Abstract
T4 DNA ligase catalyzes the template-dependent ligation of DNA. Using T4 DNA ligase under specific experimental conditions, we demonstrate that each of the four canonical nucleosides, centrally located on a template molecule such that they flank the site of ligation, can direct the ligation of nucleic acids regardless of the identity of the terminal nucleosides being covalently joined. This universal templating capability extends to those positions adjacent to the ligation junction. This is the first report, irrespective of the ligation method used or the identity of the template nucleosides (including analogs), which shows that nucleosides can act essentially as universal templates at ligation junctions in vitro. The canonical nucleosides do, however, differ in their ability to template sequence- independent ligations, with thymidine and guanosine being equally effective, yet more effective than adenosine and cytidine. Results indicate that hybridization strength surrounding the ligation junction is an important factor. The implications of this previously undiscovered property of T4 DNA ligase with canonical nucleosides are discussed.
INTRODUCTION
DNA ligases catalyze the ligation of an acceptor oligonucleotide, which contains a required 3′-OH group, and a donor oligonucleotide, which contains a required 5′-phosphate (Fig. 1), both of which typically are base paired to a nucleic acid template (1–4). Ligase is an essential enzyme in vivo, playing a critical role in recombination, replication and excision repair. High fidelity in these reactions is important for maintaining genetic integrity (5). For example, ligation of non-base-paired segments in replication would lead to mutagenesis. Ligase is also an essential enzyme in a wide variety of in vitro biotechniques, including standard cloning protocols, the ligase chain reaction (LCR) (6) and the ligase detection reaction (LDR) (7–9). High fidelity in these reactions is important for experimental accuracy.
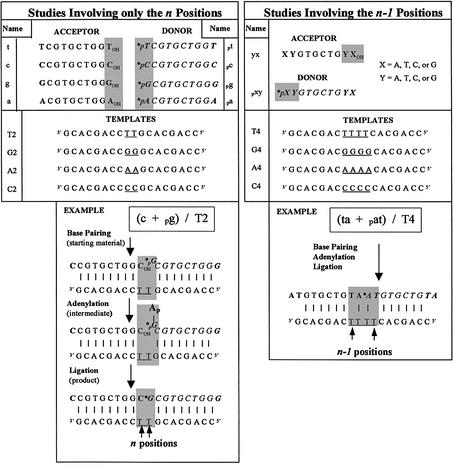
Figure 1.
Ligating two decamers using DNA ligase and templates containing tandem canonical bases at the site of ligation. (Left) Studies on only the n positions, showing both catalytic steps. (Right) Studies on the n – 1 positions, which also consist of experiments where the n positions are not base paired. The phosphorylated (represented by p) decamers are radiolabeled (represented by an asterisk) and are in italics. The names of the decamers to be ligated are represented by lower case lettering according to what nucleoside (from 5′ to 3′) is paired with the universal template positions (in the gray box). The template names are in upper case and represent the central universal template positions (underlined). Note that the adenylation step consists of a 5′→5′ phosphate bond between the phosphorylated decamer and an AMP cofactor. Unless otherwise noted, all oligonucleotides are written in the 5′→3′ direction. Be aware that, although we have no evidence to the contrary, hybridization of each oligonucleotide to the template might not be precisely as drawn here.
Ligases with low fidelity could also be useful for certain experimental strategies, including ligating together degenerate or unknown oligonucleotides for purposes such as site- directed mutagenesis or sequence identification (10,11). To this end, various ligases have been studied under a wide variety of conditions in order to analyze the fidelity of these ligation reactions. The results show that some ligases have the ability, at least in vitro, to ligate oligonucleotides that form certain mismatched base pairs with their templates, both at the ligation junction and at other positions in the resultant helices (9,10,12–22). Under no circumstances, however, has a ligase been shown to catalyze ligation reactions in a sequence-independent manner at ligation junctions, which would be required for developing new types of sequence-independent experimental strategies. Moreover, chemical methods of template-dependent oligonucleotide ligation have been developed (23–25) and have not been shown to be any more effective at catalyzing low fidelity ligation reactions (24–28).
Another route that is being actively pursued is the development of nucleoside analogs that could act as universal bases (29). By definition, universal bases should be able to bind each of the four canonical nucleosides equally, although not necessarily tightly. In this way, ligases (or chemical methods) would be expected to exploit a universal base template region at ligation junctions to catalyze sequence-independent ligations. Perhaps surprisingly, under no circumstances have known universal base analogs been shown to be effective in these low fidelity reactions, especially at ligation junctions (20,30). Moreover, nucleoside analogs are not likely to be practical for subsequent work-up experiments that rely on standard molecular biology protocols.
It is well known that the properties of enzymes depend on the experimental conditions under which reactions are conducted. With this in mind, we evaluated whether a DNA ligase, under a definable set of reaction conditions, could catalyze sequence-independent ligation reactions. For this study, we chose to evaluate T4 DNA ligase, as it can ligate oligonucleotides that contain at least some mismatches (9–13,15,17), in addition to it being one of the most useful enzymes in nucleic acids research. In addition, we analyzed whether canonical nucleosides could act essentially as universal bases in the template at the ligation junction (i.e. promoting sequence-independent ligation with T4 DNA ligase).
We now report that template-directed ligation reactions can be catalyzed in an essentially sequence-independent manner at ligation junctions. For this we exploit reaction conditions under which T4 DNA ligase becomes an extremely low fidelity enzyme. Moreover, we show that each of the four canonical nucleosides can act essentially as universal template bases at and adjacent to the ligation junction. The canonical nucleosides do, however, differ in their ability to template sequence-independent ligations, with thymidine and guanosine being equally effective, yet more effective than adenosine and cytidine. These results suggest that stable base pairing at and adjacent to the ligation junction is not fundamentally required for T4 DNA ligase activity. Hybridization strength surrounding the ligation junction, however, does appear to be important for effective ligation. The implications of this previously undiscovered property of T4 DNA ligase with canonical nucleosides are discussed.
MATERIALS AND METHODS
Oligonucleotide synthesis and preparation
Oligonucleotides were synthesized either on an Applied Biosystems 380B DNA Synthesizer or purchased from Integrated DNA Technologies (Coralville, IA). Oligo nucleotides were purified either by reverse phase HPLC (Integrated DNA Technologies) or thin layer chromatography, as described (31–33). Designated oligonucleotides were 5′-end radiolabeled by incubating 0.4 µM DNA, 0.43 µM [γ-32P]rATP (Amersham Pharmacia, Piscataway, NJ), 70 mM Tris–HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 5 mM dithiothreitol (DTT), 0.1 mM EDTA, 0.1 µM rATP, 10% glycerol and 10 U T4 polynucleotide kinase (New England Biolabs, Beverly, MA) in a reaction volume of 10 µl for 45 min at 37°C. The products were isolated and purified via a 17% native polyacrylamide gel, using 0.25× TBE as the running buffer (1× TBE is 100 mM Tris, 90 mM boric acid and 1 mM EDTA, pH 8.4). The products were then isolated from the gel matrix via overnight pulverization in 2 ml sterile water with a stir bar, followed by centrifugation to partition the gel matrix from the solution. The radiolabeled product solutions were then evaporated to a final concentration of 8 nM.
Ligation reactions
All ligation reactions consist of 3.125 pmol template, 2.5 pmol non-radiolabeled oligonucleotide (the acceptor molecule), 70 fmol radiolabeled oligonucleotide (the donor molecule) and 3 U T4 DNA ligase (Promega, Madison, WI). In all ligation reactions the template and donor molecules were mixed in an appropriate buffer solution and denatured at 90°C for 30 s. After a 5 min annealing period at room temperature, the ligase and the acceptor molecules were added to the reaction mixture (10 µl total volume). This was then incubated for 18 h at an appropriate temperature (listed in the figures and tables). The reactions were terminated by adding 7 µl stop buffer (10 mM urea, 0.1× TBE, 3 mM EDTA). Initial reactions were conducted under Promega’s standard recommended conditions, which consist of 30 mM Tris–HCl (pH 7.8), 10 mM DTT, 10 mM MgCl2 and 1 mM rATP at 30°C. These conditions were then optimized to allow for universal templating abilities by altering the MgCl2 concentration (varied from 1 to 100 mM), the ATP concentration (varied from 0.25 µM to 10 mM), time (varied from 1 to 24 h), temperature (varied from 4 to 44°C) and by including additives such as dimethyl sulfoxide (DMSO), polyethyleneglycol 8000 (PEG) and single-stranded binding protein (from 5 to 50%). The final optimized buffer conditions were 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 3 mM MgCl2, 10 µM ATP and 20% DMSO at either 30 or 22°C for 18 h. In certain cases, 1 U calf intestinal alkaline phosphatase (Promega) was added to individual reactions. Reaction products and intermediates were isolated using 11% denaturing polyacrylamide gels and the reactions were quantified using a Storm 860 Molecular Dynamics PhosphorImager. Observed rate constants were quantified by fitting the time-dependent data to a simple exponential function.
RESULTS
Experimental strategy
To test whether canonical nucleosides, in the template at the ligation junction, can direct the ligation of oligonucleotides irrespective of the identity of the terminal nucleosides being ligated, we developed a model system that allows the quantification of oligonucleotide ligation as a function of the sequence of the oligonucleotides being ligated. A schematic of this experimental strategy is shown in Figure 1. The oligonucleotides to be ligated are decamers, each of which begin and end with the same nucleoside. The acceptor molecules contain a 3′-OH group and the donor molecules contain a radiolabeled 5′-phosphate. When the decamers bind to a template, T4 DNA ligase catalyzes the formation of a covalent bond between the 5′-phosphate and 3′-OH group (example in Fig. 1). By phosphorylating the 5′-end of only one of the decamers (the donor molecule) and adding an excess of unphosphorylated decamer (the acceptor molecule), we can control the sequence of the oligonucleotides at the ligation junction to be ligated. The templates contain a centrally located region consisting of a single type of nucleoside in tandem, which will flank the ligation junction when bound to the donor and acceptor molecules. In this way, we can analyze the ability of the ligase to ligate particular oligonucleotides as a function of template sequence. In this report we focus on the sequences immediately flanking the ligation junction (the n positions in Fig. 1, left) and their adjacent positions (the n – 1 positions in Fig. 1, right).
Investigation of the n position: standard conditions
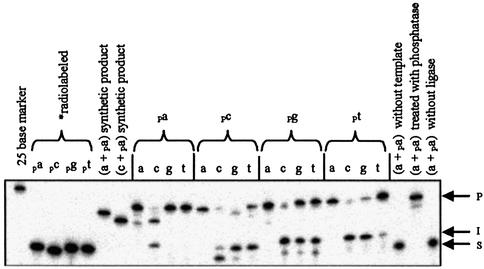
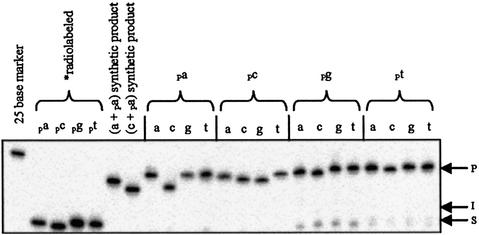
We analyzed the 16 dinucleotide n position combinations using four different templates with either A, C, T or G as the templating nucleoside at the site of ligation. For this we utilized T4 DNA ligase (Promega) under the standard manufacturer’s conditions. We found that the ratio of 1/0.8/0.02 template, non-radiolabeled acceptor and radiolabeled donor, respectively, yielded the best results (data not shown). A representative gel using the T2 template is shown in Figure 2. All experimental indications are that the assays used to study the ligation reactions are working correctly, as the numbers and sizes of the products are as expected. One unique ligation product stems from the ligation of (c + pa). With each of the four templates, this product runs noticeably lower than the other ligation products (Fig. 2). A synthetic size control of the (c + pa) product was run on the gel and was found to also migrate faster than other products. Evidently, this specific sequence is an anomaly as far as its migration behavior. Note that the (a + pa) and (c + pa) size controls run marginally faster than the actual ligation products, which is due to the presence of 5′-phosphates in the controls that are absent in the actual ligation products. A further indication that the assays are working correctly is that formation of both products and intermediates are template dependent. In addition, dephosphorylation of the product band results in a minor (if any) reduction in its intensity, which is expected if the product is only internally radiolabeled. In other words, we are not ligating together two radiolabeled donor molecules.
Figure 2.
Representative gel of ligation reactions run under standard conditions using the T2 template. The standard reaction conditions are 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 1 mM ATP, 2.5 pmol acceptor molecule, 70 fmol donor molecule, 3.125 pmol T2 template and 3 U T4 DNA ligase at 30°C for 18 h. P represents product, I represents adenylated intermediate and S represents starting material. The reaction components are shown above each lane. The first lane contains a generic 25 base size control, the next four lanes are the four unreacted donor molecules and the next two lanes are the synthetic (a + pa) and (c + pa) size controls. The next four panels show the 16 possible reactions. The next three lanes show the ligation of (a + pa) without template, with template and treated with calf alkaline intestinal phosphatase after the reaction has gone to completion, and without ligase.
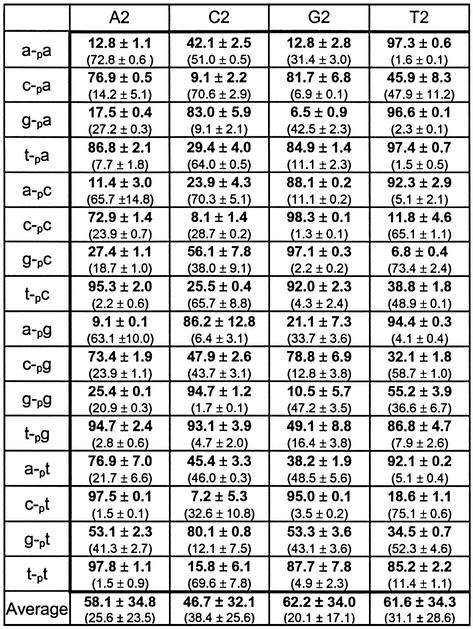
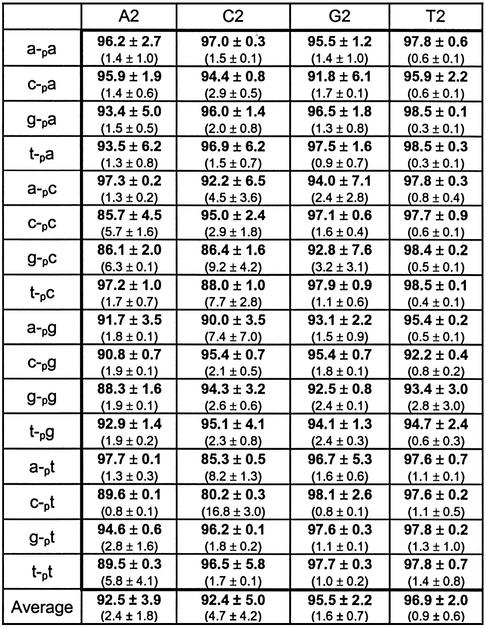
Table 1 lists the results of duplicate trials using each of the four templates. The results show that in cases of complete complementarity, the ligation extent is 95% or more, as would be expected. In cases where one mismatch occurs between the universal template region and the ligation junction, most ligations react greater than 80% (the few exceptions still reacting over 45%). In cases where no complementary base pairing occurs at the ligation junction, the results vary widely, with some dinucleotide combinations being ineffective [for example, 11% for (a-pc)/A2] while some are quite effective [for example, 77% for (c-pa)/A2]. Nevertheless, each of the canonical nucleosides display at least some ability to template the ligation of all 16 dinucleotide combinations. As simply judged by the average product generated as a function of template, the effectiveness of each template reacting with each of the 16 possible sequence combinations follows the order G2 (63%) ≈ T2 (62%) ≈ A2 (58%) > C2 (47%).
Table 1. Results of n position investigation under standard conditions.
The data show percent product formation and percent intermediate formation (in parentheses). The horizontal heading denotes template used and the vertical heading denotes the new ligated product. See Figure 1 for naming conventions of acceptors, donors and templates. All reactions were run at 30°C for 18 h. Reaction conditions were as follows: 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 1 mM ATP, 2.5 pmol acceptor, 70 fmol donor and 3.125 pmol template.
Investigation of the n position: optimized conditions
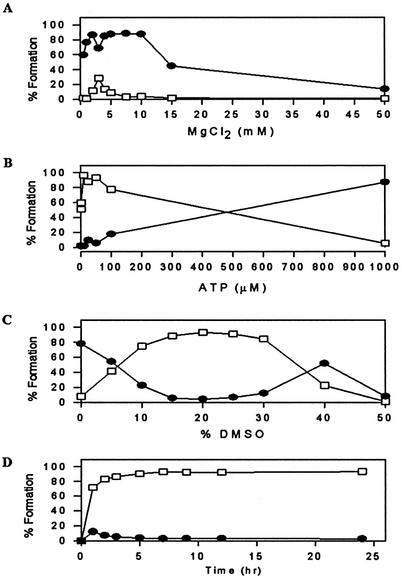
The previous results show that under standard conditions T4 DNA ligase is a sequence-dependent enzyme. Where product formation was low, however, adenylated intermediate formation was usually substantial. Therefore, to increase product formation, only the second step of the reaction had to be enhanced (the ligation step in Fig. 1). We found that altering three components of the reaction enhanced product formation. Figure 3A and B shows that optimizing the MgCl2 concentration and then the ATP concentration in the reaction enhanced product formation of (c + pc)/T2 from 12% (Table 1) to >90%. Under these conditions, however, (c + pc)/C2 ligates poorly (data not shown). Figure 3C shows that the inclusion of DMSO to this reaction enhances the ligation of (c + pc)/C2 from <10% to >90%. These reactions were optimal at 30°C, although only marginally better than at 22°C (data not shown). Figure 3D shows a typical time-dependence plot of the reaction, which indicates that under the optimized conditions products do not break down over time. The addition of PEG, which is a molecular crowding agent, does not further stimulate the reaction, nor does the inclusion of single-stranded binding protein (data not shown).
Figure 3.
Optimization of ligation reactions. (A and B) ATP and MgCl2 concentration-dependent ligations using (c + pc)/T2. The MgCl2 concentration was first optimized under standard reaction conditions of 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 1 mM ATP, 2.5 pmol acceptor molecule, 70 fmol donor molecule, 3.125 pmol template and 3 U T4 DNA ligase at 30°C for 18 h. An optimum concentration of 3 mM MgCl2 was found, which was then used in the subsequent ATP-dependent assay. (C) DMSO-dependent ligation reaction using (c + pc)/C2 in 3 mM MgCl2 and 10 µM ATP. (D) Time-dependent ligation reaction using (c + pc)/T2 in 3 mM MgCl2, 10 µM ATP and 20% DMSO. In each case, product is represented by squares and intermediate by circles.
A representative gel under final optimized conditions with the T2 template is shown in Figure 4. Table 2 shows the results of duplicate trials of all 64 combinations of decamers and template under the optimized conditions (to be compared with the standard conditions in Table 1). These optimized conditions dramatically increase the ligation of mismatched nucleosides at the site of ligation without reducing the effectiveness of ligation with matched base pairs. The template-dependent average for each of the 16 dinucleotide combinations is T2 (97%) ≈ G2 (96%) > A2 (93%) ≈ C2 (92%). Under these optimized conditions, each of the four canonical nucleosides can direct the effective ligation of all 16 possible sequence combinations at the n position. Note that C-G base pairs at the n – 1 positions are not required for effective ligation, as replacing them with A-T pairs does not reduce ligation extent (data not shown).
Figure 4.
Representative gel of ligation reactions run under the optimum conditions using the T2 template. The optimum reaction conditions are 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 3 mM MgCl2, 10 µM ATP, 20% DMSO, 2.5 pmol acceptor molecule, 70 fmol donor molecule, 3.125 pmol T2 template and 3 U T4 DNA ligase at 30°C for 18 h. P represents product, I represents adenylated intermediate and S represents starting material. See Figure 2 for a description of the individual reactions.
Table 2. Results of n position investigation under optimized conditions.
The data show percent product formation and percent intermediate formation (in parentheses). The horizontal heading denotes template used and the vertical heading denotes the new ligated product. See Figure 1 for naming conventions of acceptors, donors and templates. All reactions were run at 30°C for 18 h. Reaction conditions were as follows: 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 10 µM ATP, 20% DMSO, 2.5 pmol acceptor, 70 fmol donor and 3.125 pmol template.
Investigation of the n – 1 position: optimized conditions
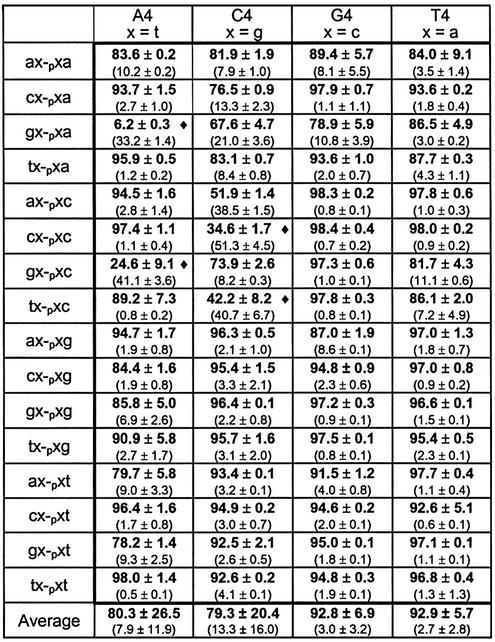
We analyzed whether these optimal experimental conditions can allow ligation of decamers that are mismatched at the positions one nucleotide away from the ligation junction (the n – 1 donor and acceptor positions in Fig. 1) when the ligation junction itself (the n position) is base paired. In these cases, the templates have a central tandem segment of four identical nucleosides. The results (Table 3) show that these optimized experimental conditions do allow the effective ligation of mismatches at positions adjacent to the ligation junction. Note that, while nearly all combinations worked well, 4 out of 64 yielded <50% product. Dropping the reaction temperature from 30 to 22°C in these instances significantly enhanced these reactions [(gt-pta)/A4 went from 6.2 to 83.1%, (gt-ptc)/A4 went from 24.6 to 87.2%, (cg-pgc)/C4 went from 34.7 to 90.9% and (tg-pgc)/C4 went from 42.2 to 97.4%]. Dropping the reaction temperature presumably aids in the hybridization of the decamers to the template and is not expected to significantly hinder the other reactions. The effectiveness of the template nucleosides follows a trend similar to that seen for the n position; T4 (92%) ≈ G4 (92%) > A4 (80%) ≈ C4 (79%).
Table 3. Results of n – 1 position investigation under optimized conditions.
The data show percent product formation and percent intermediate formation (in parentheses). The horizontal heading denotes template used and the vertical heading denotes the new ligated product. See Figure 1 for naming conventions of acceptors, donors and templates. All reactions were run at 30°C for 18 h. The x shown in the vertical heading is defined in the horizontal heading. Note that the reactions with a diamond were also run at 22°C (see Results). Reaction conditions were as follows: 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 10 µM ATP, 20% DMSO, 2.5 pmol acceptor, 70 fmol donor and 3.125 pmol template.
Investigation of mismatches at both the n and n – 1 positions
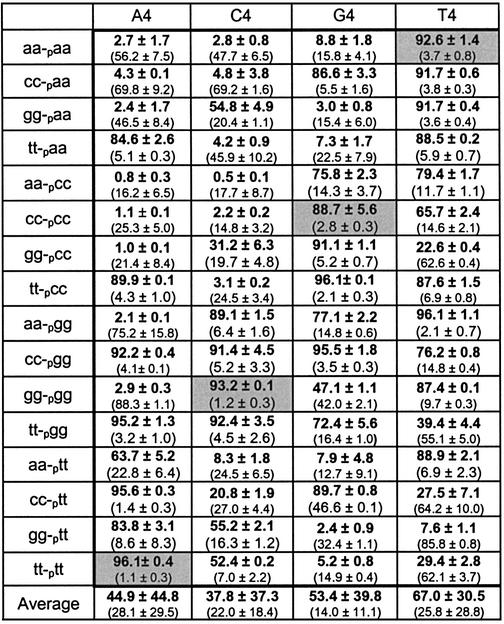
We analyzed the effects of having mismatches at both the n and n – 1 positions, either in the donor molecule, the acceptor molecule or both. Analyzing all 256 possibilities (using a uniform universal template region) is impractical, so we analyzed a subset of 16 representative sequence combinations for each template (Table 4). Note that for this system the reaction temperature was 22°C, as it appeared to work better than 30°C.
Table 4. Results of n and n – 1 mismatch investigation under optimized conditions.
The data show percent product formation and percent intermediate formation (in parentheses). The horizontal heading denotes template used and the vertical heading denotes the new ligated product. See Figure 1 for naming conventions of acceptors, donors and templates. Dark gray background denotes completely matched cases. All reactions were run at 22°C for 18 h. Reaction conditions were as follows: 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 10 µM ATP, 20% DMSO, 2.5 pmol acceptor, 70 fmol donor and 3.125 pmol template.
In cases where no mismatches occur (dark gray background in Table 4) ligation went very well (89–96%), as expected. In cases where two tandem mismatches occurred with either the donor and template or the acceptor and template, ligation generally went well (55–97%), except for (gg-pcc)/C4 (31%). In cases where all four positions contain mismatches, the extents of reaction were highly variable. Using the A4, C4 and G4 templates, some donor–acceptor combinations essentially don’t work at all (<3% ligation). Using the T4 template, however, resulted in substantial ligation for all sequence combinations. Decreasing the reaction temperature to less than 22°C did not enhance these reactions further. Decreased ligation effectiveness as the number of mismatches increases is not unexpected, as each mismatch is likely destabilizing to the entire helix and also will result in end fraying. Nevertheless, these results show that up to four consecutive mismatches flanking the ligation junction can be effectively ligated.
In all of our assays, mismatches and/or overhangs occur at the termini not being ligated. To make sure that these elements are not significantly affecting our results, we synthesized and analyzed systems that mimic (ta-pat)/T4 and (tt-ptt)/T4, except that the non-ligating terminal ends of each substrate are shortened and are completely complementary with the template. In this way, we found that the non-ligating terminal mismatches do not significantly affect our results, as the extent of ligation for (ta-pat)/T4 went from 95.6% in the end-matched system to 96.8% in the end-mismatched system. Similarly, (tt-ptt)/T4 went from 24% in the end-matched system to 29.4% in the end-mismatched system. Therefore, the presence of terminal mismatches does not significantly affect our experimental results.
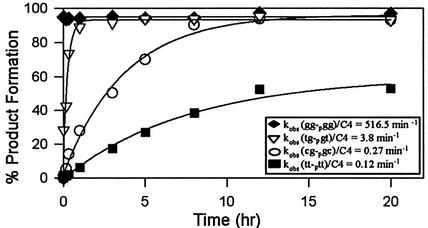
Although the ligation reactions were quantified in this report at 18 h, it is expected that the ligation rates of the various substrate–template combinations vary widely. Substrate–template combinations that are more complementary would be expected to have higher rate constants. This trend was confirmed by conducting time-dependent ligations at 22°C using the representative systems shown in Figure 5. The all matched system, (gg-pgg)/C4, reacts the fastest, with ligation being complete within 1 min. The system with four mismatches, (tt-ptt)/ C4, reacts the slowest, with ligation being complete in ∼12 h. The two systems with intermediate observed rate constants each have mismatches at the n – 1 positions. Predictably, (cg-pgc)/C4, which does not ligate effectively at 30°C (Table 3), is more than 10-fold slower at 22°C than (tg-pgt)/C4, which does ligate effectively at 30°C (Table 3). Importantly, product breakdown is not observed during the reactions, indicating that 18 h appears to be an acceptable end point for these representative systems.
Figure 5.
Time-dependent ligation reactions. The optimum reaction conditions were used, which are 30 mM Tris–HCl (pH 7.5), 10 mM DTT, 3 mM MgCl2, 10 µM ATP, 20% DMSO, 2.5 pmol acceptor molecule, 70 fmol donor molecule, 3.125 pmol C4 template and 3 U T4 DNA ligase at 22°C. The graph shows the plots and fitted curves of each of the four ligation reactions analyzed (see inset for oligonucleotide sequences). The observed rate constants (kobs) for each reaction are given on the graph. The data points are the average of two independent assays.
DISCUSSION
This is the first report showing that T4 DNA ligase can ligate oligonucleotides irrespective of the sequence of the terminal ends of the oligonucleotides being joined. In cases where mismatches only at the n position were tested, the extent of ligation was no less than 80%. In cases where mismatches only at the n – 1 position were tested, the extent of ligation was no less than 50% (after final temperature optimization). Although cases with two or more mismatches at the ligation junction can generate product in amounts above 50%, the ligation extent generally decreases as the number of mismatches increases.
Reaction conditions
The initial experiments indicate that T4 DNA ligase starts to lose fidelity under a relatively narrow range of MgCl2 concentration (between 2 and 4 mM) (Fig. 3A). The reason for the narrow range is unknown, but is not due to inactivity of the enzyme at higher MgCl2 concentrations, as intermediate formation readily occurs at up to 10 mM MgCl2. Furthermore, since intermediate formation is dependent on duplex formation (Fig. 4), this effect is also not due to the presence or absence of duplex formation at higher MgCl2 concentration (although it could relate to the stability of the helix, once formed). It has been previously noted that ligation of mismatches occurs most effectively when the melting temperature (Tm) of the pre-ligated duplex is slightly below the reaction temperature (5) and that only transient in-line attack is required for ligation (5,21). It could be that only the narrow range of divalent cation allows these steric conditions to occur. If true, ligating molecules that form more stable or less stable duplexes might require an adjustment to the MgCl2 concentration.
The optimal range of ATP concentration is 10–100 µM. Too little of this cofactor is ineffective as it will not allow for sufficient binding of ATP to the enzyme (34), leading to a lack of enzyme adenylation. At high ATP concentrations, intermediate formation is prevalent, indicating that the enzyme is active and the DNA duplex is formed; however, product formation is low. One possible explanation is that the enzyme is continuously adenylated at high ATP concentration. After the adenylated enzyme transfers the AMP to the donor molecule, the enzyme can dissociate from the complex. If the enzyme is re-adenylated before rebinding to the adenylated intermediate it can no longer catalyze the final ligation step.
The addition of 10–30% DMSO greatly enhanced the ligation of some mismatched oligonucleotides, primarily through an increase in the second step of the reaction (the ligation step in Fig. 1). It is possible, then, that nucleoside analogs developed to act as universal bases might also be more effective in this regard using buffers containing DMSO. Although the origin of the DMSO effect is unknown, it has been shown that DMSO can reduce the fidelity of nucleoside-binding restriction enzymes (35,36), which parallels our results. Nevertheless, DMSO might not be acting directly on the protein, as ligation of fully base paired nicks does not occur any faster in the presence or absence of DMSO (data not shown). Perhaps DMSO acts by interacting with the DNA in such a way as to favorably orient the mismatched components for the reaction.
That the ligation reactions are generally most effective at 30°C for the single and double mismatches and 22°C for the quadruple mismatches indicates that the drop in temperature permits more effective hybridization of the oligonucleotides to the template. Dropping the temperature even further, however, is ineffective, probably because the duplex becomes rigid or the enzyme loses activity. Increasing the temperature is also not helpful, as hybridization strength decreases, which also would be expected to increase the fidelity of the ligation reaction (26,37). Unfortunately, attempts to determine thermal stabilities of the various pre-ligated duplexes were unsuccessful due to the fact that the melting curves under our experimental conditions do not show a sigmoidal structural transition. We were able to obtain useable melting curves, however, using a standard thermodynamic analysis buffer (20 mM sodium cacodylate, 1 M NaCl and 0.5 mM Na2EDTA, pH 7.0) (38,39). We found that, at least for the pre-ligated representative duplex systems shown in Figure 5, those systems that ligate most effectively in general have the highest melting temperatures (data not shown). Apparently, those systems that are more stable (in terms of hybridization) are more effective in terms of reactivity, which suggests that hybridization strength is a key determinant of reactivity.
Canonical nucleosides as universal template bases
Each of the four canonical nucleosides act as universal template bases at both of the n positions or both of the n – 1 positions (see Fig. 1). This is also true for combined n and n – 1 position mismatches in either the donor or the acceptor (Table 4, light gray boxes), although such mismatches in the donor molecule using the C4 template are less effective than other double mismatch combinations. When mismatches occur at all four positions, however, ligation efficiencies drop off markedly.
There does not appear to be a straightforward correlation between the predicted thermodynamics of the individual mismatches (39–44) and the effectiveness of ligating oligonucleotides that contain such mismatches (at the ligation junction). For those situations where only one or two mismatches occur, all mismatches work well, irrespective of whether the individual mismatches are predicted to be stable or structured. For situations where four mismatches occur and are expected to be stable, only (gg-pgg)/G4 and (tt-ptt)/G4 work well, whereas (aa-paa)/G4 and (gg-pgg)/T4 do not. For those situations where four mismatches occur and are expected to be unstable, (tt-ptt)/C4 and (cc-pcc)/T4 work well, whereas (cc-pcc)/C4 does not. Therefore, either the presence of structure at these positions is not a factor in determining ligase activity or the different reaction conditions utilized between these reports preclude a direct comparison of the results.
In general, sequence-independent ligation can be accomplished using any of the canonical nucleosides in the template flanking the ligation junction. Note that these template nucleosides do not necessarily form hydrogen bonds with the corresponding nucleosides in the donor or the acceptor molecule. In fact, it could be the lack of any specific and required interactions that permit the sequence-independent ligase activity that we observe. Indeed, previous results show that T4 DNA ligase can ligate oligonucleotides even if there is a single nucleotide gap or an abasic residue (15) at the ligation junction. Combined with the above data, it appears that the requirements for T4 DNA ligase do not include stable base pairing at the site of ligation. As suggested previously (21), transient proximity is likely sufficient.
As a template nucleoside, thymidine consistently works as well as or better than the other nucleosides. This is highlighted by the fact that the G4 template directs the ligation of tt to ptt poorly (5%) as compared with the T4 template directing the ligation of gg to pgg (87%). There is no indication that guanosines ligate together more effectively than thymdines. Therefore, thymidine is playing a unique role, but only in the template positions (not in the donor and acceptor terminal positions). It is not because thymidine is a pyrimidine, as cytidine is the poorest universal template base. The low tendency of thymidine to form rigid structures combined with its relatively small size might give it the flexibility to allow its cross-strand partners to transiently adopt a conformation necessary for reactivity. Since most mismatches are expected to be destabilizing, however, the number of canonical universal template bases that a given template can encompass will likely be limited.
Comparison with previous reports
The most common thread throughout the literature is that mismatches, when tolerated, are more permissible at the terminal donor position than the terminal acceptor position (9,10,16,18–20,22). Under our experimental conditions we do not see this effect with n or n – 1 mismatches, as these mismatches appear to ligate similarly.
Although we have utilized enzymatic methods for analyzing low fidelity ligation reactions, evidence indicates that chemical ligation methods (23–25) can be utilized to ligate oligonucleotides that contain mismatches (24–28). A possible advantage to using chemical methods is that the rate of chemistry appears to be substantially faster than that with enzymatic methods (on the order of seconds or minutes rather than hours or days). Furthermore, chemical methods can be effective down to 0°C, which allows for more effective oligo nucleotide hybridization, while enzymes are largely inactive under these low temperature conditions. Unfortunately, when even a single mismatch is present at the ligation junction, the efficiency of chemical ligations is relatively low (and the fidelity is relatively high) in comparison with the results presented herein (24,26,28).
Implications
The results here suggest that one should be cautious performing experiments that rely on the exclusive ligation of matched pairs using T4 DNA ligase, as it can easily be turned into a relatively low fidelity enzyme. In particular, low fidelity conditions are those that consist of relatively low concentrations of MgCl2 (3 mM) and ATP (10–100 µM). This effect can be significantly enhanced by the inclusion of 10–30% DMSO.
Note that although we have shown that T4 DNA ligase can be a low fidelity enzyme in vitro, it is not expected that this is the case in vivo, as a low fidelity ligase would be highly mutagenic in terms of the genomic processes of recombination, replication and repair.
Our results show that canonical nucleosides can be utilized as universal bases in ligation templates, at least at ligation junctions. This property could potentially be exploited to develop protocols that allow for the ligation of oligonucleotides whose termini are unknown or degenerate. For example, a template molecule with tandem thymidines at the ligation junction could serve to direct the ligation of oligonucleotides of varying sequences. In such cases, if the sequence of the substrate region flanking the ligation junction is known, simple PCRs can be used to amplify ligated products for their subsequent sequencing. At its simplest, this strategy, among others, could potentially be useful for sequencing the terminal ends of oligonucleotides.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr Michael Bell and Dana Baum for helpful discussions. This work was aided by grant 85-001-13-IRG from the American Cancer Society, by an award from the Research Corporation and by the Kentucky Research Challenge Trust Fund.
REFERENCES
- 1.Weiss B. and Richardson,C.C. (1967) Enzymatic breakage and joining of deoxyribonucleic acid, I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage. Proc. Natl Acad. Sci. USA, 57, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cozzarelli N.R., Melechen,N.E., Jovin,T.M. and Kornberg,A. (1967) Polynucleotide cellulose as a substrate for a polynucleotide ligase induced by phage T4. Biochem. Biophys. Res. Commun., 28, 578––586.. [DOI] [PubMed] [Google Scholar]
- 3.Gupta N.K., Ohtsuka,E., Weber,H., Chang,S.H. and Khorana,H.G. (1968) Studies on polynucleotides, LXXXVII. The joining of short deoxyribopolynucleotides by DNA-joining enzymes. Proc. Natl Acad. Sci. USA, 60, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss B., Thompson,A. and Richardson,C.C. (1968) Enzymatic breakage and joining of deoxyribonucleic acid, VII. Properties of the enzyme-adenylate intermediate in the polynucleotide ligase reaction. J. Biol. Chem., 243, 4556–4563. [PubMed] [Google Scholar]
- 5.Lehman I.R. (1974) DNA ligase: structure, mechanism and function. Science, 260, 790–797. [DOI] [PubMed] [Google Scholar]
- 6.Barany F. (1991) Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl Acad. Sci. USA, 88, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landegren U., Kaiser,R., Sanders,J. and Hood,L. (1988) A ligase-mediated gene detection technique. Science, 241, 1077–1080. [DOI] [PubMed] [Google Scholar]
- 8.Alves A.M. and Carr,F.K. (1988) Dot blot detection of point mutation with adjacently hybridising synthetic oligonucleotide probes. Nucleic Acids Res., 18, 8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D. and Wallace,R.B. (1989) Specificity of the nick-closing activity of bacteriophage T4 DNA ligases. Gene, 76, 245–254. [DOI] [PubMed] [Google Scholar]
- 10.Cherepanove A., Yildirim,E. and de Vries,S. (2001) Joining of short DNA oligonucleotides with base pair mismatches by T4 DNA ligase. J. Biochem. (Tokyo), 129, 61–68. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov A.V. and de Vries,S. (2002) Scanning mutagenesis using T4 DNA ligase and short degenerate DNA oligonucleotides containing tri-nucleotide mismatches. J. Biochem. (Tokyo), 132, 143–147. [DOI] [PubMed] [Google Scholar]
- 12.Tsiapalis C.M. and Narang,S.A. (1970) On the fidelity of phage T4-induced polynucleotide ligase in the joining of chemically synthesized deoxyribooligonucleotides. Biochem. Biophys. Res. Commun., 39, 631–636. [DOI] [PubMed] [Google Scholar]
- 13.Sgaramella V. and Khorana,H.G. (1972) Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J. Mol. Biol., 72, 427–444. [DOI] [PubMed] [Google Scholar]
- 14.Wiaderkiewicz R. and Ruiz-Carillo,A. (1987) Mismatch and blunt to protruding-end joining by DNA ligases. Nucleic Acids Res., 15, 7831–7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goffin C., Bailly,V. and Verly,W.G. (1987) Nicks 3′ or 5′ to AP sites or to mispaired bases and one-nucleotide gaps can be sealed by T4 DNA ligase. Nucleic Acids Res., 15, 8755–8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomkinson A.E., Tappe,N.J. and Friedberg,E.C. (1992) DNA ligase I from Saccharomyces cerevisiae: physical and biochemical characterization of the CDC9 gene product. Biochemistry, 31, 11762–11771. [DOI] [PubMed] [Google Scholar]
- 17.Harada K. and Orgei,L.E. (1993) Unexpected substrate specificity of T4 DNA ligase revealed by in vitro selection. Nucleic Acids Res., 21, 2287–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuman S. (1995) Vaccinia virus DNA ligase: specificity, fidelity and inhibition. Biochemistry, 34, 16138–16147. [DOI] [PubMed] [Google Scholar]
- 19.Husain I., Tomkinson,A.E., Burkhart,W.A., Moyer,M.B., Ramos,W., Mackey,A.B., Besterman,J.M. and Chen,J. (1995) Purification and characterization of DNA ligase III from bovine testes. J. Biol. Chem., 270, 9683–9690. [DOI] [PubMed] [Google Scholar]
- 20.Luo J., Bergstrom,D.E. and Barany,F. (1996) Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res., 24, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard C.E. and Southern,E.M. (1997) Effects of base mismatches on joining of short oligodeoxynucleotides by DNA ligases. Nucleic Acids Res., 25, 3403–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sriskanda V. and Shuman,S. (1998) Specificity and fidelity of strand joining by Chlorella virus DNA ligase. Nucleic Acids Res., 26, 3536–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokolova N.I., Ashirbekova,D.T., Dolinnaya,N.G. and Shabarova,Z.A. (1988) Chemical reactions within DNA duplexes: cyanogen bromide as an effective oligodeoxyribonucleotide coupling agent. FEBS Lett., 232, 153–155. [DOI] [PubMed] [Google Scholar]
- 24.Dolinnaya N.G., Sokolova,N.I., Gryaznova,O.I. and Shabarova,Z.A. (1988) Site-directed modification of DNA duplexes by chemical ligation. Nucleic Acids Res., 16, 3721–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gryaznov S.M., Schultz,R., Chaturvedi,S.K. and Letsinger,R.L. (1994) Enhancement of selectivity in recognition of nucleic acids via chemical autoligation. Nucleic Acids Res., 22, 2366–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James K.D. and Ellington,A.D. (1997) Surprising fidelity of template-directed chemical ligation of oligonucleotides. Chem. Biol., 4, 595–605. [DOI] [PubMed] [Google Scholar]
- 27.Harada K. and Orgel,L.E. (1994) In vitro selection of optimal DNA substrates for ligation by a water-soluble carbodiimide. J. Mol. Evol., 38, 558–560. [DOI] [PubMed] [Google Scholar]
- 28.James K.D. and Ellington,A.D. (1999) The fidelity of template-directed oligonucleotide ligation and the inevitability of polymerase function. Origins Life Evol. Biosphere, 29, 375–390. [DOI] [PubMed] [Google Scholar]
- 29.Loakes D. (2001) The applications of universal DNA base analogues. Nucleic Acids Res., 29, 2437–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loakes D., Aerschot,A.V., Brown,D.M. and Hill,D. (1996) Enzymatic recognition of acyclic universal base analogues in oligonucleotides. Nucl. Nucl., 15, 1891–1904. [Google Scholar]
- 31.Testa S.M., Haidaris,C.G., Gigliotti,F. and Turner,D.H. (1997) A Pneumocystis carinii group I intron ribozyme that does not require 2′ OH groups on its 5′ exon mimic for binding to the catalytic core. Biochemistry, 36, 15303–15314. [DOI] [PubMed] [Google Scholar]
- 32.Bell M.A., Johnson,A.K. and Testa,S.M. (2002) Ribozyme-catalyzed excision of targeted sequences from within RNAs. Biochemistry, 41, 15327–15333. [DOI] [PubMed] [Google Scholar]
- 33.Johnson A.K., Baum,D.A., Tye,J., Bell,M. and Testa,S.M. (2003) Molecular recognition properties of IGS-mediated reactions catalyzed by a Pneumocystis carinii group I intron. Nucleic Acids Res., 31, 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eun H.M. (1996) Enzymology Primer for Recombinant DNA Technology. Academic Press, San Diego, CA.
- 35.Nasri M. and Thomas,D. (1986) Relaxation of recognition sequence of specific endonuclease HindIII. Nucleic Acids Res., 14, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by Type-II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 37.Herschlag D. (1991) Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn’t always better. Proc. Natl Acad. Sci. USA, 88, 6921–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia T., SantaLucia,J., Burkard,M.E., Kierzek,R., Schroeder,S.J., Jiao,X., Cox,C. and Turner,D.H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry, 42, 14719–14735. [DOI] [PubMed] [Google Scholar]
- 39.Allawi H.T. and SantaLucia,J. (1998) Nearest-neighbor thermodynamic parameters for internal G·A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 40.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J. (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G and T·T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 41.Aboul-ela F., Koh,D., Tinoco,I. and Martin,F.H. (1985) Base-base mismatches. Thermodynamics double helix formation for dCA3XA3G + dCT3YT3G (X,Y = A,C,G,T). Nucleic Acids Res., 13, 4811–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allawi H.T. and SantaLucia,J. (1997) Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 43.Allawi H.T. and SantaLucia,J. (1998) Nearest-neighbor thermodynamics of internal A·C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 44.Allawi H.T. and SantaLucia,J. (1998) Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]