Abstract
We have previously shown that replication- protein A (RPA), the heterotrimeric single-stranded DNA binding protein of eukaryotes, plays a role in Okazaki fragment processing by acting as a molecular switch between the two endonucleases, Dna2 and Fen1, to ensure the complete removal of primer RNAs in Saccharomyces cerevisiae. The stimulation of Dna2 endonuclease activity by RPA requires direct protein–protein interaction. In this report we have analyzed genetically and biochemically the interaction of Dna2 with RPA. RFA1, the gene encoding the large subunit of RPA, displayed allele-specific interactions with DNA2 that included synthetic lethality and intergenic complementation. In addition, we identified physical and functional interactions between these proteins and found that RPA binds Dna2 predominantly through its large subunit, Rpa1. Consistent with the mapping of synthetic lethal mutations, robust interaction localizes to the C-termini of these proteins. Moreover, the N-terminal domains of Dna2 and Rpa1 appear to be important for a functional interaction because the N-terminal domain of RPA1 was required to maximally stimulate Dna2 endonuclease activity. We propose that a bimodal interaction of Dna2 with Rpa1 is important for Dna2 function both in vivo and in vitro. The relevance of each interaction with respect to the function of the Dna2 endonuclease activity is discussed.
INTRODUCTION
DNA replication is a complex enzymatic process that requires a number of proteins to function in a highly coordinated manner (1–3). Among them, replication-protein A (RPA) is known to interact with many replication factors identified in yeast or human cells (reviewed in 4,5). RPA possesses intrinsic single-stranded DNA (ssDNA)-binding activity that plays a role in stabilizing ssDNA in the single-stranded conformation, an active form for many DNA transactions such as DNA replication, repair and recombination (4,5). RPA was first identified in human cells as an essential protein required for simian virus (SV) 40 DNA replication in vitro and is a heterotrimeric complex consisting of 70- (Rpa1), 34- (Rpa2) and 11-kDa (Rpa3) subunits (6–8). Yeast RPA (yRPA) is a heterotrimeric complex of 69-, 36- and 13-kDa subunits (9,10). The genes encoding yRpa1-3 are referred to as RFA1-3, respectively, and each gene is essential for viability (9,10).
There are many reports of the dynamic regulatory role played by RPA in DNA transactions. For example, interactions of RPA with SV40 large T antigen and DNA polymerase (pol) α-primase complex (11) are required for productive generation of primer RNAs for the initiation of SV40 DNA replication (12–14). RPA also interacts with proteins involved in DNA repair, transcriptional activation, as well as the cellular regulator p53 (15–19). In some cases, the functional importance of these protein–protein interactions is not well understood. In addition, RPA is modified by phosphorylation, which may further contribute to its diverse roles (4,5,20–22).
Among the three subunits of RPA, the 70 kDa subunit (Rpa1) contains the major ssDNA binding activity (23–25) and has been shown to interact with many proteins including DNA pol α-primase complex, the DNA damage recognition protein XPA, the tumor suppressor p53, and transcriptional activators like VP16 (11,16,18,19,26). Studies in yeast have demonstrated that mutations in Rpa1 affect DNA replication, repair and recombination in vivo (27–30). Rpa1 is composed of four structural domains: three DNA-binding domains are located in the C-terminal two-thirds of Rpa1 (31,32) and a related, but non-DNA binding domain (RPA1N) is located at the N-terminus (32,33). The C-terminus of Rpa1 is also necessary for the correct assembly of the heterotrimer (34). These structural and functional conclusions are supported by the fact that trypsin digestion of RPA causes initial cleavage of Rpa1 into ∼18 kDa N-terminal and ∼52 kDa C-terminal fragments (35). Although various regions of Rpa1 may be responsible for binding some proteins, the N-terminal region has been implicated in many of the known interactions (36,37). This region is apparently dispensable for in vitro activity because its deletion does not affect its ssDNA-binding activity or its ability to support SV40 replication in vitro (38,39). Surprisingly, the 18-kDa N-terminal region of Rpa1 is essential for growth in yeast (40). Although the precise biological roles of the N-terminal domain are unknown, the domain is likely to participate in RPA–protein interactions since it is not involved in binding DNA.
In previous studies, we identified RPA as a genetic suppressor that corrected the growth defect of a mutant allele of Saccharomyces cerevisiae DNA2, which encodes an enzyme directly involved in Okazaki fragment maturation (41–43). Yeast Dna2 is a 172-kDa protein with conserved DNA helicase motifs that contains DNA helicase as well as ssDNA-specific endonuclease activities (44–46). Dna2 from Archaea, the hyperthermophile Pyrococcus horikoshii, has also been shown to possess ATPase, DNA helicase and nuclease activities, although some differences in enzymatic properties were noted (47). We showed that RPA promotes the endonuclease switching between Dna2 and Fen1 based on the following observations (43,48). RPA binds the 5′ ends of synthetic Okazaki fragments as they are displaced by pol δ and recruits Dna2, resulting in the stimulation of Dna2-catalyzed cleavage of primer RNAs. The cleavage reaction causes RPA to dissociate from the shortened flap, permitting Fen1 to access the remaining flap (43). This mechanism suggests that Dna2 is an endonuclease that processes the long flap DNA generated by displacement DNA synthesis by pol δ (43). Consistent with this, recent studies confirmed that short 5′ flaps were efficiently matured by pol δ and Fen1, and Dna2 did not stimulate maturation, while maturation of long 5′ flaps to which RPA can bind required both Dna2 and Fen1 (49,50).
We also showed that Dna2 consists of at least three functional and structural domains: the N-terminal domain which is dispensable for enzymatic activities, but is required in vivo for growth at 37°C; the central domain required for endonuclease activity; and the C-terminal domain required for helicase activity (46,51). We proposed that the N-terminus of Dna2 might regulate the function of Dna2 in vivo by mediating specific protein–protein interactions (51).
In order to investigate the possibility that RPA may exert its regulatory effect via its interaction with the N-terminal region of Dna2 and understand how RPA influences the activity of Dna2, we analyzed their genetic and physical interactions. As expected for proteins that directly interact, we find that certain mutations in Dna2 and Rpa1 display synthetic lethality or suppression of mutant phenotypes. Deletion derivatives were then used to map interaction domains between Dna2 and Rpa1. The primary interaction occurred through the C-termini of Dna2 and Rpa1, while a secondary interaction was identified between their N-terminal domains. We show that the N-terminal domain of Rpa1 is required for maximal stimulation of Dna2 activity in vitro. These results indicate that the bimodal interaction of Dna2 with Rpa1 is important for Dna2 function, and provide insight into why the N-terminal domains of Rpa1 and Dna2, each of which is dispensable in vitro, are essential in vivo.
MATERIALS AND METHODS
Strains, oligonucleotides, nucleoside triphosphates and enzymes
The synthetic lethal screen and the cloning and characterization of slr157 were performed as described (37). The host strain used for yeast two-hybrid analysis was S.cerevisiae pJ69-4A (Clontech). All oligonucleotides used to construct DNA substrates were synthesized commercially (Genotech, Daejeon, Korea) and gel-purified prior to use. Nucleoside triphosphates were obtained from Boehringer Mannheim, and [γ-32P]ATP (>5000 Ci/mmol) was purchased from Amersham Pharmacia Biotech. Restriction endonucleases and T4 kinase were from Promega.
Purification of recombinant RPA subunits
The genes encoding Rpa1, Rpa2, Rpa3, Rpa11–180 (the N-terminal 180 amino acid fragment of Rpa1), Rpa1Δ180N (Rpa1 lacking the N-terminal 180 amino acids) and Rpa1I14S (Rpa1 containing an Ile→Ser substitution at amino acid position 14) were expressed in Escherichia coli using the pET system (Novagen). All recombinant proteins were expressed as fusion proteins containing polyhistidine residues at their N-termini. Expression of each protein, preparation of extracts and Ni2+-NTA-agarose purification were carried out as recommended (Qiagen). After this step, Rpa2, Rpa3 and Rpa11–180 were >95% pure and used for enzyme-linked immmunosorbent assays (ELISAs) without further purification. Rpa1, Rpa1Δ180N and Rpa1I14S were purified further using two additional steps to remove contaminating proteins. The peak fractions obtained from the Ni2+-NTA-agarose column were pooled and dialyzed for 2 h against T buffer (25 mM Tris–HCl/pH 7.5, 1 mM EDTA, 10% glycerol, 1 mM DTT, 0.1 mM PMSF, 0.15 µg/ml leupeptin and antipain each) containing 100 mM NaCl. The dialysate was loaded onto a ssDNA–cellulose column equilibrated with buffer T plus 100 mM NaCl. After washing with five column volumes of T buffer plus 100 mM NaCl, the column was eluted with T buffer + 2 M NaCl and 40% ethylene glycol. The fractions containing Rpa1 were pooled, concentrated 10-fold and then loaded onto a linear glycerol gradient (15–35%). The gradient was subjected to centrifugation for 24 h at 45 000 r.p.m. in a Beckman SW55 Ti rotor. Fractions (220 µl) were collected from the bottom of the gradient and used for ELISAs. These two additional purification steps resulted in Rpa1 preparations >90% in purity.
Purification of recombinant RPA complex
For the purification of the recombinant RPA complex, the RFA1, 2 and 3 genes were co-expressed in E.coli using the pET system (Novagen). Escherichia coli BL21(DE3) containing the RPA expression plasmid was grown in 6 l of LB broth containing 50 µg/ml kanamyicin at 30°C to an A600 of 0.5. IPTG was added (final 1 mM) and the cells were grown for an additional 3 h. Cells were collected by centrifugation, resuspended in 600 ml of T buffer + 1 M NaCl and sonicated. The supernatant obtained after centrifugation was adjusted to 0.5 M NaCl with T buffer (780 ml, 2.5 g of total protein) and applied to a 120-ml Blue Sepharose column, pre-equilibrated with T buffer + 0.5 M NaCl. This column was washed with two column volumes of T buffer + 0.8 M NaCl and RPA was eluted with two column volumes of T buffer + 2 M NaCl and 40% ethylene glycol. The eluted protein peak was pooled (50 ml, 780 mg), diluted to 0.5 M NaCl with T buffer, and applied to a 30-ml ssDNA cellulose column pre-equilibrated with T buffer + 0.5 M NaCl. This column was washed with three column volumes of T buffer + 0.8 M NaCl and two column volume of T buffer + 2 M NaCl and 40% ethylene glycol. The peak of eluted protein was pooled (30 ml, 8.4 mg) and dialyzed against 1 l of T buffer + 0.2 M NaCl containing 20% sucrose. The dialyzate was diluted to 50 mM NaCl with T buffer and applied to a 5-ml phosphocellulose column. This column was washed with 20 ml of T buffer + 50 mM NaCl and eluted with a 30-ml linear gradient from 50 to 500 mM NaCl in T buffer. This procedure yielded 1.7 mg of recombinant RPA complex of 95% purity. Glycerol gradient centrifugation was carried out as above for further purification. Approximately 95% of protein was obtained as a single protein peak.
RPA–Rpa1Δ180N (RPA containing Rpa1Δ180N as the large subunit) was purified from E.coli using the same procedure described for the isolation of the wild-type RPA complex from E.coli, except that a Resource Q column (1 ml, Pharmacia) was used instead of phosphocellulose. Unlike the recombinant RPA complex, the RPA–Rpa1Δ180N complex did not bind to phosphocellulose column. RPA–Rpa1Δ180N was bound and eluted from the Resource Q column using the same buffers as described in the phosphocellulose column chromatographic step.
Native RPA was also purified directly from yeast extracts as described earlier (23).
Purification of recombinant Dna2 proteins: HX-Dna2, HX-Dna2Δ405N and HX-Dna2N in insect cells
Recombinant Dna2 proteins including HX-Dna2 (wild type), HX-Dna2Δ405N (lacking the N-terminal 405 amino acids of Dna2), and HX-Dna2-405N (the N-terminal 405 amino acid fragment of Dna2) were expressed in insect cells and purified as described (45,51).
Enzyme-linked immunosorbent assay
ELISA was used to examine interactions between Dna2 and RPA. RPA was diluted to a concentration of 10 nM in carbonate buffer (16 mM Na2CO3/34 mM NaHCO3, pH 9.6). RPA was then added to the appropriate wells of a 96-well ELISA plate (100 µl/well) and incubated for 2 h at 25°C. In control wells, BSA was substituted for RPA in the coating step. Wells were aspirated and washed three times with wash buffer (Tris–HCl/pH 7.5,137 mM NaCl, 2.5 mM KCl, 0.05% Tween 20). Blocking buffer (Tris–HCl/pH 7.5,137 mM NaCl, 2.5 mM KCl, 0.05% Tween 20 and 3% BSA) was added and incubated for 2 h at 25°C. Wells were aspirated and washed once with the blocking buffer. Dna2 was serially diluted as indicated in binding buffer (50 mM Tris–HCl/pH 7.5, 100 µg/ml BSA, 125 mM NaCl and 0.02% NP-40). The diluted Dna2 protein was then added to RPA-coated wells of the ELISA plate (100 µl/well) and allowed to bind to RPA by incubating for 30 min at 25°C. Wells were aspirated and washed three times with the binding buffer. Primary antibody (rabbit polyclonal antibodies against Dna2 protein) was diluted 1000-fold in the blocking buffer, added to appropriate wells, and incubated for 1 h at 25°C. Wells were aspirated and washed four times with the blocking buffer. Secondary antibody was diluted 5000-fold in conjugate buffer (50 mM Tris–HCl/pH 7.5, 150 mM NaCl, 0.05% Tween 20, 1% BSA), added to appropriate wells, and incubated for 30 min at 25°C. Wells were aspirated and washed five times with the conjugate buffer. Dna2–RPA complexes were detected using the ABTS substrate (Roche Molecular Biochemicals) for the coloring reaction. ELISAs used to examine the interaction between Dna2 and RPA or RPA derivatives were performed at least four times and the resulting error bars are shown in each figure. Absorbance readings at each point were corrected by subtracting background absorbance generated with BSA-coated wells.
Preparation of DNA substrate and Dna2 endonuclease assay
The procedures used in DNA substrate preparations and their labeling were as described previously (42). The structure of DNA substrates and position of radioisotopic labels are indicated in each figure. The reaction mixtures and conditions used to examine endonuclease activity were as described (42).
Yeast two-hybrid analyses
The DNA fragment encoding the full-length DNA2 was cloned into pGBDU-C vector for GAL4 DNA binding domain fusion. Each DNA fragment encoding Rpa1, Rpa11–180 and Rpa1Δ180N was prepared and cloned into pGAD-C for GAL4 activation domain fusion. Yeast two-hybrid assays were carried out as described (41) and transformations were performed as described (52).
RESULTS
Isolation of dna2-157
We have previously described a synthetic lethal screen using the rfa1-Y29H allele (37). Briefly, we constructed an rfa1Δ ade2 ade3 strain containing a stably integrated rfa1-Y29H allele and plasmid pJM195 (RFA1/URA3/ADE3/CEN). Approximately 1 × 105 ethyl methanesulfonate-mutagenized colonies were screened for strains that require pJM195 for viability. These strains produced unsectored red colonies and were sensitive to 5-fluoroorotic acid (5-FOA) as it selects against pJM195. Three recessive slr (synthetically lethal with rfa1) mutants were isolated: slr51, slr157 and slr44. The slr157 mutation was found to be recessive, complemented by a plasmid carrying DNA2 and genetically linked to DNA2 (data not shown). This allele is hereafter referred to as dna2-157.
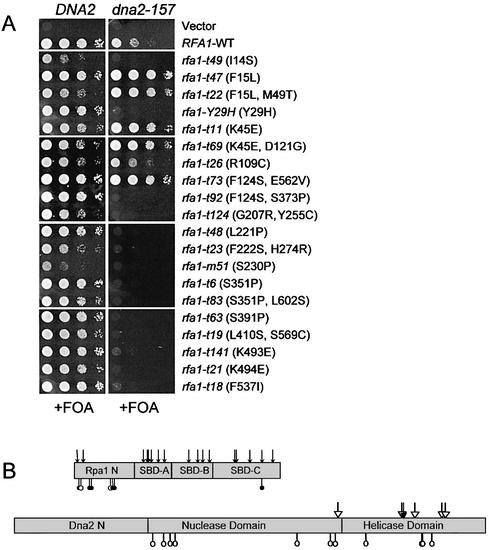
Allele-specific synthetic lethality between RFA1 and DNA2
To test the specificity of the rfa1–dna2 interaction, strain HSY741 (dna2-157 rfa1Δ plus pJM195) was individually transformed with 19 different RFA1 alleles on a LEU2 vector. These RFA1 alleles were previously isolated based on their sensitivity to MMS (27). Freshly growing cells were scraped from a plate lacking leucine and tested for growth in the presence of 5-FOA. As shown in the right-hand panels of Figure 1, dna2-157 was lethal in combination with almost all RFA1 alleles that contain amino acid substitutions in the ssDNA binding domains (SBD-A, B and C) of Rpa1. In contrast, most RFA1 mutations that map to the N-terminal domain of Rpa1 were viable in the presence of dna2-157. The exceptions to this rule included rfa1-Y29H and rfa1-t49 which were shown by western blot analysis to express ∼10% of the level of Rpa1 protein found in wild-type cells (data not shown). This low expression was likely to contribute to the synthetic lethal phenotype of these mutants. The rfa1-t73 allele is the only viable mutation with a change in the ssDNA binding domain (E562V) and is discussed further below. In a wild-type (WT) DNA2 background, the synthetically lethal rfa1 alleles had either no effect on growth or produced a mild slow growth phenotype (Fig. 1A, left). We conclude that dna2-157 requires the wild-type ssDNA-binding domains of Rpa1 for viability.
Figure 1.
Allele-specific interaction between RFA1 and DNA2. (A) Strains HSY636 (DNA2 rfa1Δ with pJM195, left column) and HSY741 (dna2-157 rfa1Δ with pJM195, right column) were transformed with the indicated rfa1 mutants on a LEU2-based vector. Transformants were resuspended in water at equal concentrations and 5 µl of 1/10 serial dilutions were transferred to plates containing 5-FOA to measure synthetic lethality. (B) Summary of the genetic interactions between RFA1 and DNA2. In the schematic diagram of Rpa1, arrows indicate the position of mutations encoded by rfa1 alleles that are synthetically lethal with dna2-157. Open circles indicate the position of mutations encoded by rfa1 alleles that are viable with dna2-157. Closed circles indicate the position of mutations encoded by rfa1 alleles that suppress the growth defect of dna2-157. The F124S mutation is assumed to have no effect on this interaction, as it is present in the viable rfa1-t73 allele. In the lower diagram, open arrows indicate the position of mutations encoded by dna2 alleles that do not complement synthetic lethality of the dna2-157 rfa1-Y29H double mutant. Open circles indicate the position of mutations in dna2 that do complement this lethality. The closed arrow indicates the position of the mutation encoded by dna2-157 (C1255Y).
To determine if RFA1 interacted with specific DNA2 alleles, the strain HSY738 (dna2-157 rfa1-Y29H plus pJM195) was transformed with 12 different DNA2 alleles (53) on plasmids. We then tested whether these DNA2 mutations could complement the synthetic lethality of the rfa1-Y29H dna2-157 strain by selecting against pJM195 on 5-FOA. As shown in Table 1, most of the dna2 mutations that mapped to the helicase domain could not complement the synthetic lethality. In contrast, most mutations that mapped to the nuclease domain were viable in the presence of rfa1-Y29H. These results are summarized in Figure 1B. Consistent with these results, dna2-157 was found to have a point mutation in one of the conserved helicase motifs: a G to A mutation at nucleotide 3765 causing a cysteine to tyrosine change at residue 1255. This result raised the possibility that the RPA heterotrimer physically interacts with Dna2 through the ssDNA binding domains of Rpa1 and helicase domain of Dna2.
Table 1. Complementation of rfa1-Y29H dna2-157 synthetic lethality by DNA2 alleles.
| Mutations | Complementation |
|---|---|
| DNA2-wt | ++++ |
| dna2-15 (G446A, R521K) | ++++ |
| dna2-5 (H471Y) | +++ |
| dna2-3 (P504S) | + |
| dna2-7 (G913D) | ++++ |
| dna2-4 (D1015N, P1031L) | ++++ |
| dna2-14 (P1040L) | – |
| dna2-8 (H1129Y) | +++ |
| dna2-2 (R1253Q) | – |
| dna2-11 (E1297K, G1397S) | – |
| dna2-13 (P1311L, T1312I) | ++++ |
| dna2-16 (G1350E) | + |
| dna2-19 (E1387K) | – |
Allele-specific suppression of dna2-157 by rfa1 mutants
As described above, six of eight rfa1 N-terminal domain mutants were viable in combination with dna2-157. We noticed that five of these six viable double mutants were healthier than the dna2-157 single mutant (Fig. 1A, right). Thus, these rfa1 alleles are suppressors of the dna2-157 slow-growth defect. We asked if these mutations could suppress other dna2-157 phenotypes. The dna2-157 single mutant is sensitive to the DNA damaging agents MMS and UV. As shown in Figure 2A, rfa1-t47 and rfa1-t73 suppressed the MMS sensitivity of dna2-157 whereas three other mutants, which were themselves MMS sensitive, did not. Interestingly, rfa1-t73 also suppressed the UV sensitivity of dna2-157 (Fig. 2B). The suppression of these dna2-157 phenotypes by rfa1 provides further evidence of an interaction between Rpa1 and Dna2.
Figure 2.
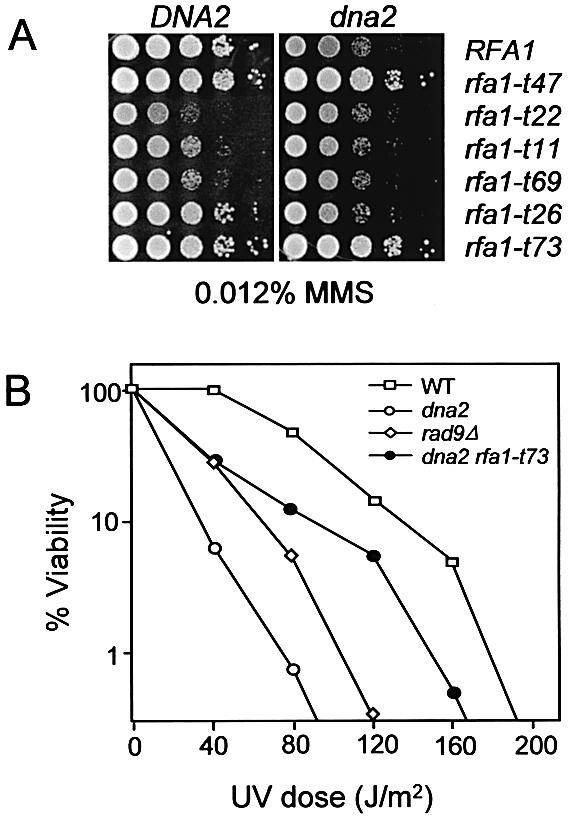
Suppression of MMS and UV sensitivity of dna2-157 by rfa1 mutants. (A) The indicated rfa1 single and rfa1 dna2 double mutants were resuspended in water at equal concentrations and 5 µl of 1/10 serial dilutions were transferred onto YPD plates containing 0.012% MMS. (B) Strains HSY636 (WT), HSY741 (dna2-157), HSY891 (dna2-157 rfa1-t73) and HSY1200 (rad9Δ) were grown to early log phase in YPD. Approximately 500 cells were plated on YPD and irradiated with the indicated doses of UV light. The number of viable cells was determined by counting colonies following 3 days of growth.
Expression and isolation of RPA and its mutant derivatives
The above genetic results suggested that the RPA heterotrimer physically interacts with Dna2, perhaps through the ssDNA binding domains of Rpa1 and helicase domain of Dna2. In order to test this idea, we attempted to confirm this observation in vitro. For this purpose, each subunit of RPA (Rpa1, Rpa2 and Rpa3; Fig. 3, lanes 1–3) was expressed and purified from E.coli. In addition, we isolated several mutant forms of Rpa1; these included Rpa11–180, the N-terminal 180 amino acid fragment of Rpa1 (Fig. 3, lane 4); Rpa1Δ180N, lacking the N-terminal 180 amino acids (Fig. 3, lane 5); and Rpa1I14S, containing an Ile→Ser substitution at amino acid position 14 (Fig. 3, lane 6). We were unable to obtain Rpa1Y29H since expression of this allele in E.coli was extremely poor. We also prepared wild-type RPA and mutant RPA–Rpa1Δ180N, in which the large subunit was substituted with Rpa1Δ180N (Fig. 3, lanes 7 and 8, respectively). The Rpa1 polypeptides contained polyhistidine tags at their N-termini. As previously reported (40), most proteins were insoluble when expressed individually in E.coli (data not shown). However, we found that substantial amounts of each subunit (∼10–30%) were soluble. The presence of hydrophilic polyhistidine residues at their N-termini may have improved the solubility of these proteins. All recombinant proteins were purified to near homogeneity (>90%; Fig. 3), as described in Materials and Methods.
Figure 3.
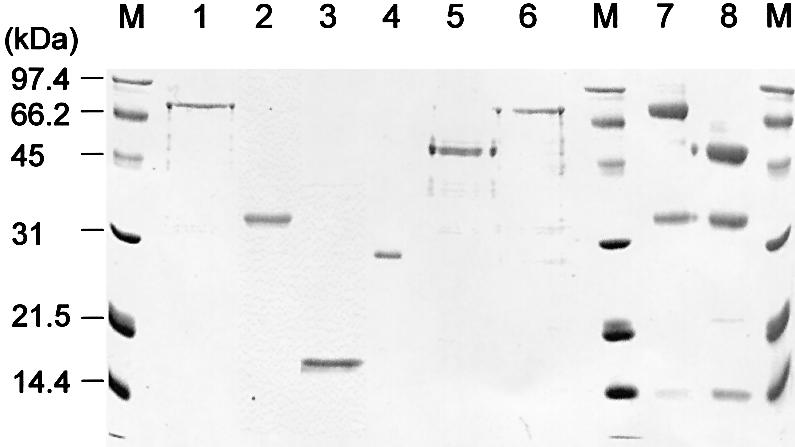
Purification of RPA subunits and mutant derivatives. Coomassie blue staining of a protein gel containing Rpa1 (lane 1), Rpa2 (lane 2), Rpa3 (lane 3), Rpa11–180 (lane 4), Rpa1Δ180N (lane 5), Rpa1I14S (lane 6), wild-type RPA complex (lane 7) and RPA–Rpa1Δ180N complex (lane 8) that were resolved by a 12% SDS–PAGE. Each lane was loaded with ∼2 µg of proteins, based on the value obtained by Bradford assays (Bio-Rad). M indicates molecular size markers and the numbers to the left of the figure denote molecular weights (kDa).
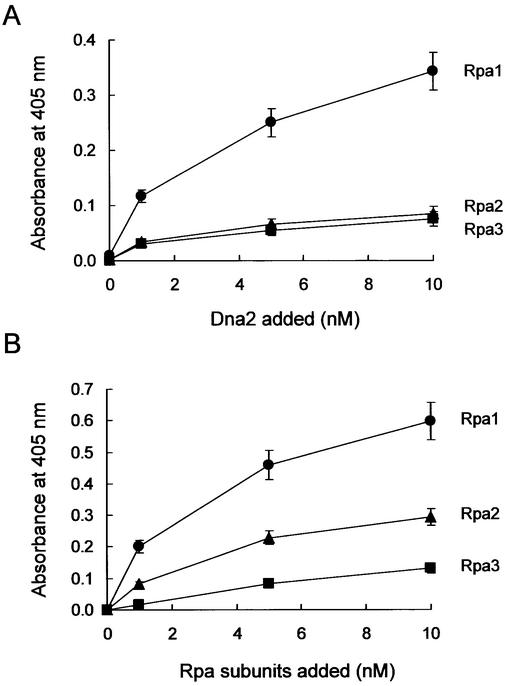
Dna2 interacts preferentially with Rpa1
In order to determine which subunit of RPA contributed to its interaction with Dna2, we carried out ELISA. Each subunit (1 pmol each) of RPA was first adsorbed to a microtiter plate, and then incubated with Dna2 in the presence of increasing concentrations (1–10 nM) of purified Dna2. After the plate was washed, adsorbed Dna2 was detected with polyclonal antibodies specific to Dna2. As shown in Figure 4A, interaction between purified Rpa1 and Dna2 was observed, which increased as a function of the level of Dna2 added. This finding confirmed the genetic observations of a direct interaction between Rpa1 and Dna2. When Rpa2 or Rpa3 was used in place of Rpa1, their interactions with Dna2 were much weaker (∼20%) than that observed with Rpa1.
Figure 4.
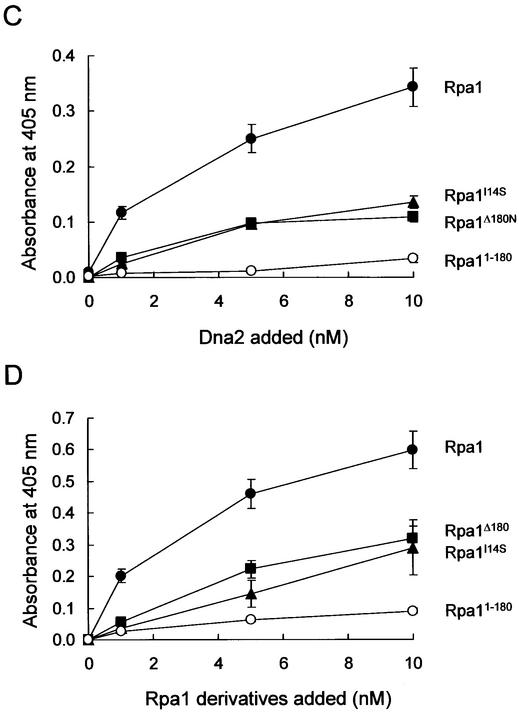
Dna2 interacts with RPA through Rpa1. (A) RPA subunit-coated wells were incubated with increasing concentrations (0, 1, 5 and 10 nM) of Dna2. The RPA subunit used is indicated to the right of the figure. Wells were aspirated, washed three times, and bound Dna2 was detected by ELISA using rabbit polyclonal antibodies against Dna2. (B) Dna2-coated wells were incubated with increasing concentrations (0, 1, 5 and 10 nM) of each RPA subunit. The RPA subunit used is indicated to the right of the figure. The bound RPA subunit protein was detected by ELISA using rabbit polyclonal antibodies raised against each RPA subunit. (C) Interaction of Dna2 with immobilized Rpa1 derivatives was measured with ELISA as described in (A). The Rpa1 derivatives are as indicated. (D) Interaction of Rpa1 derivatives with immobilized Dna2 was measured with ELISA as described in (B). Absorbance readings at each point were corrected by subtracting background absorbance generated with BSA-coated wells.
In order to further confirm this conclusion, we carried out reciprocal ELISA. We first adsorbed Dna2 to the plate and measured its ability to interact with each RPA subunit (Fig. 4B). To minimize variations in the reciprocal ELISA that may arise from differences in antibody efficiency, the antisera were diluted so that different RPA subunits, at the same molar concentration, yielded an identical adsorption value at A405. Consistent with the results presented in Figure 4A, Rpa1 interacted more strongly with Dna2 than Rpa2 and Rpa3 (Fig. 4B). One noticeable difference, however, is that Dna2 interacted more efficiently (2-fold) with Rpa2 than Rpa3 in the reciprocal ELISA. These results, together with those presented above, suggest that Rpa2 and 3 may also contribute to the overall interaction of RPA with Dna2. The significance of the interactions observed between Dna2 and Rpa2 or Rpa3 remains to be defined by other means.
Two domains within Rpa1 interact with Dna2
Since Dna2 interacted with RPA largely through Rpa1, we analyzed mutant forms of Rpa1 (described above, Fig. 3) to map the region required for binding Dna2. As shown in Figure 4C, Rpa11–180 barely interacted with Dna2, whereas Rpa1Δ180N bound Dna2, ∼3-fold less efficiently than wild-type Rpa1. Rpa1I14S, which was synthetically lethal with dna2-157 (Fig. 1A), interacted with Dna2 as efficiently as Rpa1Δ180N. This suggests that the C-terminal three-fourths of Rpa1 is an important region for the interaction with Dna2. Deletion of the N-terminal 180 amino acid domain of Rpa1 or a mutation within this region reduced significantly (∼70%) its ability to interact with Dna2 in vitro. The N-terminal region of Rpa1, however, does appear to contribute to the interaction when it is an integral part of Rpa1 because the interaction between Dna2 and Rpa1 was more robust than the sum of the interactions of the Rpa1Δ180N and Rpa11–180 with Dna2. This indicates that the N- and C-terminal domains of RPA1 interact with Dna2 in a synergistic fashion. As shown in Figure 4D, identical results were obtained in a reciprocal ELISA (binding of the Rpa1 derivatives to wells coated with Dna2).
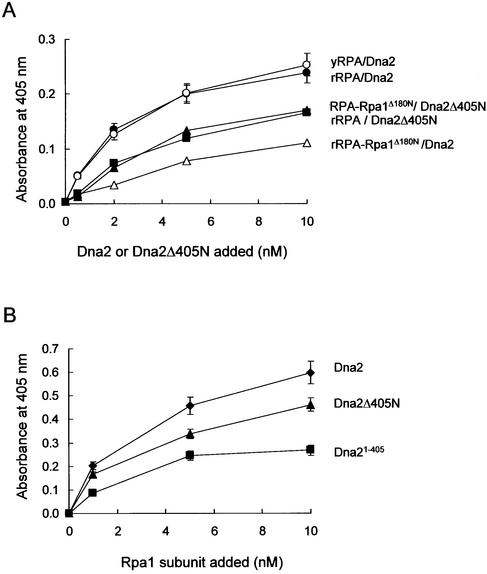
The N-terminal 45-kDa region of Dna2 is involved in the interaction with Rpa1
We tested whether modifications such as phosphorylation affected the binding of Dna2 to RPA. For this purpose, we isolated native RPA (yRPA) from yeast extracts or recombinant RPA (rRPA) from E.coli and examined their ability to bind full-length Dna2 (Fig. 5A). The interactions were nearly identical suggesting that modifications in yeast RPA do not appear to affect its binding to Dna2 (Fig. 5A). RPA over-expression suppresses the temperature-sensitive growth defect caused by deletion of the N-terminal 45-kDa region of Dna2 (43). One interpretation of this result is that deletion of this N-terminal region of Dna2 may reduce affinity of Dna2 for RPA, thereby requiring elevated levels of RPA to restore wild-type growth. In order to test this possibility, we examined the abilities of Dna2Δ405N and wild-type Dna2 to bind RPA and its mutant derivatives.
Figure 5.
The N-terminal region of Rpa1 also contributes to the interaction with Dna2. (A) RPA complex-coated wells were incubated with increasing concentrations (0, 1, 2, 5 and 10 nM) of either Dna2 or Dna2ΔN405. The combination of RPA and Dna2 proteins used is indicated to the right of the figure. Bound Dna2 protein was detected with ELISA by using rabbit polyclonal antibodies against the Dna2 protein. yRPA is RPA complex isolated from yeast, and the recombinant RPA (rRPA) complex was expressed and isolated from E.coli. (B) ELISAs with Rpa1 and Dna2 derivatives were carried out as described in Figure 4D. Absorbance readings at each point were corrected by subtracting background absorbance obtained with BSA-coated wells.
The binding of Dna2 to rRPA–Rpa1Δ180N (in which the Rpa1 subunit lacks the N-terminal 180 amino acids) was significantly (∼40%) lower than the wild-type rRPA (Fig. 5A). This is consistent with the results obtained with Rpa1Δ180N alone as shown in Figure 4C and D, and supports the notion that efficient binding of Dna2 to RPA requires the N-terminal 180 amino acid domain of Rpa1. In parallel, we also examined the interaction between Dna2Δ405N and wild-type RPA and found that it was also significantly impaired (∼40% compared to full-length Dna2) (Fig. 5A). Interestingly, Dna2Δ405N bound rRPA–Rpa1Δ180N and wild-type RPA with identical efficiency (Fig. 5A). This result demonstrates that the N-terminal 180 amino acid domain of Rpa1 cannot participate in the protein–protein interaction with Dna2 when the N-terminal 405 amino acid domain of Dna2 is absent. These results, together with those described in Figure 4, indicate that both N-terminal domains of Dna2 and Rpa1 are required for the maximal interaction between Dna2 and Rpa1. Consistent with this, Dna2Δ405N binds Rpa1 at ∼70–80% of the level observed with wild-type Dna2 (Fig. 5B). Moreover, Dna21–405, the N-terminal 45-kDa fragment of Dna2, was capable of interacting with Rpa1, but with reduced efficiency (∼40% compared to that observed between Dna2 and Rpa1) (Fig. 5B). In support of these in vitro findings, yeast two-hybrid assays revealed similar interactions between Dna2 and Rpa1 in vivo (Fig. 6). The interaction between Rpa1Δ180N and Dna2 was stronger than that observed for Rpa11–180 and Dna2. These two interactions, however, were weaker than that between wild-type Rpa1 and Dna2.
Figure 6.
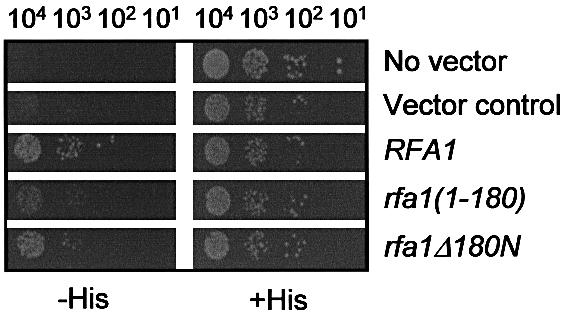
In vivo interaction between DNA2 and RFA1. Yeast strain pJ69-4A expressing wild-type DNA2 as bait was transformed with either prey vector alone (Vector control) or plasmids expressing either RFA1, rfa1(1–180) or rfa1Δ180N as indicated to the right of the figure. ‘No vector’ control denotes pJ69-4A expressing wild-type DNA2 as bait. The resulting transformants were examined for their ability to express the HIS3 reporter gene by spotting cells (104, 103, 102 and 10) on synthetic minimal media in the presence (+His) or absence (–His) of histidine.
The N-terminal region of Rpa1 is important for stimulating Dna2 endonuclease activity
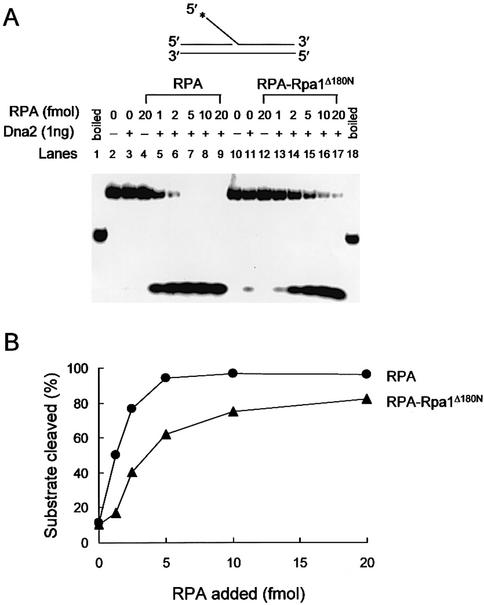
In order to test whether the physical and genetic interactions observed between RPA and Dna2 have physiological relevance to the enzymatic properties of Dna2, we examined the influence of RPA or RPA–Rpa1Δ180N on the endonuclease activities of Dna2 and Dna2Δ405N. In an earlier study, we showed that RPA recruits Dna2 to a flap containing primer RNA (generated by displacement DNA synthesis by pol δ), and stimulates the cleavage of the flap by Dna2 endonuclease (43). We first examined whether RPA–Rpa1Δ180N was able to stimulate the endonuclease activity of Dna2. As previously reported (43), the amount of flap-structured substrate cleaved by wild-type Dna2 increased markedly with increasing levels of wild-type RPA (Fig. 7). On the other hand, RPA–Rpa1Δ180N was less efficient than wild-type RPA in stimulating Dna2-catalyzed cleavage of flap DNA substrates (20–80% of that observed with wild-type RPA). At the lowest level (1 fmol) of RPA added, stimulation was marginal (1.4-fold) with RPA–Rpa1Δ180N, whereas strong (∼5-fold) stimulation was observed with wild-type RPA. At saturating levels of RPA (5–10 fmol), however, stimulation did not differ significantly (6–7-fold and 9-fold, respectively, for RPA–Rpa1Δ180N and wild-type RPA). This result indicates that the N-terminal 180 amino acids of Rpa1 is important for the interaction that leads to the stimulation of the Dna2 endonuclease activity.
Figure 7.
Effect of the N-terminal 180 amino acid deletion of Rpa1 on endonuclease activity of Dna2. (A) The 5′ end-labeled substrate is shown at the top of the figure. The asterisk indicates the position of the 32P label in the substrate. The standard reaction mixture (20 µl) contained 50 mM Tris–HCl/pH 7.8, 2 mM DTT, 5 mM MgCl2, 0.25 mg/ml BSA, 100 mM NaCl and 15 fmol of the flap DNA substrate. The amount of Dna2 was as indicated. Increasing amounts (1, 2, 5, 10 and 20 fmol) of RPA or RPA–Rpa1Δ180N were added. The reactions were incubated at 37°C for 2 min and cleavage products were analyzed using 10% acrylamide low- resolution gel. (B) Quantitation of the results in (A).
In the absence of the N-terminal 45-kDa domain of Dna2, RPA–Rpa1Δ180N is more efficient in stimulating Dna2 endonuclease activity
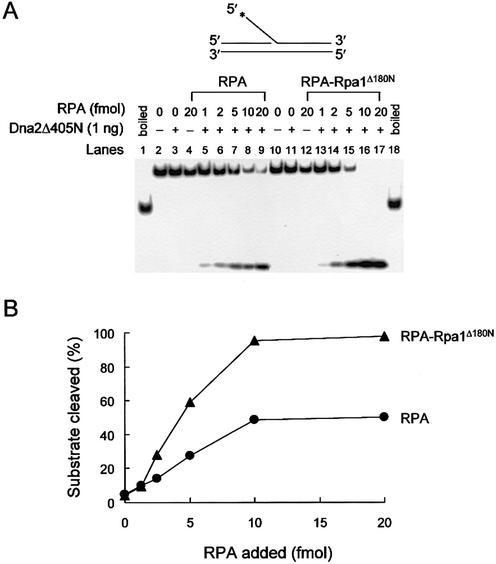
We then examined whether RPA could stimulate the endonuclease activity of Dna2Δ405N, given that this protein is partially impaired in its ability to bind Rpa1 (Fig. 5A and B). The endonuclease activity of Dna2Δ405N was stimulated by RPA, but the stimulation was significantly lower (2-fold) than that observed with wild-type Dna2 (compare Figs 7 and 8, lanes 2–9). This suggests that deletion of the N-terminal domain of Dna2 impairs its functional interaction with RPA. In our previous report, the Dna2Δ405N protein was fully active in vitro, but cells carrying a single copy of this mutation showed temperature-dependent growth (51). It is worthwhile to mention that the specific activities of endonuclease and ATPase of Dna2Δ405N were significantly higher (∼3–5-fold) than the full-length Dna2 activities (51). This, however, was observed only with a ΦX174-based DNA substrate containing bulk ssDNA. When we used the oligonucleotide-based substrate here, the specific activities of the two enzymes were quantitatively similar (data not shown). The growth defect caused by the N-terminal deletion of 405 amino acids of Dna2 was rescued by the increasing copy number of either the dna2Δ405N allele or RFA1 (43,51). Therefore, elevated levels of RPA may further stimulate Dna2Δ405N endonuclease activity, providing an explanation for the suppression of the dna2Δ405N growth defect. Unexpectedly, Dna2Δ405N was stimulated more efficiently by RPA–Rpa1Δ180N than by wild-type RPA, approaching the level of stimulation observed with the wild-type proteins (compare Figs 7 and 8). This observation will be further discussed (see Discussion) in light of a bimodal interaction of Dna2 with Rpa1.
Figure 8.
Effect of RPA or RPA–Rpa1Δ180N on endonuclease activity of Dna2Δ405N. (A) The 5′ end-labeled substrate and the reaction condition was the same as described in Figure 7. The amount of Dna2Δ405N was as indicated. Increasing amounts (1, 2, 5, 10 and 20 fmol) of RPA or RPA–Rpa1Δ180N were added. (B) Quantitation of the results in (A).
DISCUSSION
We have previously shown that RPA plays a role in targeting Dna2 to flap-structured ssDNA for Okazaki fragment processing, which results in the stimulation of the endonuclease activity of Dna2 (43). Biochemical analyses indicate that both targeting and stimulation of Dna2 in this process involves the interaction between the two proteins for the following reasons: (i) a ternary complex composed of RPA, Dna2 and the DNA substrate appears to be an intermediate for the cleavage reaction; (ii) RPA stimulates the formation of a complex of Dna2 with the DNA substrate in a synergistic fashion; (iii) RPA accelerates the rate of Dna2-catalyzed cleavage of ssDNA in a manner that requires RPA binding to substrate DNA. In order to understand these functional interactions at the protein level, we searched for biochemical and genetic interactions between Dna2 and the three subunits of RPA. We have discovered that Dna2 primarily interacts with Rpa1, the large subunit of RPA. In support of our conclusion that Rpa1 and Dna2 show direct protein–protein interaction, we identified allele-specific interactions between RPA1 and DNA2 (Fig. 1).
Based upon the following genetic and biochemical observations, we suggest that Dna2 and Rpa1 interact with each other in a bimodal fashion. (i) Mutations in RFA1 that are synthetically lethal with dna2-157 largely map to the C-terminal ssDNA-binding domains of Rpa1. (ii) The affinity of full-length Dna2 for the C-terminal two-thirds of Rpa1 is significantly less than that of the full-length proteins (Fig. 4C and D). (iii) Although the binding of the isolated N-terminal domain of Rpa1 to full-length Dna2 was barely detectable (Fig. 4C and D), optimal interaction required both N-terminal and C-terminal regions of Dna2 and Rpa1 (Figs 4 and 5). (iv) Deletion of the N-terminal region of either Rpa1 or Dna2 reduced the extent of stimulation of Dna2 endonuclease activity (Fig. 7), even though the stimulation of Dna2 endonuclease activity was observed regardless of the presence of N-terminal regions of both Rpa1 and Dna2. This suggests that critical interactions occur between the C-termini of Rpa1 and Dna2. (vi) In the absence of both N-termini of Rpa1 and Dna2, the extent of endonuclease stimulation was similar to that observed with both wild-type proteins in vitro (Fig. 8). Although counterintuitive, such a result might be expected if the two N-terminal domains regulate a transiently formed complex in vivo. For example, the complete abrogation of the N-terminal interaction might destabilize the complex formed and allow more rapid turnover of Dna2, which in turn results in an elevated rate of cleavage of the substrate in vitro. We were not able to test whether rfa1Δ180N could rescue the growth defect of dna2Δ405N in vivo since rfa1Δ180N cells are lethal.
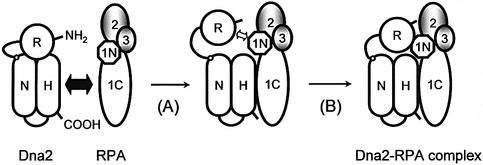
The bimodal interaction between Dna2 and Rpa1 (Fig. 9) is likely to play an important role in the recruitment of Dna2 to RPA-bound flaps and the subsequent stimulation of the endonuclease activity of Dna2. We now know that deletion of the N-terminal region of either Dna2 or RPA affects cell growth and impairs the physical interactions between these proteins. These two effects could be related if the lethality caused by the N-terminal 180 amino acid deletion of Rpa1, which supports SV40 DNA replication in vitro, is due to a failure of the mutant RPA to interact optimally with Dna2 in vivo. In our bimodal interaction model, the primary mechanism of recruiting Dna2 to ssDNA-bound RPA is the interaction between C-termini of these proteins. This interaction is followed by a secondary interaction between the two N-terminal regions, further stabilizing their overall interaction. The secondary interaction between the two N-termini of Dna2 and Rpa1, however, appears to be auxiliary in vivo and in vitro, since cells carrying the dna2Δ405N allele are viable, at least at 30°C. The secondary interaction may be required to control the optimal rate of Dna2-catalyzed cleavage of the RPA-containing flap, by affecting a conformational change of Dna2 or RPA before and after the cleavage reaction. Alternatively, it may be required for the timely dissociation of either Dna2 or RPA or both proteins from substrates after cleavage occurs. At present, we cannot rule out the possibility that the N-termini of Rpa1 and Dna2 may be important for a functional protein conformation required for each protein action rather than the direct protein–protein interaction. This model does not rule out the possibility that the essential role of the N-terminus of RPA is to mediate Dna2 interactions via a third protein that was absent from our in vitro assays. Although two major interactions were detected between domains of Dna2 and Rpa1 in vitro, we cannot formally rule out the possibility that there is an interaction between the N-terminal domain of Rpa1 and the C-terminal domain of Dna2 since some mutations in the N-terminal domain of Rpa1 can suppress the growth defect of dna2-157 (Fig. 1B). If this is the case, the N-terminal domain of Rpa1 could interact with one other domain in addition to the N-terminal domain of Dna2.
Figure 9.
The bimodal interaction of Dna2 and Rpa1. The primary interaction (as indicated by a thick closed arrow with double arrowheads) between the two C-terminal domains of Dna2 and Rpa1 helps form an initial complex, leading to conformational changes in the two proteins (step A). This in turn allows the secondary interaction (indicated by an open arrow with double arrowheads) to occur between the N-terminal domains of the two proteins (step B), forming a functional Dna2–RPA complex. This bimodal interaction is likely to play a role for optimal stimulation of Dna2-catalyzed cleavage of DNA substrates and the subsequent disassembly of the protein–DNA complex formed with the flap DNA. The domain structure of Dna2 is described elsewhere (51). Each subunit of RPA is indicated as 1 (Rpa1), 2 (Rpa2) or 3 (Rpa3). The N-terminal and C-terminal domains of Rpa1 are depicted as 1N and 1C, respectively. See text for further details.
How the interaction between RPA and Dna2 stimulates the function of Dna2 is presently unclear. The interaction, at least between the two N-terminal domains, is analogous to the inter-subunit interaction between Rpa1 and Rpa2. The N-terminal region of Rpa1 is positively charged, appears to resemble a ssDNA-binding domain (32,33), and may contribute to the stability of the ssDNA–RPA complex. Recently, it has been suggested that this region of Rpa1 is able to interact with the phosphorylated form of the N-terminal region of Rpa2 (M. Wold, personal communication). It is known that the N-terminal 180 amino acid region is relatively flexible (33). The flexibility of this domain may confer on it the ability to interact with several replication proteins such as DNA polymerase α and Dna2. It is noteworthy that the N-terminal 45-kDa region of Dna2 is also unstructured and highly sensitive to protease digestion (51). Therefore, the flexibility and relatively unstructured nature of both N-termini might allow the two proteins to interact more readily and regulate the enzymatic activities of Dna2. This property may account for why the N-terminal 180 amino acids of Rpa1 is essential in vivo but is not required to support SV40 DNA replication in vitro. The essential role of this domain in vivo may be to regulate the critical functions of Dna2.
Interactions between Dna2 and Rpa2 or Rpa3 were also observed although weaker (20–30%) than those detected with Rpa1, raising the possibility that Dna2 also interacts with RPA through these two subunits in vivo. The significance of this interaction, however, is not clear at present since we used purified subunits of Rpa2 and Rpa3 rather than the three-subunit complex. This may reflect a simple electrostatic interaction between Dna2 and Rpa2 or Rpa3 in vitro. The role of these two subunits in governing the interaction of RPA and Dna2 and its effect on the processing of Okazaki fragments remains to be explored in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Drs Richard Kolodner (University of California at San Diego) and Tim Formosa (University of Utah, Salt Lake City) for plasmids. We also thank Dr Jerard Hurwitz (Sloan-Kettering Institute, New York) for critical reading of the manuscript. This work was supported by a grant from the Creative Research Initiatives of the Korean Ministry of Science and Technology to Y.-S.S. and by grant GM55583 from the NIH to S.B.
REFERENCES
- 1.Bambara R.A., Murante,R.S. and Henricksen,L.A. (1997) Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- 2.Waga S. and Stillman,B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- 3.Seo Y.S. and Hübscher,U. (2001) Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cell, 12, 149–157. [PubMed] [Google Scholar]
- 4.Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 5.Iftode C., Daniely,Y. and Borowiec,J.A. (1999) Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Cell Biol., 34, 141–180. [DOI] [PubMed] [Google Scholar]
- 6.Wobbe C.R., Weissbach,L., Borowiec,J.A., Dean,F.B., Murakami,Y., Bullock,P. and Hurwitz,J. (1987) Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl Acad. Sci. USA, 84, 1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wold M.S. and Kelly,T. (1988) Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl Acad. Sci. USA, 85, 2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairman M.P. and Stillman,B. (1988) Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J., 7, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyer W.D., Rao,M.R., Erdile,L.F., Kelly,T.J. and Kolodner,R.D. (1990) An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J., 9, 2321–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brill S.J. and Stillman,B. (1991) Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev., 5, 1589–1600. [DOI] [PubMed] [Google Scholar]
- 11.Dornreiter I., Erdile,L.F., Gilbert,I.U., von Winkler,D., Kelly,T.J. and Fanning,E. (1992) Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J., 11, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins K.L. and Kelly,T.J. (1991) Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol. Cell. Biol., 11, 2108–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami Y., Eki,T. and Hurwitz,J. (1992) Studies on the initiation of simian virus 40 replication in vitro: RNA primer synthesis and its elongation. Proc. Natl Acad. Sci. USA, 89, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melendy T. and Stillman,B. (1993) An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem., 268, 3389–3395. [PubMed] [Google Scholar]
- 15.Matsuda T., Saijo,M., Kuraoka,I., Kobayashi,T., Nakatsu,Y., Nagai,A., Enjoji,T., Masutani,C., Sugasawa,K., Hanaoka,F., Yasui,A. and Tanaka,K. (1995) DNA repair protein XPA binds replication protein A (RPA). J. Biol. Chem., 270, 4152–4157. [DOI] [PubMed] [Google Scholar]
- 16.He Z., Brinton,B.T., Greenblatt,J., Hassell,J.A. and Ingles,C.J. (1993) The transactivator proteins VP16 and GAL4 bind replication factor A. Cell, 73, 1223–1232. [DOI] [PubMed] [Google Scholar]
- 17.He Z., Henricksen,L.A., Wold,M.S. and Ingles,C.J. (1995) RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature, 374, 566–569. [DOI] [PubMed] [Google Scholar]
- 18.Li R. and Botchan,M.R. (1993) The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell, 73, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 19.Dutta A., Ruppert,J.M., Aster,J.C. and Winchester,E. (1993) Inhibition of DNA replication factor RPA by p53. Nature, 36, 579–582. [DOI] [PubMed] [Google Scholar]
- 20.Dutta A. and Stillman,B. (1992) cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA and activate DNA replication. EMBO J., 11, 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fotedar R. and Roberts,J.M. (1992) Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J., 11, 2177–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang F. and Newport,J.W. (1993) Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J. Cell Sci., 106, 983–994. [DOI] [PubMed] [Google Scholar]
- 23.Brill S.J. and Stillman,B. (1989) Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature, 342, 92–95. [DOI] [PubMed] [Google Scholar]
- 24.Kenny M.K., Schlegel,U., Furneaux,H. and Hurwitz,J. (1990) The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem., 265, 7693–7700. [PubMed] [Google Scholar]
- 25.Erdile L.F., Heyer,W.D., Kolodner,R. and Kelly,T.J. (1991) Characterization of a cDNA encoding the 70-kDa single-stranded DNA-binding subunit of human replication protein A and the role of the protein in DNA replication. J. Biol. Chem., 266, 12090–12098. [PubMed] [Google Scholar]
- 26.Li L., Lu,X., Peterson,C.A. and Legerski,R.J. (1995) An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol., 15, 5396–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umezu K., Sugawara,N., Chen,C., Haber,J.E. and Kolodner,R.D. (1998) Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics, 148, 989–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longhese M.P., Plevani,P. and Lucchini,G. (1994) Replication factor A is required in vivo for DNA replication, repair and recombination. Mol. Cell. Biol., 14, 7884–7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firmenich A.A., Elias-Arnanz,M. and Berg,P. (1995) A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol. Cell. Biol., 15, 1620–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith J. and Rothstein,R. (1995) A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol. Cell. Biol., 15, 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brill S.J. and Bastin-Shanower,S. (1998) Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol. Cell. Biol., 18, 7225–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochkareva E., Belegu,V., Korolev,S. and Bochkarev,A. (2001) Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J., 20, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs D.M., Lipton,A.S., Isern,N.G., Daughdrill,G.W., Lowry,D.F., Gomes,X. and Wold,M.S. (1999) Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR, 14, 321–331. [DOI] [PubMed] [Google Scholar]
- 34.Bochkareva E., Frappier,L., Edwards,A.M. and Bochkarev,A. (1998) The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem., 273, 3932–3936. [DOI] [PubMed] [Google Scholar]
- 35.Gomes X.V., Henricksen,L.A. and Wold,M.S. (1996) Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry, 35, 5586–5595. [DOI] [PubMed] [Google Scholar]
- 36.Braun K.A., Lao,Y., He,Z., Ingles,C.J. and Wold,M.S. (1997) Role of protein–protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry, 36, 8443–8454. [DOI] [PubMed] [Google Scholar]
- 37.Kim H.S. and Brill,S.J. (2001) Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 3725–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomes X.V. and Wold,M.S. (1996) Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry, 35, 10558–10568. [DOI] [PubMed] [Google Scholar]
- 39.Kim D.K., Stigger,E. and Lee,S.H. (1996) Role of the 70-kDa subunit of human replication protein A (I). Single-stranded dna binding activity, but not polymerase stimulatory activity, is required for DNA replication. J. Biol. Chem., 271, 15124–15129. [DOI] [PubMed] [Google Scholar]
- 40.Philipova D., Mullen,J.R., Maniar,H.S., Lu,J., Gu,C. and Brill,S.J. (1996) A hierarchy of SSB protomers in replication protein A. Genes Dev., 10, 2222–2233. [DOI] [PubMed] [Google Scholar]
- 41.Kang H.Y., Choi,E., Bae,S.H., Lee,K.H., Gim,B.S., Kim,H.D., Park,C., MacNeill,S.A. and Seo,Y.S. (2000) Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics, 155, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae S.H. and Seo,Y.S. (2000) Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. [DOI] [PubMed] [Google Scholar]
- 43.Bae S.H., Bae,K.H., Kim,J.A. and Seo,Y.S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412, 456–461. [DOI] [PubMed] [Google Scholar]
- 44.Budd M.E. and Campbell,J.L. (1995) A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl Acad. Sci. USA, 92, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bae S.H., Choi,E., Park,J.S., Lee,S.H., Lee,K.H. and Seo,Y.S. (1998) Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem., 273, 26880–26890. [DOI] [PubMed] [Google Scholar]
- 46.Lee K.H., Kim,D.W., Bae,S.H., Kim,J.A., Ryu,G.H., Kwon,Y.N., Kim,K.A., Koo,H.S. and Seo,Y.S. (2000) The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res., 28, 2873–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higashibata H., Kikuchi,H., Kawarabayasi,Y. and Matsui,I. (2003) Helicase and endonuclease activities of hyperthermophile Pyrococcus horikoshii Dna2 inhibited by substrates with RNA segments at 5′-end. J. Biol. Chem., 278, 15983–15990. [DOI] [PubMed] [Google Scholar]
- 48.MacNeill S.A. (2001) DNA replication: partners in the Okazaki two-step. Curr. Biol., 11, R842–R844. [DOI] [PubMed] [Google Scholar]
- 49.Ayyagari R., Gomes,X.V., Gordenin,D.A. and Burgers,P.M.J. (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between Fen1 and Dna2. J. Biol. Chem., 278, 1618–1625. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y.H., Ayyagari,R., Resnick,M.A., Gordenin,D.A. and Burgers,P.M.J. (2003) Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease of pol δ in the creation of a ligatable nick. J. Biol. Chem., 278, 1626–1633. [DOI] [PubMed] [Google Scholar]
- 51.Bae S.H., Kim,J.A., Choi,E., Lee,K.H., Kang,H.Y., Kim,H.D., Kim,J.H., Bae,K.H., Cho,Y., Park,C. and Seo,Y.S. (2001) Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res., 29, 3069–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Formosa T. and Nittis,T. (1999) Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol alpha accessory factor and show that full Dna2 helicase activity is not essential for growth. Genetics, 151, 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]