Abstract
Antigenes, which are substances that inhibit gene expression by binding to double-stranded DNA (dsDNA) in a sequence-specific manner, are currently sought for the treatment of various gene-related diseases. As such antigenes, we developed new nuclease-resistant oligopyrimidine nucleotides that are partially modified with 2′-O,4′-C-ethylene nucleic acids (ENA), which are constrained in the C3′-endo conformation and can form a triplex with dsDNA at physiological pH. It was found that these oligonucleotides formed triplexes similarly to those partially modified with 2′-O,4′-C-methylene nucleic acids (2′,4′-BNA or LNA), as determined by UV melting analyses, electromobility shift assays, CD spectral analyses and restriction enzyme inhibition assays. In our studies, oligonucleotides fully modified with ENA have δ torsion angle values that are marginally higher than those of 2′,4′-BNA/LNA. ENA oligonucleotides present in 10-fold the amount of dsDNA were found to be favorable in forming triplexes. These results provide useful information for the future design of triplex-forming oligonucleotides fully modified with such nucleic acids constrained in the C3′-endo conformation considering that oligonucleotides fully modified with 2′,4′-BNA/LNA do not form triplexes.
INTRODUCTION
Oligopyrimidine nucleotides can bind to the major groove of double-stranded DNA (dsDNA) in parallel with the corresponding purine strand of dsDNA (1,2). These complexes are known as triplexes and the oligonucleotides of the third strand are called triplex-forming oligonucleotides (TFOs). Thymine and cytosine in the TFO strand can form hydrogen bonds with the adenine of the A:T base pair and the guanine of the G:C base pair in Hoogsteen fashion, to give the triple bases, T*A:T and C*G:C, respectively. To make stable triplexes, the N-3 of the cytosine in C*G:C should be protonated under acidic conditions. Incorporation of 5-methylcytosine in the TFO gives us a more stable triplex, because of the increase in pKa for 5-methylcytosine (4.3) compared with cytosine (4.15) (3,4).
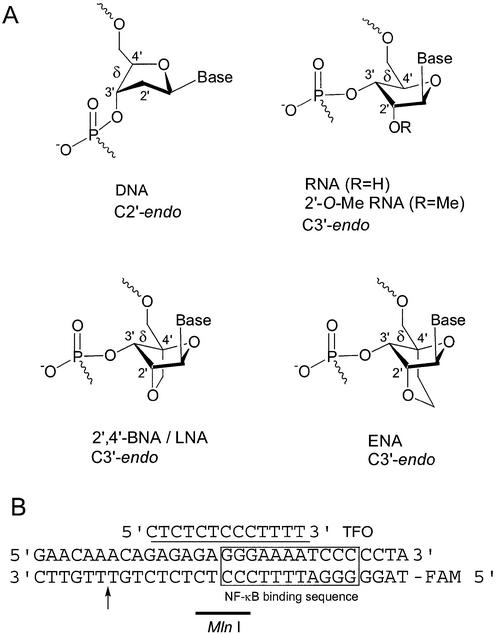
Recently, we have reported that the thermal stability of a triplex containing oligopyrimidine nucleotides partially modified with 2′-O,4′-C-methylene nucleosides, which are known as bridged nucleic acids (2′,4′-BNA) or locked nucleic acids (LNA), and fixed in the C3′-endo conformation (Fig. 1A) was greater than that of a triplex containing natural 2′-deoxyribose units, at physiological pH (5–7). Further, based on our kinetic evaluation of the 2′,4′-BNA/LNA-DNA mixmers, an increase in the association constant, which is due to a decrease in the dissociation rate, was observed. However, fully modified 2′,4′-BNA/LNA did not bind to target dsDNA (8).
Figure 1.
(A) Structures of 2′,4′-BNA/LNA and ENA and (B) a sequence of a triplex. The arrow indicates the cleavage site of MlnI, a restriction enzyme. The 5′ -end of a pyrimidine-rich strand of the dsDNA was labeled with fluorescein (FAM).
In addition, we have synthesized novel nucleosides, 2′-O,4′-C-ethylene nucleosides, as shown in Figure 1A (9). 1H-NMR and X-ray structure analyses showed that the 2′-O,4′-C-ethylene linkage of these nucleosides restricts the sugar puckering to the C3′-endo conformation, as is the case for the linkage of 2′,4′-BNA/LNA (5,7,10,11). Moreover, it has been reported that 2′,4′-BNA/LNA and ethylene-bridged nucleic acids (ENA) show a high binding affinity for the complementary RNA strand and that ENA are more nuclease-resistant than natural DNA and 2′,4′-BNA/LNA (9).
From NMR and X-ray structure analyses of a variety of natural and modified nucleosides and oligonucleotides, it has become clear that the sugar–phosphate backbone torsion angle δ influences the C3′-endo or C2′-endo conformation of nucleosides (12). Recently, three types of triplex structures, those with DNA, RNA or 2′-O-Me-RNA as the third strand, have been solved by NMR analyses and it has been shown that the sugar puckerings of the nucleosides in the third strand are in the C2′-endo and C3′-endo conformation (13,14). We have shown that 2′,4′-BNA/LNA and ENA residues have C3′-endo conformation and do not change their conformation to the C2′-endo conformation (5,7,10,11). They are also structurally completely different from flexible, non-bridged RNA or DNA. Taking into consideration the NMR data of the above-mentioned triplexes for 2′,4′-BNA/LNA-DNA mixmers, such as BNA-1 and BNA-2 (Table 1), as they contain some flexible, natural DNA residues, which have the more preferable C2′-endo conformation, it can be presumed that they can adjust their tertiary structure and form triplexes. It has generally been assumed that fully modified 2′,4′-BNA/LNA oligonucleotides having no natural ribose do not allow triplex formation with dsDNA because all the 2′,4′-BNA/LNA residues are fixed in the C3′-endo conformation.
Table 1. Tm values of triplexes containing ENA or 2′,4′-BNA/LNA at pH 6.6 and pH 7.2.
| TFO | Sequence (5′→3′) | Modification | Tm (°C) | |
|---|---|---|---|---|
| pH 6.6 | pH 7.2 | |||
| DNA-1 | TCTCTCTCCCTTTT | Natural | NDa | NDa |
| BNA-1 | TCTCTCTCCCTTTT | BNA/LNA | 41a | NDa |
| ENA-1 | TCTCTCTCCCTTTT | ENA | 52 | ND |
| BNA-2 | TCTCTCTCCCTTTT | BNA/LNA | 41a | NDa |
| ENA-2 | TCTCTCTCCCTTTT | ENA | 39 | ND |
| BNA-3 | TCTCTCTCCCTTTT | BNA/LNA | NDa | NDa |
| ENA-3 | TCTCTCTCCCTTTT | ENA | 42 | ND |
| T5mC | TCTCTCTCCCTTTT | Natural, 5-Me-C | 28a | ND |
| BNA-4 | TCTCTCTCCCTTTT | BNA/LNA, 5-Me-C | 54a | 37a |
| ENA-4 | TCTCTCTCCCTTTT | ENA, 5-Me-C | nd | ND |
| BNA-5 | TCTCTCTCCCTTTT | BNA/LNA, 5-Me-C | 55a | 33a |
| ENA-5 | TCTCTCTCCCTTTT | ENA, 5-Me-C | 58 | ND |
| BNA-6 | TCTCTCTCCCTTTT | BNA/LNA, 5-Me-C | NDa | NDa |
| ENA-6 | TCTCTCTCCCTTTT | ENA, 5-Me-C | 57 | 41 |
Modified nucleotides are underlined. 5-Me-C, 5-methylcytidine analogs; ND, not detected; nd, not determined because the transition from the triplex to the duplex, although present, was not clear in the melting curve. The buffer was 10 mM phosphate buffer (pH 6.6 and 7.2) and 100 mM NaCl.
aData from Obika et al. (5).
In this paper, in order to develop ideal antigene molecules for intracellular studies, we investigated whether our oligonucleotides modified partially or fully with 2′-O,4′-C-ethylene nucleosides in the C3′-endo conformation could be used as TFOs at physiological pH by comparing the triplex-forming ability of ENA oligonucleotides with that of 2′,4′-BNA/LNA oligonucleotides based on UV melting analyses, electromobility shift assays, CD spectral analyses and restriction enzyme inhibition assays.
MATERIALS AND METHODS
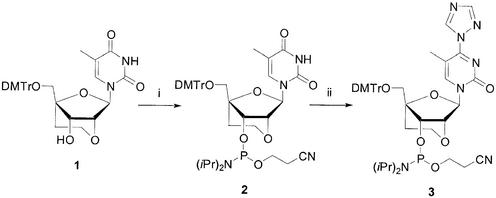
1-[3′-O-[2-Cyanoethoxy(diisopropylamino)phosphino]-5′-O-(4,4′-dimethoxytrityl)-2′-O,4′-C-ethylene-β-d-ribofuranosyl]-5-methyl-4-(1,2,4-triazole-1-yl)pyrimidinone (3)
To a stirred suspension of 1,2,4-triazole (2.78 g, 40 mmol) in acetonitrile (50 ml) POCl3 was added slowly POCl3 (820 µl, 8.8 mmol) followed by triethylamine (0.84 ml, 6 mmol) at 0°C. After stirring for 2 h at 0°C, a solution of 5′-O- (4,4′-dimethoxytrityl)-2′-O,4′-C-ethylene thymidine-3′-O-(2-cyanoethyl)-N,N′-diisopropylphosphoramidite 2 (990 mg, 1.26 mmol) (9) in acetonitrile was added slowly. After stirring for another 2 h at 0°C, the reaction mixture was warmed to room temperature and stirred for 1.5 h. The reaction was quenched by the addition of saturated NaHCO3 solution and then the mixture was extracted with CH2Cl2. The residue was purified by silica gel column chromatography (AcOEt/n-hexane, 4:1) to afford compound 3 (470 mg, 47%). 1H-NMR (400 MHz, CDCl3): 1.0–1.3 (m, 15H), 2.06–2.13 (m, 1H), 2.34–2.38 (m, 1H), 2.54–2.59 (m, 1H), 3.27–4.05 (m, 15H), 4.45–4.50 (m, 1H), 4.55–4.58 (m, 1H), 6.19 (s, 1H), 6.83–6.89 (m, 4H), 7.26–7.47 (m, 9H), 8.05, 8.06 (ss, 1H), 8.52, 8.55 (ss, 1H), 9.25, 9.26 (ss, 1H). FAB-MS (mNBA): [M+H]+ 839.
Synthesis of modified oligonucleotides containing 2′-O,4′-C-ethylene nucleosides
Modified oligonucleotides containing 2′-O,4′-C-ethylene nucleosides were prepared by solid-phase phosphoramidite chemistry using an Applied Biosystems DNA/RNA Synthesizer 392. Reagent solutions were purchased from Applied Biosystems. The coupling of 2′-O,4′-C-ethylene nucleoside-3′-phosphoramidite was performed under standard synthesis conditions except for a longer coupling time (15 min). In the synthesis of a fluorescein-modified oligonucleotide, 6-FAM amidite (Applied Biosystems) was coupled with the oligonucleotide at the 5′ end. After the coupling reaction, the controlled pore glass (CPG) were treated with concentrated aqueous ammonia at 60°C for 5 h. The crude products were purified by C18 silica gel column (Cosmosil 75 C18-OPN, 8 × 100 mm; Nacalai Tesque, Japan) chromatography with an acetonitrile gradient in 50 mM triethylammonium bicarbonate, pH 7.5 treated with 80% acetic acid in water for 20 min and then purified by reverse phase HPLC (Wakosil DNA, 10 × 250 mm; Wako Pure Chemical Industries, Japan) with an acetonitrile gradient in 0.1 M triethylammonium acetate, pH 7.0. The structures of the modified oligonucleotides were determined by negative ion ESI mass spectroscopy. ENA-1, calculated 4358.94, found 4358.84; ENA-2, calculated 4358.94, found 4358.61; ENA-3, calculated 4653.20, found 4653.47; ENA-4, calculated 4442.09, found 4442.93; ENA-5, calculated 4442.09, found 4442.62; ENA-6, calculated 4778.39, found 4778.87.
UV melting analysis
Measurements of the melting temperature (Tm) of triplexes were performed according to a procedure previously described (5).
CD spectra of TFO–dsDNA complexes
TFO and dsDNA were separately dissolved in a buffer containing 10 mM cacodylate (pH 6.8), 200 mM NaCl and 20 mM MgCl2. Equal volumes of the two solutions were mixed to give the TFO–dsDNA complexes. TFO of 5 or 50 µM and dsDNA of 5 µM were used. The complexes were heated at 60°C for 10 min and left at room temperature for 1 or 24 h. The CD spectra of the complexes were obtained with a JASCO J-500C spectropolarimeter.
Electromobility shift assay
The fluorescein-labeled pyrimidine-rich oligonucleotide was mixed with a purine-rich oligonucleotide to give labeled dsDNA. TFO and the labeled dsDNA were separately dissolved in an MlnI reaction buffer containing 10 mM Tris–HCl (pH 7.2), 50 mM NaCl, 10 mM MgCl2 and 0.1 mg/ml bovine serum albumin. Equal volumes of the two solutions were mixed to give the TFO–dsDNA complexes. TFO of 5 or 50 µM and dsDNA of 5 µM were used. The complexes were heated at 60°C for 10 min and left at room temperature for 60 min. Then, the loading solution (30% glycerol in water, 0.025% bromophenol blue and 0.025% xylene cyanol FF) was added to the complexes. The complex formation was analyzed by non-denaturing 10% PAGE in 50 mM Tris–HCl (pH 7.2) and 10 mM MgCl2 at 20°C. The bands on the gel were visualized with a Molecular Imager FX Fluoresent Imager system (Bio-Rad) and quantified with Quantity One software (Bio-Rad). The image of the gel was optimized using Adobe Photoshop.
Restriction enzyme inhibition assay
TFO and the labeled dsDNA were separately dissolved in the MlnI reaction buffer described above. Equal volumes of the two solutions were mixed to give the TFO–dsDNA complexes. TFO of 5 µM and dsDNA of 0.5 µM were used. The complexes were heated at 60°C for 10 min and left at room temperature for 5 min. Then, 1 U MlnI (MBI Fermentas) was added and the reaction mixture was incubated at 37°C for 1 h. The reaction was terminated by ethanol precipitation in the presence of 0.25 M NH4Ac and 1.4 µg/ml glycogen. The samples were resuspended in 50% formamide solution containing 0.025% bromophenol blue and 0.025% xylene cyanol FF and analyzed by denaturing 10% PAGE in 0.89 M Tris–borate–EDTA solution. The bands on the gel were visualized and quantified as described above.
Stability of oligonucleotides
The stability of the oligonucleotides was measured according to a procedure previously described (15). A modified oligonucleotide, 5′-U(dC)8-3′, where U is 2′-O-t-butyldimethylsilyl uridine and dC is 2′-deoxycytidine, was used as an internal standard for the HPLC analysis.
RESULTS AND DISCUSSION
Chemistry
The synthesis of 2′-O,4′-C-ethylene-bridged pyrimidine nucleosides as building blocks was previously reported (9). To prepare ENA oligonucleotides containing 2′-O,4′-C-ethylene 5-methylcytidine units, 2′-O,4′-C-ethylene thymidine-3′-O-phosphoramidite 2 was transformed to 4-triazo derivative 3 (Scheme 1), which was then converted to 5-methylcytidine by treatment with an ammonia solution at the time of deprotection of the oligonucleotides. Oligonucleotides modified with ENA residues were synthesized by solid-phase phosphoramidite chemistry using a DNA/RNA synthesizer and purified by reverse-phase HPLC. The structures of the oligonucleotides were confirmed by negative ion ESI mass spectroscopy.
Scheme 1. Synthesis of 4-triazole thymidine derivative. Reagents and conditions: (i) [(iPr)2N]2P(OC2H4CN), N,N-diisopropylammonium tetrazolide; (ii) 1,2,4-triazole, POCl3, triethylamine, CH3CN.
UV melting temperature analysis
An NF-κB binding sequence in the promoter region of mouse inducible nitric oxide synthase was selected as a target oligopurine-oligopyrimidine sequence (Fig. 1B) (16). We measured the UV Tm of this dsDNA combined with a series of TFOs containing 2′,4′-BNA/LNA or ENA residues. It is known that not all natural DNA as TFOs show a Tm value at pH 6.6 and 7.2 (Table 1) (5). However, Tm values of partially modified TFOs, ENA-1 and ENA-2, and a fully modified TFO, ENA-3, were observed and were 52, 39 and 42°C, respectively, which were similar to or higher than those of 2′,4′-BNA/LNA TFOs, BNA-1, BNA-2 and BNA-3, as shown in Table 1. In particular, the stability of the triplex containing ENA-3, a TFO modified fully with ENA, was greater than that of a mixture of dsDNA and an oligonucleotide modified fully with 2′,4′-BNA/LNA, which failed to bind to the dsDNA. Partially modified ENA-5 and fully modified TFO ENA-6, with 5-methylcytidine instead of cytidine, showed much higher Tm values, of 58 and 57°C, respectively. These Tm values were superior to those of 2′,4′-BNA/LNA oligonucleotides (BNA-5 and BNA-6) (5). Unfortunately, it was hard to evaluate and determine the Tm value of ENA-4 with dsDNA. This is probably because the Tm for the triplex and duplex are very close (the Tm value of the duplex was 65°C). At pH 7.2, the Tm values of the triplexes formed between these ENA oligonucleotides and dsDNA could not be observed except for ENA-6 (Tm = 41°C) because the transition from the triplex to the duplex, although present, was not clear in the melting curve. In comparing TFOs containing ENA and 2′,4′-BNA/LNA oligonucleotides, differences in the Tm values of these triplexes of +11°C (ENA-1/BNA-1), –2°C (ENA-2/BNA-2) and +3°C (ENA-5/BNA-5) (Table 1) were observed. Such a variation in Tm value differences may have occurred due to the incorporation of 2′-O,4′-C-ethylene thymidine residues rather than the cytidine residues into the TFOs. It may also be due to a partial modification of the TFOs owing to the incorporation of ENA residues at odd numbered positions from the 5′ end, which increases the Tm values. Furthermore, for BNA-4, BNA-5 and ENA-6, the increase in Tm per pH unit change was >25°C (Table 1), which was unexpectedly large. Repeating the experiment produced similar results (data not shown). This large increase in Tm values is probably due to the novel properties of these bridged nucleic acids. Although we could not elucidate the reasons from the data we obtained, further investigations such as the evaluation of the Tm values of triplexes composed of pyrimidine-rich, modified oligonucleotides may shed more light on these matters. Since a series of 2′,4′-BNA/LNA and ENA with 5-methylcytidine units showed better properties in forming triplexes than oligonucleotides with cytidine units, ENA-4, ENA-5 and ENA-6 and BNA-4, BNA-5 and BNA-6 were selected for further analyses in order to verify their triplex formation at neutral pH.
Mobility gel shift analysis
In mobility gel shift analyses, triplexes move more slowly than duplexes on the gel (5,8,17). We investigated the binding activity of ENA oligonucleotides to dsDNA by gel analysis. Each TFO was incubated with the dsDNA at a ratio of 1:1 or 10:1, for 10 min at 60°C. After they were left for 60 min at room temperature, they were subjected to 10% PAGE with a neutral buffer at pH 7.2 at 20°C. Although the mixmers, ENA-4 and ENA-5, formed triplexes even at a ratio of 1:1, as did BNA-4 and BNA-5, in the case of fully modified ENA-6 only a faint band indicating triplex formation was observed (Fig. 2A). At a ratio between TFO and dsDNA of 10:1, fully modified ENA-6 formed a triplex. However, the fully modified 2′,4′-BNA/LNA oligonucleotide BNA-6 completely failed to bind to dsDNA (Fig. 2B). These results demonstrate that oligonucleotides partially modified with ENA can form triplexes as well as 2′,4′-BNA/LNA and that fully modified ENA oligonucleotides can also be used as TFOs, as opposed to the fully modified 2′,4′-BNA/LNA oligonucleotides, which fail to form a triplex, as previously reported (5).
Figure 2.
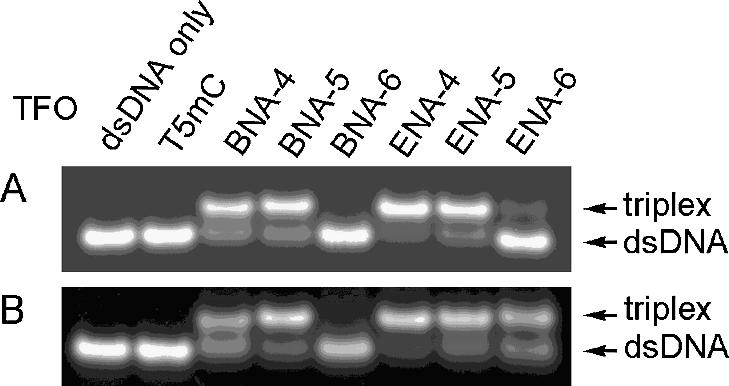
Electrophoretic mobility shift assay of triplex formation at pH 7.2. (A) dsDNA:TFO, 1:1; (B) dsDNA:TFO, 1:10. dsDNA of 5 µM and TFO of 5 or 50 µM were used. A solution of 50 mM Tris–HCl (pH 7.2 at 20°C) and 10 mM MgCl2 was used as the running buffer. MlnI reaction buffer described in Materials and Methods was used as the reaction buffer.
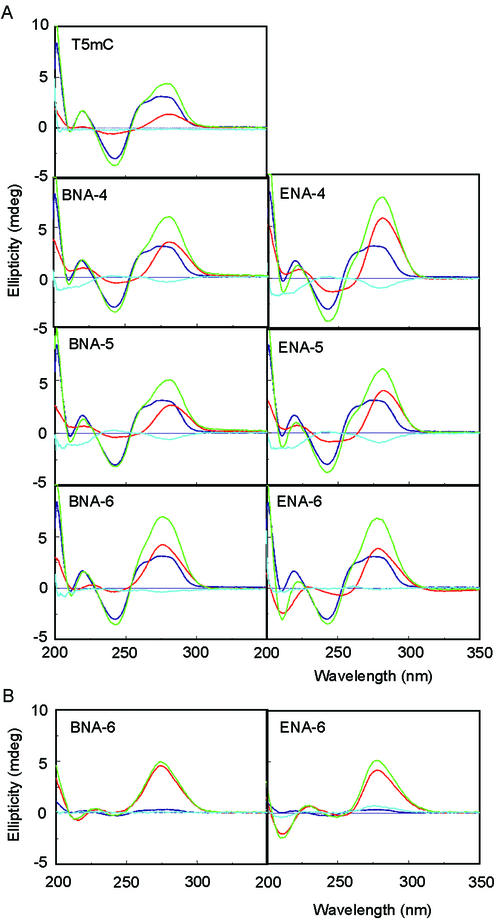
Circular dichroism (CD) analysis
It was reported that a negative cotton effect was observed at ∼215 nm in the CD spectra of the triplex (17,18). We evaluated the CD spectra of mixtures of dsDNA and a TFO and searched for triplex formation based on the data obtained by subtracting the CD spectra of the complex from those of the dsDNA and TFO alone (blue lanes in Fig. 3). When dsDNA was mixed with a variety of TFOs at a ratio of 1:1 at pH 6.8, the CD spectra of the complexes with partially modified ENA oligonucleotides, ENA-4 and ENA-5, and partially modified 2′,4′-BNA/LNA oligonucleotides, BNA-4 and BNA-5, showed negative bands, which indicate the formation of a triplex, at ∼220 nm. However, fully modified ENA-6 did not show any negative bands, as was the case with fully modified BNA-6 (Fig. 3A). At a ratio between dsDNA and TFO of 1:10, the negative cotton effect was observed at ∼220 nm in the CD spectra of the complex with fully modified ENA-6 (Fig. 3B). In the case of fully modified BNA-6, no negative band was observed. Furthermore, when the CD spectra of the above dsDNA–oligonucleotide complexes were compared with CD spectra taken 24 h later at room temperature, the results were almost identical (data not shown). This shows that these complexes, which had been incubated for 1 h, were in equilibrium.
Figure 3.
CD spectra of triplexes. (A) dsDNA:TFO, 1:1; (B) dsDNA:TFO, 1:10. The buffer was 10 mM sodium cacodylate (pH 6.8), 200 mM NaCl and 20 mM magnesium chloride. Red, TFO only; purple, dsDNA only; green, complex between TFO and the dsDNA; blue, CD spectra of the complex between TFO and the dsDNA (green) minus that of the dsDNA (purple) and TFO (red).
Restriction enzyme MlnI analysis
The selected NF-κB binding sequences have a recognition site that is identified by restriction enzyme MlnI (Fig. 1B). If a TFO binds to this recognition site of the dsDNA, the MlnI reaction would be inhibited. At pH 7.2, each TFO was incubated with dsDNA at a ratio of 10:1 for 10 min at 60°C and left for 5 min at room temperature. This was followed by the addition of MlnI and incubation for 1 h at 37°C. Finally, the resulting mixture was analyzed by denaturing 10% PAGE. A natural DNA TFO, T5mC, did not inhibit MlnI cleavage at all. However, partially modified 2′,4′-BNA/LNA TFOs, BNA-4 and BNA-5, had weak inhibitory activity (Fig. 4). The ENA-containing TFOs, ENA-4, ENA-5 and ENA-6, clearly showed a higher activity than the 2′,4′-BNA/LNA TFOs, BNA-4, BNA-5 and BNA-6. In particular, fully modified ENA-6 inhibited MlnI cleavage but the fully modified 2′,4′-BNA/LNA TFO BNA-6 did not. These results show that ENA oligonucleotides form triplexes more efficiently than 2′,4′-BNA/LNA oligonucleotides.
Figure 4.
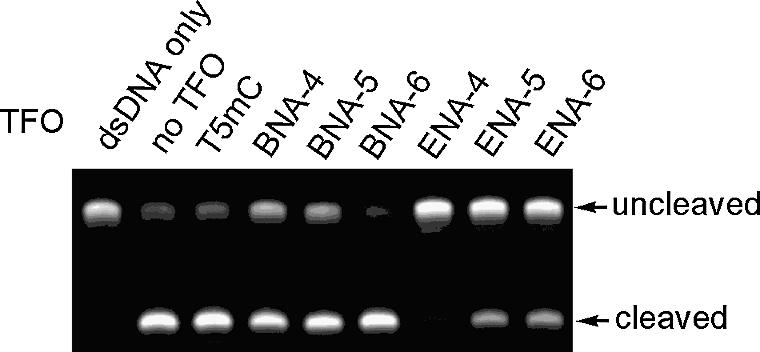
Inhibition of MlnI cleavage due to triplex formation.
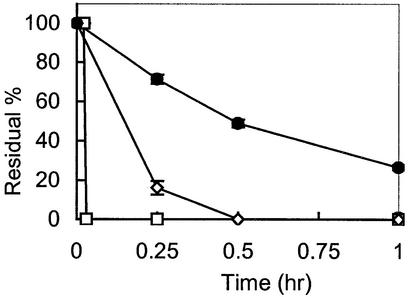
Stability of partially modified ENA oligonucleotides
We showed that oligonucleotides partially modified with 2′,4′-BNA/LNA or ENA had triplex-forming ability. The stability of partially modified oligonucleotides in approximately 100% rat plasma was measured as shown in Figure 5. Natural DNA [oligo(dT9)] was degraded rapidly when incubated in rat plasma at 37°C. A partially modified ENA oligonucleotide (5′-TTTTTTTTT-3′, where T is 2′-O,4′-C-ethylene thymidine) was much more stable than a 2′,4′-BNA/LNA oligonucleotide with the same modification pattern. These data confirmed the results of the in vitro nuclease assay reported previously (9).
Figure 5.
Stability of oligo(dT9) and mixmers with ENA or 2′,4′-BNA/LNA residues in rat plasma. Closed circles, 5′-TTTTTTTTT-3′, where T is 2′-O,4′-C-ethylene thymidine; open diamonds, T is 2′-O,4′-C-methylene thymidine; open squares, natural DNA oligo(dT9).
Modification of oligonucleotides for triplex formation
Antisense oligonucleotides have been widely used to inhibit the translation of mRNA via duplex formation with and without RNase H-mediated degradation of the mRNA (19,20). As well, in the antigene approach, antigenes are used to inhibit gene expression by binding to dsDNA in a sequence-specific fashion. A large number of modified oligonucleotides have been synthesized and their binding affinity has been tested to evaluate their use as antisense and antigene oligonucleotides (21). Recently, oligonucleotides containing modified nucleosides in the C3′-endo sugar conformation, 2′-O-alkylnucleosides (22–24), phosphoramidate (3′-NP DNA) (25–27), 2′,4′-BNA/LNA (6,7,11,28) and ENA (9), which were synthesized as antisense oligonucleotides, have been successfully demonstrated to show stable duplex formation. Such oligonucleotides had less entropy loss in forming duplexes. Some of them greatly inhibited mRNA expression in a true antisense manner. Furthermore, oligonucleotides with 3′-NP DNA (25,29) or nucleosides constrained conformationally, such as 2′,4′-BNA/LNA (5), amino-BNA (30) and tricyclo-DNA (31), form stable triplexes. As far as we know, only two types of modified DNA, 3′-NP DNA and tricyclo-DNA, can act as fully modified triplex-forming oligonucleotides. In this report, we have shown that ENA oligonucleotides can function as TFOs under near physiological conditions, even fully modified ones, whereas fully modified 2′,4′-BNA/LNA oligonucleotides cannot (5). The difference between 2′,4′-BNA/LNA and ENA is one carbon in the linkage that forms the five-membered and six-membered ring, respectively. Based on a detailed conformational analysis of ENA (32) and 2′,4′-BNA/LNA nucleosides (11), the reason for the difference in triplex formation of the two types of fully modified oligonucleotides was investigated. When we analyzed the crystal structures of a variety of bridged nucleosides, such as 2′,4′-BNA/LNA and ENA, we found that the average δ values were 66° and 77°, respectively, and that their difference was marginal (Table 2). In the case of fully modified oligonucleotides, it is considered that the marginal difference for each nucleoside unit in sum may cause a large difference in the end. More detailed structural information on the oligonucleotides based on NMR or X-ray analysis is necessary to explain the difference in their ability to form triplexes.
Table 2. Torsion angles of bridged nucleosides.
| Type | Bridged nucleoside | δ angle | Average δ angle |
|---|---|---|---|
| 2′,4′-BNA/LNA | 2′-O,4′-C-methylene adenosine | 62.5° | 66° |
| 2′-O,4′-C-methylene thymidine | 68.5°, 66.9° | ||
| 2′-O,4′-C-methylene uridine | 66.2° | ||
| ENA | 2′-O,4′-C-ethylene adenosine | 75.7° | 77° |
| 2′-O,4′-C-ethylene-N2-isobutyrylguanosine | 78.9° |
Binding constants of ENA oligonucleotides to dsDNA as TFOs
It has been reported that the binding constants (Ka) of partially modified 2′,4′-BNA/LNA oligonucleotides and 3′-NP DNA as TFO were ∼106 M–1 at near physiological pH (8,29). On the basis of the results of the gel analysis in Figure 2, we can speculate that the Ka of partially modified ENA oligonucleotides used as TFOs might be similar to that of 2′,4′-BNA/LNA, and that the Ka of fully modified ENA oligonucleotides might be less by approximately 10-fold. These values are lower than that of another type of TFO, oligopurine nucleotides (∼109 M–1) (33). However, these oligopurine TFOs consist of guanine-rich sequences that form a guanine quartet structure leading to the undesired binding to proteins such as thrombin (34), HIV gp120 (15,35) and HIV integrase (36). Therefore, an oligopyrimidine TFO is considered to have better properties than an oligopurine TFO. In relation to binding constants of proteins to dsDNA, which are generally >108 M–1 (37), the binding activity of 2′,4′-BNA/LNA and ENA as TFOs is relatively too weak to compete with binding proteins and/or suppress gene expression. Indeed, a high concentration (1 × 10–6 M) of 2′,4′-BNA/LNA oligonucleotides as TFOs was necessary to prevent the binding of NF-κB (p50) to target dsDNA in vitro (5). To improve the binding ability of 2′,4′-BNA/LNA and ENA to dsDNA, introduction of crosslinkers or intercalaters as reported previously (38,39) at the 3′ or 5′ end of the 2′,4′-BNA/LNA TFO or ENA TFO could be effective.
Therapeutic application of antigene technology
One of the most important properties of antisense and antigene oligonucleotides in their use as therapeutics is their nuclease resistance. Phosphorothioate oligonucleotides are the most common type of oligonucleotides having relatively high nuclease resistance and have been introduced to the market as drugs against cytomegalovirus-induced retinitis (40). Oligonucleotides with an ENA residue at the second position from the 3′ end show much higher nuclease resistance than those with an LNA residue at the same position (9). Although Kurreck et al. reported that LNA oligonucleotides were stable in human serum (41), in this study, partially modified ENA oligonucleotides were much more nuclease-resistant than LNA oligonucleotides in rat plasma, as shown in Figure 5. Moreover, oligonucleotides contiguously modified with ENA residues at the 3′ and 5′ end show more stability than those partially modified (data not shown). Thus, ENA oligonucleotides have high potential as antisense and antigene agents that can be used in vivo.
Conclusion
We showed that partially modified oligopyrimidine ENA oligonucleotides as well as partially modified 2′,4′-BNA/LNA oligonucleotides can be used as TFOs at near physiological pH. Moreover, although no triplex is formed with fully modified 2′,4′-BNA/LNA, oligonucleotides fully modified with ENA, which have torsion angle δ values that are marginally higher than those of 2′,4′-BNA/LNA, were found to have high triplex formation ability. We also found that a TFO fully modified with ENA, which are all in the C3′-endo conformation, combined with dsDNA at one-tenth the TFO amount and formed a triplex, which has never been observed before in structure analysis studies. These results provide useful information for designing fully modified TFOs consisting of modified nucleosides that are all in the C3′-endo conformation. We believe that these nuclease-resistant oligopyrimidine ENA oligonucleotides with triplex-forming ability are promising new antigene agents. Further investigation on the properties of such triplex-forming ENA oligonucleotides is in progress.
REFERENCES
- 1.Moser H.E. and Dervan,P.B. (1987) Sequence-specific cleavage of double helical DNA by triple helix formation. Science, 238, 645–650. [DOI] [PubMed] [Google Scholar]
- 2.Le Doan T., Perrouault,L., Praseuth,D., Habhoub,N., Decout,J.L., Thuong,N.T., Lhomme,J. and Hélène,C. (1987) Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[α]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res., 15, 7749–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Povsic T.J. and Dervan,P.B. (1989) Triple helix formation by oligonucleotides on DNA extended to the physiological pH range. J. Am. Chem. Soc., 111, 3059–3061. [Google Scholar]
- 4.Plum G.E., Park,Y.W., Singleton,S.F., Dervan,P.B. and Breslauer,K.J. (1990) Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc. Natl Acad. Sci. USA, 87, 9436–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obika S., Uneda,T., Sugimoto,T., Nanbu,D., Minami,T., Doi,T. and Imanishi,T. (2001) 2′-O,4′-C-Methylene bridged nucleic acid (2′,4′-BNA): synthesis and triplex-forming properties. Bioorg. Med. Chem., 9, 1001–1011. [DOI] [PubMed] [Google Scholar]
- 6.Obika S., Hari,Y., Sekiguchi,M. and Imanishi,T. (2001) A 2′,4′-bridged nucleic acid containing 2-pyridone as a nucleobase: efficient recognition of a C.G interruption by triplex formation with a pyrimidine motif. Angew. Chem. Int. Ed. Engl., 40, 2079–2081. [DOI] [PubMed] [Google Scholar]
- 7.Koshkin A.A., Singh,S.K., Nielsen,P., Rajwanshi,V.K., Kumar,R., Meldgaard,M., Olsen,C.E. and Wengel,J. (1998) LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation and unprecedented nucleic acid recognition. Tetrahedron, 54, 3607–3630. [Google Scholar]
- 8.Torigoe H., Hari,Y., Sekiguchi,M., Obika,S. and Imanishi,T. (2001) 2′-O,4′-C-methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiological pH: thermodynamic and kinetic studies. J. Biol. Chem., 276, 2354–2360. [DOI] [PubMed] [Google Scholar]
- 9.Morita K., Hasegawa,C., Kaneko,M., Tsutsumi,S., Sone,J., Ishikawa,T., Imanishi,T. and Koizumi,M. (2002) 2′-O,4′-C-Ethylene-bridged nucleic acids (ENA): highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett., 12, 73–76. [DOI] [PubMed] [Google Scholar]
- 10.Egli M., Minasov,G., Teplova,M., Kumar,R. and Wengel,J. (2001) X-ray crystal structure of a locked nucleic acid (LNA) duplex composed of a palindromic 10-mer DNA strand containing one LNA thymine monomer. Chem. Commun., 651–652. [Google Scholar]
- 11.Obika S., Nanbu,D., Hari,Y., Morio,K., In,Y., Ishida,T. and Imanishi,T. (1997) Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3′-endo sugar puckering. Tetrahedron Lett., 8735–8738. [Google Scholar]
- 12.de Leeuw H.P.M., Haasnoot,C.A.G. and Altona,C. (1980) Empirical correlation between conformational parameters in β-D-furanoside fragments derived from a statistical survey of crystal structures of nucleic acid constituents. Full description of nucleoside molecular geometries in terms of four parameters. Israel J. Chem., 20, 108–126. [Google Scholar]
- 13.Gotfredsen C.H., Schultze,P. and Feigon,J. (1998) Solution structure of an intramolecular pyrimidine-purine-pyrimidine triplex containing an RNA third strand. J. Am. Chem. Soc., 120, 4281–4289. [Google Scholar]
- 14.Asensio J.L., Carr,R., Brown,T. and Lane,A.N. (1999) Conformational and thermodynamic properties of parallel intramolecular triple helices containing a DNA, RNA, or 2′-OMeDNA third strand. J. Am. Chem. Soc., 121, 11063–11070. [Google Scholar]
- 15.Koizumi M., Koga,R., Hotoda,H., Momota,K., Ohmine,T., Furukawa,H., Agatsuma,T., Nishigaki,T., Abe,K., Kosaka,T., Tsutsumi,S., Sone,J., Kaneko,M., Kimura,S. and Shimada,K. (1997) Biologically active oligodeoxyribonucleotides—IX. Synthesis and anti-HIV-1 activity of hexadeoxyribonucleotides, TGGGAG, bearing 3′- and 5′-end-modification. Bioorg. Med. Chem., 5, 2235–2243. [DOI] [PubMed] [Google Scholar]
- 16.Xie Q. and Nathan,C. (1994) The high-output nitric oxide pathway. J. Leukoc. Biol., 56, 576–582. [DOI] [PubMed] [Google Scholar]
- 17.Manzini G., Xodo,L.E., Gasparotto,D., Quadrifoglio,F., van der Marel,G.A. and van Boom,J.H. (1990) Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J. Mol. Biol., 213, 833–843. [DOI] [PubMed] [Google Scholar]
- 18.Johnson K.H., Gray,D.M. and Sutherland,J.C. (1991) Vacuum UV CD spectra of homopolymer duplexes and triplexes containing A.T or A.U base pairs. Nucleic Acids Res., 19, 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlingensiepen R. and Schlingensiepen,K.H. (1997) Antisense oligodeoxynucleotides—highly specific tools for basic research and pharmacotherapy. Schlingensiepen,R., Brysch,W. and Schlingensiepen,K.H. (eds), Antisense—From Technology to Therapy. Blackwell Science, Berlin, Germany, pp. 3–28.
- 20.Bennett C.F. and Cowsert,L.M. (1999) Antisense oligonucleotides as a tool for gene functionalization and target validation. Biochim. Biophys. Acta, 1489, 19–30. [DOI] [PubMed] [Google Scholar]
- 21.Freier S.M. and Altmann,K.H. (1997) The ups and downs of nucleic acid duplex stability: structure–stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res., 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monia B.P., Johnson,J.F., Sasmor,H. and Cummins,L.L. (1996) Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J. Biol. Chem., 271, 14533–14540. [DOI] [PubMed] [Google Scholar]
- 23.Griffey R.H., Monia,B.P., Cummins,L.L., Freier,S., Greig,M.J., Guinosso,C.J., Lesnik,E., Manalili,S.M., Mohan,V., Owens,S. et al. (1996) 2′-O-aminopropyl ribonucleotides: a zwitterionic modification that enhances the exonuclease resistance and biological activity of antisense oligonucleotides. J. Med. Chem., 39, 5100–5109. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H., Cook,J., Nickel,J., Yu,R., Stecker,K., Myers,K. and Dean,N.M. (2000) Reduction of liver Fas expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nat. Biotechnol., 18, 862–867. [DOI] [PubMed] [Google Scholar]
- 25.Gryaznov S.M., Lloyd,D.H., Chen,J.K., Schultz,R.G., DeDionisio,L.A., Ratmeyer,L. and Wilson,W.D. (1995) Oligonucleotide N3′→P5′ phosphoramidates. Proc. Natl Acad. Sci. USA, 92, 5798–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tereshko V., Gryaznov,S. and Egli,M. (1998) Consequences of replacing the DNA 3′-oxygen by an amino group: high-resolution crystal structure of a fully modified N3′→P5′ phosphoramidite DNA dodecamer duplex. J. Am. Chem. Soc., 120, 269–283. [Google Scholar]
- 27.Faria M., Spiller,D.G., Dubertret,C., Nelson,J.S., White,M.R.H., Scherman,D., Hélène,C. and Giovannangeli,C. (2001) Phosphoramidate oligonucleotides as potent antisense molecules in cells and in vivo. Nat. Biotechnol., 19, 40–44. [DOI] [PubMed] [Google Scholar]
- 28.Wahlestedt C., Salmi,P., Good,L., Kela,J., Johnsson,T., Hökfelt,T., Broberger,C., Porreca,F., Lai,J., Ren,K. et al. (2000) Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA, 97, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torigoe H. (2001) Thermodynamic and kinetic effects of N3′→P5′ phosphoramidate modification on pyrimidine motif triplex DNA formation. Biochemistry, 40, 1063–1069. [DOI] [PubMed] [Google Scholar]
- 30.Obika S., Onoda,M., Morita,K., Andoh,J., Koizumi,M. and Imanishi,T. (2001) 3′-Amino-2′,4′-BNA: novel bridged nucleic acids having an N3′→P5′ phosphoramidite linkage. Chem. Commun., 1992–1993. [DOI] [PubMed] [Google Scholar]
- 31.Steffens R. and Leumann,C.J. (1999) Synthesis and thermodynamic and biophysical properties of tricyclo-DNA. J. Am. Chem. Soc., 121, 3249–3255. [Google Scholar]
- 32.Morita K., Takagi,M., Hasegawa,C., Kaneko,M., Tsutsumi,S., Sone,J., Ishikawa,T., Imanishi,T. and Koizumi,M. (2003) Synthesis and properties of 2′-O,4′-C-ethylene-bridged nucleic acids (ENA) as effective antisense oligonucleotides. Bioorg. Med. Chem., 11, 2211–2226. [DOI] [PubMed] [Google Scholar]
- 33.Durland R.H., Kessler,D.J. and Hogan,M. (1991) Antiparallel triplex formation at physiological pH. In Wickstrom,E. (ed.), Prospects for Antisense Nucleic Acid Therapy of Cancer and AIDS. Willy-Liss, New York, NY, pp. 219–226.
- 34.Bock L.C., Griffin,L.C., Latham,J.A., Vermaas,E.H. and Toole,J.J. (1992) Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature, 355, 564–566. [DOI] [PubMed] [Google Scholar]
- 35.Wyatt J.R., Vickers,T.A., Roberson,J.L., Buckheit,R.W.,Jr, Klimkait,T., DeBaets,E., Davis,P.W., Rayner.B., Imbach,J.L. and Ecker,D.J. (1994) Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl Acad. Sci. USA, 91, 1356–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing N and Hogan,M.E. (1998) Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem., 273, 34992–34999. [DOI] [PubMed] [Google Scholar]
- 37.Schmid J.A., Birbach,A., Hofer-Warbinek,R., Pengg,M., Burner,U., Furtmüller,P.G., Binder,B.R. and de Martin,R. (2000) Dynamics of NF κB and IκBα studied with green fluorescent protein (GFP) fusion proteins. Investigation of GFP-p65 binding to DNA by fluorescence resonance energy transfer. J. Biol. Chem., 275, 17035–17042. [DOI] [PubMed] [Google Scholar]
- 38.Faruqi A.F., Seidman,M.M., Segal,D.J., Carroll,D. and Glazer,P.M. (1996) Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell. Biol., 16, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagatsugi F., Matsuyama,Y., Maeda,M. and Sasaki,S. (2002) Selective cross-linking to the adenine of the TA interrupting site within the triple helix. Bioorg. Med. Chem. Lett., 12, 487–489. [DOI] [PubMed] [Google Scholar]
- 40.Geary R.S., Henry,S.P. and Grillone,L.R. (2002) Fomivirsen: clinical pharmacology and potential drug interactions. Clin. Pharmacokinet., 41, 255–260. [DOI] [PubMed] [Google Scholar]
- 41.Kurreck J., Wyszko,E., Gillen,C. and Erdmann,V.A. (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res., 30, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]