.5 Å resolution (26) was most helpful. Several criteria have to be taken into account in order to identify a suitable domain. To obtain a potential inhibitory effect with a binder, a domain should be spotted that is functional, or at least next to a functional site, in order to prevent the protein from interacting with its natural binding partners. Secondly, the domain should be on the surface of the protein, so that a resulting Spiegelmer does not only recognise the isolated peptide domain but also the same peptide segment in the context of the whole protein. For the same reason, the peptide domain should be inherently structurally stable. Structural motifs such as β-sheets, α-helices or loops are appropriate. Other structural elements like disulphide bridges can stabilise such domains physically. Even the introduction of artificial cysteine residues may help to stabilise β-loops (27). Peptide targets that display a basic pI are preferred in order to achieve reasonable affinity of the oligonucleotides. In Figure 1, the X-ray structure of the SEB protein is depicted. It is remarkable that one loop structure on the surface of the protein is protruding. The loop is closed and naturally stabilised with a disulphide bridge between Cys93 and Cys113. Furthermore, Cys113 also interacts via a hydrogen bond with Tyr89. Therefore, the most ideal peptide domain was rationally defined between amino acids Tyr89 and Cys113. The statistical pI of this 25mer subsequence is 8.5 which should facilitate the identification of oligonucleotide binders (1). As known from the structure determination of cocrystals with the MHC class II and the T-cell receptor, respectively (28,29), the region of the protein surrounding the 25mer subsequence can interact with MHC class II as well as the T-cell receptor. This feature seems to be of biological relevance for SEB to act as a superantigen. Binding of a Spiegelmer to this region may antagonise the superantigenic effects of SEB.
Figure 1.
Three-dimensional representation of the SEB crystal structure according to Papageorgiou et al. (26). The protruding loop structure (yellow) which is closed by a disulphide bridge was identified as a promising selection target. As depicted, the target was synthesised in its mirror-image configuration.
In vitro selection of d-SEB peptide binding DNA ligands
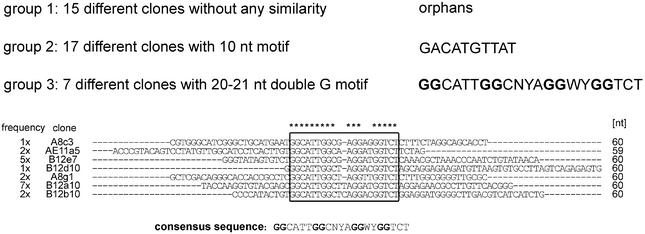
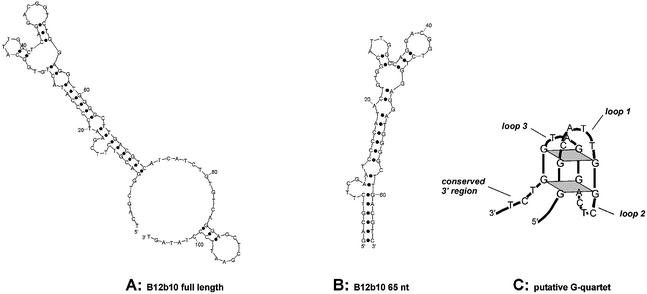
A single-stranded DNA library of ∼3 × 1014 variants containing 60 random positions internally was used for the isolation of aptamers. For partitioning, the library was incubated with the 25mer d-peptide immobilised via an N-terminal biotin modification to NeutrAvidin agarose. After washing the matrix, bound molecules were affinity-eluted with a 10–20-fold molar excess of free, unbiotinylated d-peptide. The isolated binding molecules subsequently served as a template for the PCR amplification step, in order to generate an enriched single-stranded DNA pool for the following selection round. During the course of the selection, the stringency was increased by gradually decreasing the concentration of immobilised peptide to 0.7 µM. After 12 rounds of selection and amplification, ∼9% of the pool was retained by the derivatised peptide matrix. Double-stranded pool DNA from the 12th round was cloned and 65 sequences were determined. The different clones were classified into three groups (Fig. 2) using the multiple alignment program Clustal X (30) and a motif search program (motiv 111; mantik bioinformatic GmbH, Berlin, Germany). In the first group, 15 orphan sequences are listed without any similarity in terms of their primary sequence. Some of them appeared 2-fold (1×), 3-fold (2×) or even 6-fold (1×). The second group combines 17 sequences (three of them appeared 3-fold) that share a common motif of 10 nt. Seven sequences are pooled in group 3 as they show a 20–21 nt motif that is not related to the shorter motif of the second group. This motif is mainly characterised by four double guanosine nucleotides. Representatives of each group were tested for binding to the d-peptide on a Biacore 2000. Compared to clones of the third group, clones of the orphan group as well as clones of group 2 showed only poor binding. Therefore, further analysis was concentrated on clones of group 3. Comparative ranking revealed clone B12b10 as the best binder. According to the secondary structure calculation (31) of the 106 nt long B12b10 molecule (Fig. 3A), the dangling 3′ and 5′ ends were truncated at the predicted helix end G-8:C-72 (Fig. 3B). In comparison to the full-length sequence, the resulting 65mer (B12b10_65) showed similar binding behaviour, whereas shorter molecules significantly lost binding activity. The four double G motifs presumably form a G-quartet, a structural element that cannot be predicted by conventional secondary structure analysis programs (31). G-quartets have been reported for various DNA aptamers such as the thrombin (32) and the ATP/adenosine (33) binders. A proposed secondary structure model of the potential G-quartet region of B12b10_65 is presented in Figure 3C. However, additional experimental studies would be necessary to verify the G-quartet motif in the SEB binding DNA Spiegelmer.
Figure 2.
Sequence groups identified by selection for d-SEB peptide. Thirty-nine clones with different sequences were placed into three groups. Whereas group 1 contains 15 clones without an obvious motif (orphans), 17 clones of group 2 and seven clones of group 3 share a common motif of either 10 conserved or 20–21 conserved nucleotides, respectively. For each clone of group 3, the variable region is shown and the frequency of occurrence is indicated. The conserved motif with four double Gs is boxed.
Figure 3.
Predicted secondary structure for ligand B12b10. (A) According to the predicted folding, the full-length clone B12b10 forms an extended helix with dangling 3′ and 5′ ends which presumably are not involved in binding. (B) Truncation of parts of the 3′ and 5′ end revealed B12b10_65, a functional 65 nt long molecule. (C) Successive G stretches within the primary sequence may indicate a putative G-quartet that cannot be predicted by standard folding programs. A proposed structure of the potential G-quartet region is depicted.
Binding characteristics of SEB peptide and SEB ligands
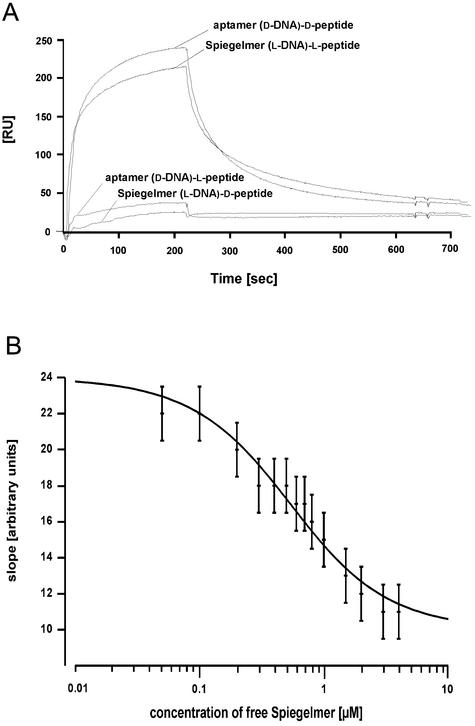
The truncated ligand B12b10_65 was synthesised both in the d- and the l-configuration. The affinities to the respective enantiomers of the SEB peptide were measured on the Biacore instrument (Fig. 4A). The aptamer B12b10_65 binds specifically to the d-SEB peptide and not to the l-SEB peptide, whereas the Spiegelmer B12b10_65 showed the exact opposite binding reaction, thus demonstrating reciprocal chiral specificity. The dissociation constant for binding of the B12b10_65 DNA ligands to the respective SEB peptides was determined to be 200 ± 20 nM in both cases, assuming a 1:1 binding event.
Figure 4.
Binding analysis of B12b10_65 using surface plasmon resonance. (A) Specific binding of the truncated sequence B12b10_65 in either the d- or l-configuration to its respective SEB peptides are shown. No binding at all was observed with the non-cognate interaction partners, thus indicating reciprocal substrate specificity of the oligonucleotide ligands. (B) Binding of Spiegelmer B12b10_65 to full-length SEB protein. In a competition assay (Friguet analysis), binding of free SEB protein to immobilised Spiegelmer B12b10_65 on the chip surface was competed with increasing concentrations of B12b10_65 in solution.
However, the ultimate test for Spiegelmer activity is its recognition of full-length SEB protein. Due to the observed unspecific binding of SEB to surfaces, a competition assay using immobilised Spiegelmer and free SEB was performed on the Biacore instrument (24,25). Therefore, a constant concentration of free SEB protein was rinsed over the immobilised Spiegelmer and competed with increasing concentrations of free Spiegelmer. This experimental setting allows the determination of the Spiegelmer SEB dissociation constant in solution as shown in Figure 4B. Assuming a 1:1 binding reaction, a Kd of ∼420 nM was determined. The dissociation constant of the Spiegelmer to full-length SEB is in good accordance with the constant calculated for the interaction with the l-peptide. The difference may reflect steric hindrance of the Spiegelmer binding to the peptide segment in the context of the full-length protein.
The specificity of Spiegelmer binding to the SEB peptide or SEB protein was tested and analysed in the presence of an excess of an extrinsic protein (Table 1). As a model protein, highly concentrated casein (10 µg/µl) was used. For competition experiments, 500 pmol of the l-SEB peptide was immobilised to NeutrAvidin agarose to ∼15 ng/µl. In a control reaction without casein, 21.5% of the Spiegelmer was binding to the immobilised peptide (Table 1, a). Nearly the same amount of Spiegelmer binding (21%) was observed in the presence of 10 µg/µl casein, confirming that the binding to the peptide is not affected by casein (Table 1, b). Furthermore, Spiegelmer binding to the l-SEB peptide in the presence of 10 µg/µl casein was reduced to 17.2 and 10.6% by competing with 250 and 500 pmol of free SEB protein, respectively (Table 1, c and d). The competition data confirms that the Spiegelmer B12b10_65 specifically recognises the SEB protein in solution and that binding to the whole protein is also not affected by casein at all.
Table 1. Specificity analysis of Spiegelmer–SEB protein binding.
| SEB (pmol) | Casein (10 µg/µl) | Binding to 500 pmol of immobilised l-SEB peptide (%) | |
|---|---|---|---|
| a | 0 | – | 21.5 |
| b | 0 | + | 21.0 |
| c | 250 | + | 17.2 |
| d | 500 | + | 10.6 |
Our results demonstrate for the first time the identification of a Spiegelmer ligand to a protein target by a domain approach, even though the protein or the respective domain does not naturally bind to nucleic acids. The principles of this approach can definitely enlarge the target accessibility for mirror-image in vitro selection.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jens Wientges, Susanne Leva and Lothar Rink for valuable hints and discussion and Dirk Eulberg and Lara Zilberkweit for critical reading of the manuscript. This work was supported by Grant Biotech 01/99 from the Bundeministerium für Bildung, Wissenschaft, Foschung und Technolgie (BMFT).
REFERENCES
- 1.James W. (2000) Aptamers. In Meyers,R. (ed.), Encyclopedia of Analytical Chemistry. John Wiley and Sons Ltd, Chichester, UK, pp. 4848–4871.
- 2.Blank M., Weinschenk,T., Priemer,M. and Schluesener,H. (2001) Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem., 276, 16464–16468. [DOI] [PubMed] [Google Scholar]
- 3.Nolte A., Klussmann,S., Bald,R., Erdmann,V.A. and Furste,J.P. (1996) Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat. Biotechnol., 14, 1116–1119. [DOI] [PubMed] [Google Scholar]
- 4.Klussmann S., Nolte,A., Bald,R., Erdmann,V.A. and Furste,J.P. (1996) Mirror-image RNA that binds D-adenosine. Nat. Biotechnol., 14, 1112–1115. [DOI] [PubMed] [Google Scholar]
- 5.Williams K.P., Liu,X.H., Schumacher,T.N., Lin,H.Y., Ausiello,D.A., Kim,P.S. and Bartel,D.P. (1997) Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc. Natl Acad. Sci. USA, 94, 11285–11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leva S., Lichte,A., Burmeister,J., Muhn,P., Jahnke,B., Fesser,D., Erfurth,J., Burgstaller,P. and Klussmann,S. (2002) GnRH binding RNA and DNA Spiegelmers: a novel approach toward GnRH antagonism. Chem. Biol., 9, 351–359. [DOI] [PubMed] [Google Scholar]
- 7.Wlotzka B., Leva,S., Eschgfaller,B., Burmeister,J., Kleinjung,F., Kaduk,C., Muhn,P., Hess-Stumpp,H. and Klussmann,S. (2002) In vivo properties of an anti-GnRH Spiegelmer: an example of an oligonucleotide-based therapeutic substance class. Proc. Natl Acad. Sci. USA, 99, 8898–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher T.N., Mayr,L.M., Minor,D.L.,Jr, Milhollen,M.A., Burgess,M.W. and Kim,P.S. (1996) Identification of D-peptide ligands through mirror-image phage display. Science, 271, 1854–1857. [DOI] [PubMed] [Google Scholar]
- 9.Lerner R.A. (1984) Antibodies of predetermined specificity in biology and medicine. Adv. Immunol., 36, 1–44. [DOI] [PubMed] [Google Scholar]
- 10.Geysen H.M., Barteling,S.J. and Meloen,R.H. (1985) Small peptides induce antibodies with a sequence and structural requirement for binding antigen comparable to antibodies raised against the native protein. Proc. Natl Acad. Sci. USA, 82, 178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W. and Ellington,A.D. (1996) Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope. Proc. Natl Acad. Sci. USA, 93, 7475–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proske D., Gilch,S., Wopfner,F., Schatzl,H.M., Winnacker,E.L. and Famulok,M. (2002) Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem, 3, 17–25. [DOI] [PubMed] [Google Scholar]
- 13.Huang I.Y. and Bergdoll,M.S. (1970) The primary structure of staphylococcal enterotoxin B. III. The cyanogen bromide peptides of reduced and aminoethylated enterotoxin B and the complete amino acid sequence. J. Biol. Chem., 245, 3518–3525. [PubMed] [Google Scholar]
- 14.Marrack P. and Kappler,J. (1990) The staphylococcal enterotoxins and their relatives. Science, 248, 705–711. [DOI] [PubMed] [Google Scholar]
- 15.Howell M.D., Diveley,J.P., Lundeen,K.A., Esty,A., Winters,S.T., Carlo,D.J. and Brostoff,S.W. (1991) Limited T-cell receptor beta-chain heterogeneity among interleukin 2 receptor-positive synovial T cells suggests a role for superantigen in rheumatoid arthritis. Proc. Natl Acad. Sci. USA, 88, 10921–10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uematsu Y., Wege,H., Straus,A., Ott,M., Bannwarth,W., Lanchbury,J., Panayi,G. and Steinmetz,M. (1991) The T-cell-receptor repertoire in the synovial fluid of a patient with rheumatoid arthritis is polyclonal. Proc. Natl Acad. Sci. USA, 88, 8534–8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breuer K., Wittmann,M., Bosche,B., Kapp,A. and Werfel,T. (2000) Severe atopic dermatitis is associated with sensitization to staphylococcal enterotoxin B (SEB). Allergy, 55, 551–555. [DOI] [PubMed] [Google Scholar]
- 18.Bunikowski R., Mielke,M., Skarabis,H., Herz,U., Bergmann,R.L., Wahn,U. and Renz,H. (1999) Prevalence and role of serum IgE antibodies to the Staphylococcus aureus-derived superantigens SEA and SEB in children with atopic dermatitis. J. Allergy Clin. Immunol., 103, 119–124. [DOI] [PubMed] [Google Scholar]
- 19.Herz U., Bunikowski,R., Mielke,M. and Renz,H. (1999) Contribution of bacterial superantigens to atopic dermatitis. Int. Arch. Allergy Immunol., 118, 240–241. [DOI] [PubMed] [Google Scholar]
- 20.Crass B.A. and Bergdoll,M.S. (1986) Involvement of staphylococcal enterotoxins in nonmenstrual toxic shock syndrome. J. Clin. Microbiol., 23, 1138–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman A., Kappler,J.W., Marrack,P. and Pullen,A.M. (1991) Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol., 9, 745–772. [DOI] [PubMed] [Google Scholar]
- 22.Misfeldt M.L. (1990) Microbial ‘superantigens’. Infect. Immun., 58, 2409–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams K.P. and Bartel,D.P. (1995) PCR product with strands of unequal length. Nucleic Acids Res., 23, 4220–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieba L., Krebber,A. and Plückthun,A. (1996) Competition BIAcore for measuring true affinities: large differences from values determined from binding kinetics. Anal. Biochem., 234, 155–165. [DOI] [PubMed] [Google Scholar]
- 25.Friguet B., Chaffotte,A.F., Djavadi-Ohaniance,L. and Goldberg,M.E. (1985) Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods, 77, 305–319. [DOI] [PubMed] [Google Scholar]
- 26.Papageorgiou A.C., Tranter,H.S. and Acharya,K.R. (1998) Crystal structure of microbial superantigen staphylococcal enterotoxin B at 1.5 A resolution: implications for superantigen recognition by MHC class II molecules and T-cell receptors. J. Mol. Biol., 277, 61–79. [DOI] [PubMed] [Google Scholar]
- 27.Rizo J. and Gierasch,L.M. (1992) Constrained peptides: models of bioactive peptides and protein substructures. Annu. Rev. Biochem., 61, 387–418. [DOI] [PubMed] [Google Scholar]
- 28.Reinherz E.L., Tan,K., Tang,L., Kern,P., Liu,J., Xiong,Y., Hussey,R.E., Smolyar,A., Hare,B., Zhang,R., Joachimiak,A., Chang,H.C., Wagner,G. and Wang,J. (1999) The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science, 286, 1913–1921. [DOI] [PubMed] [Google Scholar]
- 29.Jardetzky T.S., Brown,J.H., Gorga,J.C., Stern,L.J., Urban,R.G., Chi,Y.I., Stauffacher,C., Strominger,J.L. and Wiley,D.C. (1994) Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature, 368, 711–718. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.SantaLucia J. Jr, (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macaya R.F., Schultze,P., Smith,F.W., Roe,J.A. and Feigon,J. (1993) Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA, 90, 3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huizenga D.E. and Szostak,J.W. (1995) A DNA aptamer that binds adenosine and ATP. Biochemistry, 34, 656–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.