Abstract
A nonviral vector for highly efficient site-specific integration would be desirable for many applications in transgenesis, including gene therapy. In this study we directly compared the genomic integration efficiencies of piggyBac, hyperactive Sleeping Beauty (SB11), Tol2, and Mos1 in four mammalian cell lines. piggyBac demonstrated significantly higher transposition activity in all cell lines whereas Mos1 had no activity. Furthermore, piggyBac transposase coupled to the GAL4 DNA-binding domain retains transposition activity whereas similarly manipulated gene products of Tol2 and SB11 were inactive. The high transposition activity of piggyBac and the flexibility for molecular modification of its transposase suggest the possibility of using it routinely for mammalian transgenesis.
Keywords: gene therapy, site-specific, transposase, cancer, mutagenesis
DNA transposons and retrotransposons constitute a major component of repetitive sequences in eukaryotes (1, 2). Their movement around the host genome has played a crucial role in shaping current genomes. Owing to their natural property of mobilizing around genomes via a “cut-and-paste” mechanism, DNA transposons have been routinely used as tools for genetic manipulation in lower organisms (3–6). Only recently have scientists realized the potential value of transposons in mammalian transgenesis because most naturally occurring transposons are inactive and possibly were overlooked until recently because of the success of integrating viral vectors. Examples of transposon systems used in mammalian gene transfer are (i) hAT-like Tol2, the only naturally active vertebrate transposon, isolated from the genome of the Japanese medaka fish; (ii) two Tc1-like transposons, Sleeping Beauty (SB) and Frog Prince, reconstructed from inactive transposons of fish and frog genomes, respectively; and (iii) the founding member of the piggyBac family, piggyBac, isolated from the cabbage looper moth Trichoplusia ni, recently shown to transpose efficiently in mice (7–11). Since its awakening, SB has gained the leading position in transposon-mediated mammalian transgenesis and mutagenesis. For example, SB was used to act as a somatic insertional mutagen to identify genes involved in solid tumor formation (12–14).
Many applications in mammalian transgenesis would benefit from a vector that integrates with high efficiency and site specificity. Transposon-based vectors appear applicable to human gene therapy because of the risks associated with viral-based vectors. These risks have been highlighted by the death of a young patient from a systemic inflammatory response to the viral vector used and insertional mutagenesis resulting in the development of leukemia in another trial (15, 16).
SB has been explored in preclinical models (17–19). However, two obstacles hinder the immediate advancement of SB to human clinical trials. First, the integration rate of the SB transposon is much lower than viral-based vector transgene transfer. The second obstacle to SB is nonspecific integration of the transgene with potential undesired consequences (20). The incorporation of exogenous DNA-binding domains (DBD) into the transposition machinery can potentially increase transformation efficiency and result in site-specific integration (21). To test feasibility, SB transposase was fused to zinc finger DBD (22). However, these SB fusion transposases exhibit dramatically reduced transposition efficiency, thus tempering the potential for SB targeted integration.
To overcome the limitations of SB, this study sought to identify a transposon system that mediates gene transfer with high efficiency while possessing flexibility for molecular modifications in mammalian cells. We explored four different transposons, SB11 (a hyperactive version of SB), Tol2, piggyBac, and Mos1, directly comparing their transposition activity in four different mammalian cell lines. In every cell line tested piggyBac demonstrated the highest transposition activity, whereas no activity was detected with Mos1. Like Tc1/mariner transposons, the transposition of piggyBac is reduced as the amount of transposase DNA increases beyond a threshold. In addition, piggyBac transposase with a GAL4 DBD fused to its N terminus retains significant transposition activity, whereas such modification essentially abolishes Tol2 and SB11 transposition. Given its flexibility for molecular engineering and its relatively high transposition activity as demonstrated in this study, piggyBac is a promising nonviral vector for mammalian transgenesis, with possibilities for targeted integration.
Results
piggyBac, Tol2, and SB11 Are Active in Mammalian Cells.
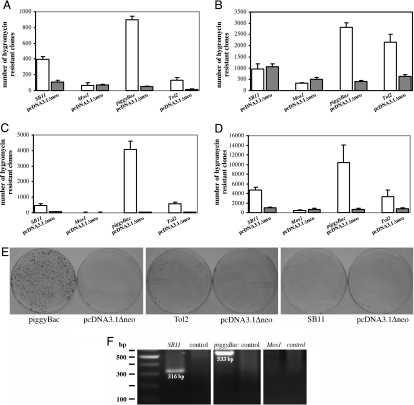
Because each transposon system has been independently developed and tested in different laboratories, it is difficult to draw conclusions regarding their relative efficiency only on the basis of published literature. Thus, a direct comparison of transposition activity was needed to identify the most promising transposon(s). To address this issue, we constructed four transposons using the two-component system: a helper plasmid containing the transposase driven by the CMV promoter and a donor plasmid with the terminal repeats (TR) bearing a cassette with hygromycin resistance and kanamycin resistance genes to facilitate selection in eukaryotes and prokaryotes, respectively, and a ColE1 replication origin for plasmid propagation in bacteria (Fig. 1). The four transposon systems constructed were Mos1, SB11, Tol2, and piggyBac. Additionally, the efficiency of chromosomal integration might vary among the transposons depending on chromatin organization and/or host factors. Therefore, transposition activity of the four transposons was determined in four mammalian cell lines, HeLa (human cervical carcinoma), HEK293 (human embryonic kidney cell), H1299 (human lung carcinoma), and CHO (Chinese hamster ovarian carcinoma). For each transposon system, 200 ng of donor plasmids with 200 ng of helper plasmids (or control) was transfected. Resistant clones were counted after 14 days of hygromycin selection. Similar to a previous report, we observed that colonies <0.5 mm in diameter often failed to be subcloned in the presence of hygromycin; therefore, these colonies may simply be “feeder colonies” (23). In this study only colonies >0.5 mm in diameter were counted. As shown in Fig. 2A–E, piggyBac and Tol2 demonstrated activity in all cell lines tested. SB11 displayed slight transposition activity in CHO, HeLa, and HEK293 cells, whereas it was inactive in H1299 cells. No transposition activity was detected with Mos1 in the four cell lines used (Fig. 2 A–E).
Fig. 1.
Schematic representations of the two-component transposon systems. (A) Helper plasmids. The expression of four transposases was driven by the CMV promoter. The plasmid backbone for all transposases is pcDNAΔneo except SB11, which was cloned in pCMV-SB11. All transposases are wild type except SB11, which is a hyperactive version. (B) Donor plasmids. A cassette with hygromycin and kanamycin resistance genes and a bacterial ColE1 replication origin was subcloned into the donor plasmid of the four transposons.
Fig. 2.
Transposition activity of SB11, Mos1, piggyBac, and Tol2 transposons in different mammalian cells. A total of 1 × 105 cells per individual well in 24-well plates were transfected with 200 ng of donor plus 200 ng of helper plasmid. The pcDNA3.1Δneo vector served as a control for the absence of transposase. Transposition activity was measured by counting hygromycin-resistant colonies after a 2-week selection period. Data are shown as mean values with SD (n = 3). (A) HeLa cells. (B) H1299 cells. (C) HEK293 cells. (D) CHO cells. (E) An example of HEK293 cells transfected with piggyBac, Tol2, and SB11 transposon systems and their controls. Colonies were stained with methylene blue after 2 weeks of hygromycin selection. (F) Excision assays in HEK293. Shown is PCR analysis of excision assays performed in HEK293 transfected with donor plus helper or the control (pcDNA3.1Δneo).
The type of DNA transposition described herein involves a two-step action: (i) excision of the transposable element from the donor plasmid and (ii) integration of the excised fragment into its DNA target (i.e., the host chromosome in our system). Therefore, the numbers of hygromycin-resistant colonies are the result of both excision and integration events. Although no activity was detected in cells transfected with Mos1, it was still possible that successful excision occurred but that integration did not. To exclude this possibility, we performed a plasmid-based excision assay using PCR. As a consequence of excision, a short version of the donor plasmid should be produced. No excision-dependent PCR product was detected in cells transfected with donor and helper plasmids for Mos1, whereas excision-dependent PCR products with sizes of 533 bp for SB11 and 316 bp for piggyBac were detected (Fig. 2F).
piggyBac Is the Most Efficient Transposon Tested.
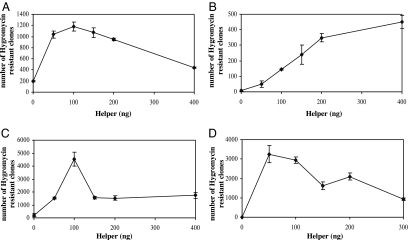
piggyBac displayed the highest transposition activity as compared with Tol2 and SB11 in all four mammalian cell lines when equal amounts of donor and helper plasmids (200 ng of DNA for each) were introduced into the cells (Fig. 2 A–E). To further confirm that piggyBac is the most efficient transposase, we performed a chromosomal integration assay by transfecting HEK293 with a fixed amount of donor (200 ng) plus varying amounts of helper for piggyBac, Tol2, and SB11. As shown in Fig. 3A–C, the lowest number of hygromycin-resistant colonies for piggyBac was ≈1,500, which was still significantly higher than the highest number of resistant colonies observed for both Tol2 (490) and SB11 (1,180). Furthermore, piggyBac achieves its highest transposition activity (4,535) when 200 ng of donor and 100 ng of helper plasmids were introduced into cells. Therefore, piggyBac consistently demonstrated the highest transposition activity in the four mammalian cell lines used in this study.
Fig. 3.
The transposition activity of SB11 (A), Tol2 (B), and piggyBac (C and D) at various ratios of donor and helper. Transposition activity was measured under a fixed amount of donor plasmid (200 ng in A–C and 50 ng in D) with increasing amounts of helper plasmid cotransfected into HEK293 cells. pcDNA3.1Δneo was used to adjust the total amount of DNA transfected in each sample. Data are shown as mean values with SD (n = 3).
As observed in Fig. 2 A–D, the transposition activity of piggyBac, Tol2, and SB11 varies in different cells. For example, ≈1,000 hygromycin-resistant colonies were detected with both the controls and SB11 in H1299, suggesting a lack of transposition activity of SB11 in this cell line, whereas in HEK293 cells only ≈500 hygromycin-resistant colonies were detected in the presence of SB11 transposase, which represents an 8-fold increase as compared with the control. We therefore used two parameters, relative fold and percentage of transposition, to assess the transposition activity of the different transposons. The relative fold is obtained by dividing the number of resistant colonies detected in cells transfected by donor plus helper with the colony number that resulted from random integration (i.e., control without transposase added). The percentage of transposition, hereafter designated as transposition rate, is calculated by subtracting the number of hygromycin-resistant colonies detected in the controls from the number of resistant colonies in the presence of transposase, dividing by 1 × 105 (the number of cells originally seeded before transfection), and finally multiplying by 100. The transposition rate represented here, however, was not normalized by the transfection efficiency in various cell lines. As summarized in Table 1, the relative fold ranges for the three transposons in different cell lines were as follows: (i) SB11 from 1 (no difference) in H1299 to 8.1 in HEK293, (ii) piggyBac from 5.7 in H1299 to 114 in HEK293, and (iii) Tol2 from 3.3 in CHO to 93.9 in HEK293. The transposition rate ranges were as follows: (i) SB11 from 0% in H1299 to 2.9% in CHO, (ii) piggyBac from 0.7% in HeLa to 7.0% in CHO, and (iii) Tol2 from 0.08% in HeLa to 1.8% in CHO. Once again piggyBac displayed the highest transposition activity among the three active transposon systems tested as judged by both the transposition rate and relative fold. The transposition rate of Tol2 is higher than SB11 in H1299 and HEK293 but not in CHO and HeLa cells. Owing to the relatively high integration rate of the SB11 control, the relative fold seen in all four cell lines for Tol2 was higher than that of SB11.
Table 1.
Summary of the transposition efficiency of SB11, piggyBac, and Tol2 transposons
| Cells |
SB11 |
piggyBac |
Tol2 |
|||
|---|---|---|---|---|---|---|
| Relative fold | Percentage of transposition | Relative fold | Percentage of transposition | Relative fold | Percentage of transposition | |
| CHO | 2.9 ± 1.6 | 2.3 ± 1.5 | 9.8 ± 6.4 | 7.0 ± 3.9 | 3.3 ± 1.7 | 1.8 ± 1.2 |
| HeLa | 2.7 ± 1.0 | 0.2 ± 0.1 | 14.5 ± 3.5 | 0.7 ± 0.2 | 5.4 ± 3.2 | 0.08 ± 0.03 |
| H1299 | 1.0 ± 0.16 | 0 | 5.7 ± 1.3 | 2.2 ± 0.2 | 4.1 ± 0.8 | 1.4 ± 0.3 |
| HEK293 | 8.1 ± 2.1 | 0.4 ± 0.1 | 114.6 ± 81.6 | 2.8 ± 1.4 | 93.9 ± 75.8 | 0.9 ± 0.3 |
Relative fold values indicate the relative fold of hygromycin-resistant clones as compared with controls (n = 6). Percentage of transposition values indicate the percentage of true transposition from 1 × 105 cells seeded.
piggyBac Transposition Declines as Helper Levels Increase.
Transposition efficiency depends on the availability of transposon (donor) and transposase (helper) in cells. It was shown elsewhere that, over a certain threshold, SB11 transposition declines with increasing transposase, a phenomenon known as overproduction inhibition (24). Conversely, Tol2 transposition was directly proportional to the levels of transposase and did not appear to exhibit overproduction inhibition (25). We also observed overproduction inhibition for SB11, whereas Tol2 transposition was directly proportional to the amount of transposase DNA (Fig. 3 A and B) (25, 26). Like SB11, piggyBac also showed peak activity at a ratio of 2:1 (donor:helper). However, unlike SB11, which demonstrated a gradual reduction of activity above this ratio, the activity of piggyBac declined rapidly (Fig. 3C). These findings suggest that piggyBac exhibits overproduction inhibition.
To further address the issue we performed a chromosomal integration assay for piggyBac using 50 ng of donor with increasing amounts of helper ranging from 50 to 300 ng. As seen in Fig. 3D, increasing amounts of helper beyond the ideal ratio (2:1) resulted in a gradual reduction in transposition. These data further support the hypothesis that overproduction inhibition occurs with piggyBac.
Activity of a GAL4-piggyBac Fusion Is Similar to That of Wild Type.
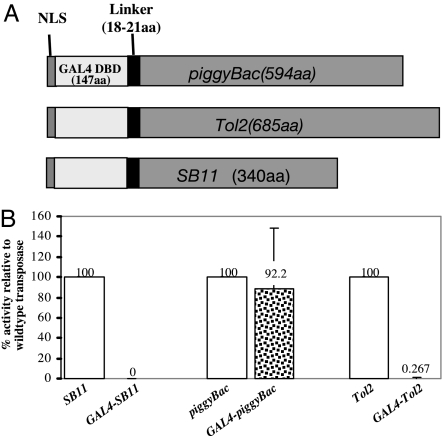
Directing transgene integration to a unique and safe site on the host chromosome would overcome the inherent hazards of insertional mutagenesis that can result with currently used integrating vectors. To achieve target specificity, engineering a chimeric transposase fused with a DBD to specifically target a unique sequence was proposed (21) and later demonstrated in mosquito embryos by targeting a unique site in a plasmid (27). A transposon-based gene delivery system ideally requires a custom-engineered transposase with high integration activity and target specificity. SB transposase modifications to engineer target specificity have resulted in dramatic reductions in transposition activity (22). Therefore, we assessed the potential for modifications of SB11, Tol2, and piggyBac transposases by fusing GAL4 DBD to their N terminus (Fig. 4A). The transposition activities of these chimeras were determined by using a chromosome integration assay in HEK293 cells. GAL4-piggyBac transposase demonstrated transposition activity similar to that of wild type, whereas GAL4-Tol2 and GAL4-SB11 transposases possessed negligible activity, even though GAL4-SB11 protein was detected by Western blot with use of a monoclonal antibody (Fig. 4B and data not shown). We were unable to detect GAL4-piggyBac or GAL4-Tol2 fusions with either monoclonal or polyclonal anti-GAL4 antibodies, perhaps because of masking/alteration of the GAL4 epitopes when fused to the transposases. Alternatively, the amount of fusion transposases may not have been sufficient for detection because of a reduction in half-life as compared with GAL4-SB.
Fig. 4.
Transposition activity of GAL4 transposases. (A) A schematic representation of engineered chimeric transposases. A GAL4 DBD was fused in-frame to the N terminus of transposases, with a linker of 18–21 aa placed between GAL4 DBD and the transposases to facilitate proper domain folding. (B) The percentage of transposition activity relative to wild-type transposase in HEK293 cells. To compare the activity of each chimeric transposase to its wild-type counterpart, the wild-type transposition efficiency was normalized to 100%. Data are shown as mean values with SD (n = 6).
piggyBac inserts into the tetranucleotide site TTAA, which is duplicated upon insertion (11, 28). To test whether fusing GAL4 to the N terminus of piggyBac transposase alters its preference for TTAA sites, we performed plasmid rescue experiments to retrieve the sequence information of the target sites using genomic DNAs isolated from individual hygromycin-resistant clones in CHO cells. Six independent genomic sequences were recovered from four drug-resistant clones. As shown in Table 2, all of these sequences contained genomic DNA with the signature TTAA sequence at the integration site. This experiment demonstrates that the chromosomal integrations observed in cells transfected with GAL4-piggyBac is mediated by a true transposition event with the same insertion preference for TTAA sites. Thus, piggyBac is potentially the ideal DNA transposon for development of a highly efficient, site-specific integrating nonviral vector because it exhibits high integration efficiencies in many cell types, and modifications to alter site specificity are unlikely to significantly reduce activity.
Table 2.
Analysis of transposon–chromosomal junction mediated by GAL4-piggyBac in CHO cells
| Independent isolated clone | Donor plasmid TR | Chromosomal insertion site | Flanking chromosomal sequence |
|---|---|---|---|
| G8-2 | 5′-TGATTATCTTTCTAGGG | TTAA | GCTCGGGCCGGCCGCGTCGCCGCTTC-3′ |
| G25-2 | TGATTATCTTTCTAGGG | TTAA | CAATCAATAAGATAAACATACACAGA |
| G25-3 | TGATTATCTTTCTAGGG | TTAA | CACCACATTTAACTTGCTCTTTGATA |
| G28-1 | TGATTATCTTTCTAGGG | TTAA | TAGAGTGCTGAGATTTGGGACATTGC |
| G29-1 | TGATTATCTTTCTAGGG | TTAA | GGCGTTGGTGGCACACAACTTTAAGT |
| G34-2 | TGATTATCTTTCTAGGG | TTAA | TAAGACAATGTATGACTTTGTCCCAT |
Six clones were analyzed, and all demonstrate that GAL4-piggyBac mediated integration into a TTAA site. The raw sequence data are provided in Fig. 5.
Discussion
A highly efficient site-specific nonviral vector would reduce the potential hazards associated with insertional mutagenesis and the uncertainty of transgene expression resulting from positional effects. We have previously demonstrated that a GAL4-piggyBac transposase can direct integration to a single target site on a plasmid in mosquito embryos (27). In this study we show that piggyBac is more efficient than Tol2 and SB11 in different mammalian cell lines and is amenable to further molecular modification without reducing its activity in mammalian cells. Thus, piggyBac has the potential for use in mammalian site-directed transgenesis and mutagenesis.
Mos1 has been shown to function in vitro, suggesting that host factors are not required for transposition (29). Surprisingly, our chromosomal assay detected no transposition activity for Mos1 in the four mammalian cell lines tested. It is possible that host factors may interfere or that the complexity of chromatin organization in mammalian cells may be too high to render notable chromosomal integrations mediated by Mos1. Structural analysis of the SB TR reveals four transposase binding sites (30). Thus, the optimal molar ratio of transposase:transposon is expected to be 4:1. However, instead of reaching a plateau, transposition activity decreases with increasing amounts of SB transposase, a phenomenon known as overproduction inhibition (Fig. 3A) (24). On the contrary, and consistent with a previous observation in mouse embryonic stem cells, increasing transposition activity was seen in HEK293 cells as the amount of Tol2 transposase increased (Fig. 3C) (25). The Tol2 transposable element has several inverted repeats, including terminal, subterminal, and internal TR, which are presumably required for binding of Tol2 transposase (31). Hence, the abundance of inverted repeats present in the Tol2 transposon may titrate out the transposase, resulting in the absence of overproduction inhibition at the ratios and doses of donor and helper plasmid tested. Alternatively, it is possible that overproduction inhibition does not occur at all in this case. Supporting this possibility, a sandwich system using the SB transposon demonstrated that increasing the number of SB binding sites in the transposon improved transposition of large-sized transgenes (26). However, whether the improvement of transposition in the SB sandwich system is a consequence of alleviating overproduction inhibition or facilitating transpososome formation remains elusive. Overproduction inhibition of piggyBac was also observed in both sets of experiments with transposon (donor) fixed at 50 ng and 200 ng (Fig. 3 C and D, respectively). We observed that piggyBac transposition was more dosage-sensitive with increasing levels of helper when the amount of donor was fixed at 200 ng rather than at 50 ng.
Different transposon families display various DNA insertion sequence preferences. For example, the Tc1/mariner family targets the dinucleotide TA whereas the piggyBac family prefers the tetranucleotide TTAA. Thus, nonmodified transposons are not site-specific. Methods to target the transposon to a unique genomic site have been proposed (21). Theoretically, transposon site-specific integration can be achieved by increasing the DNA binding specificity of its transposase (21, 27). It appears that the most feasible way to achieve targeted transposition is to fuse a DBD recognizing a unique chromosomal sequence to the transposase (or possibly just the catalytic domain). Recently, a chimeric piggyBac transposase with GAL4 DBD attached to its N terminus resulted in targeting a specific site upstream of the UAS (GAL4-binding) site in a plasmid assay system in Aedes aegypti embryos (27). Herein we show that GAL4-piggyBac retains activity as compared with the wild type without changing its TTAA insertion preference in CHO cells. It is plausible that we may see increased transposition activity as compared with the wild type if the cell contains an upstream activation sequence tandem array, as previously demonstrated in a plasmid assay system in mosquito embryos (27). It would be of interest to determine whether the GAL4-piggyBac transposase results in targeted genomic integration in engineered mammalian cells with a UAS site or to develop a piggyBac transposase fused with a zinc finger DBD to target an endogenous genomic location. Furthermore, it is theoretically possible to ablate the nonspecific DNA-binding interaction of a chimeric transposase to enhance site specificity. Site-directed or random mutagenesis of the chimeric transposase and insulators could increase site specificity and decrease positional effects of integration, respectively.
SB is the most popular transposon system currently used for genetic manipulations in mammals. Ding et al. (11) recently demonstrated that piggyBac could efficiently integrate in human and mouse cells as compared with controls; however, no direct comparison with SB, thought to be the most efficient transposon for mammalian transgenesis, was made. In our study piggyBac demonstrated higher transposition activity and transposase flexibility than SB. The fact that piggyBac efficiently transposes in a variety of organisms indicates its utility for a variety of genetic studies in both invertebrate and vertebrate systems. Our findings suggest that piggyBac could be the ideal transposon for mammalian transgenesis and holds promise for preclinical gene therapy experiments.
Materials and Methods
Cell Culture and Transposition Assay.
Unless otherwise stated, all cell lines used were maintained in MEMα medium (HyClone, Logan, UT) containing 5% FBS (HyClone). For the transposition assays, cells at 80% confluence were harvested, and 1 × 105 cells were seeded into individual wells of 24-well plates 18 h before transfection. A total of 400 ng (Fig. 2) or 600 ng (Fig. 3) of DNA was used for each transfection with FuGENE 6 (Roche, Florence, SC). For each cell line, 1/10th of the transfected cells were transferred to 100-mm plates followed by hygromycin selection for 14 days. The concentration of hygromycin B used in HeLa, HEK293, H1299, and CHO cells was 200, 100, 400, and 400 μg/ml, respectively. To count the clones, cells were fixed with PBS containing 4% paraformaldehyde for 10 min and then stained with 0.2% methylene blue for 1 h. After 14 days of hygromycin selection only colonies >0.5 mm in diameter were counted.
Transposition Assays for Various Molar Ratios of Transposon and Transposase.
Transposition assays were performed in HEK293 cells following the procedure described above. For the experiments with donor plasmids fixed at 200 ng, various amounts (0, 50, 100, 150, 200, and 400 ng) of helper plasmids (transposases) were cotransfected into cells. For the experiments with piggyBac donor fixed at 50 ng, various amounts of piggyBac transposase (0, 50, 100, 150, 200, and 300 ng) were cotransfected. In all experiments pcDNA3.1Δneo was used to normalize the total amount of DNA introduced into the cells.
Plasmid Excision Assays.
One million HEK293 cells were seeded onto 60-mm plates 18 h before transfection. One microgram each of donor and helper plasmid was transfected into cells. Plasmids were recovered by using the Hirt method 72 h after transfection (32). Plasmids isolated were used as templates for nested PCR by using the following primers to detect the presence of donor plasmids that underwent excision: piggyBac first round, 5Bac-1(TCGCCATTCAGGCTGCGC)/3Bac-1(TGTTCGGGTTGCTGATGC); piggyBac second round, 5Bac-2(CCTCTTCGCTATTACGCC)/3Bac-2 (TGACCATCCGGAACTGTG); SB first round, F1-ex (CCAAACTGGAACAACACTCAACCCTATCTC)/o-lac-R(GTCAGTGAGCGAGGAAGCGGAAGAG); SB second round, KJC031(CGATTAAGTTGGGTAACGCCAGGGTTT)/i-lac-R(AGCTCACTCATTAGGCACCCCAGGC); Mos1 first round, 5mos-1 (TCCATTGCGCATCGTTGC)/3mos-1 (AGTACTAGTTCGAACGCG); Mos1 second round, 5mos-2 (ACAGCGTTGTTCCACTGG)/3mos-2 (AAGCTGCATCAGCTTCAG).
Plasmid Constructions.
The sequences of PCR-based constructs were confirmed by DNA automatic sequencing (Applied Biosystems, Foster City, CA). With the exception of pCMV-SB11 (a gift from Perry Hackett, University of Minnesota, Minneapolis, MN), all of the helper plasmids were constructed by placing the transposase cDNA under the control of the CMV promoter in the pcDNA3.1Δneo plasmid derived from pcDNA3.1 (Invitrogen, Carlsbad, CA) by removing most of the neomycin resistance gene.
Selection Cassette.
The fragment containing the hygromycin resistance gene driven by the SV40 promoter was excised from pcDNA3.1/hygro/LacZ vector (Invitrogen). After XmnI and SapI digestion, the fragment was cloned into the SmaI site of pBlueScript SKII to complete the construction of pBS-hygro. To further insert the kanamycin resistance gene and the ColE1 origin of replication, the ApoI/AflIII fragment of pZErO-2.1 (Invitrogen) was cloned into the EcoRV site of pBS-hygro to complete the construction of the pBS cassette.
Mos1 Transposon System.
For the Mos1 transposase expression construct, the SacII/BamHI fragment isolated from pIE-Hr5mos-ORF was blunted and cloned into the EcoRV site of pcDNA3.1Δneo. The donor plasmid was built by inserting the cassette fragment into the blunted XbaI site of pELHY6-0 vector (a gift from Stephen Beverley, Washington University, St. Louis, MO).
piggyBac Transposon System.
For the piggyBac transposase expression construct, the blunted SacII/BamHI fragment from pIE-Hr5-piggyBac was cloned into the EcoRV site of pcDNA3.1Δneo. Regarding the GAL4 fusion, a PCR fragment containing GAL4 DBD with a linker (primer pairs gatcgaattcaccATGACCCCCCCCAAGAAGAAGC and CTCTAATAGTCCTCTGTGGC) was cloned into the N terminus of pcDNA3.1Δneo-piggyBac. The donor plasmid was built by inserting the selection cassette into the SmaI/EcoRV sites of pXLBacIIPUbnlsEGFP, derived from pBSII-ITR1 (33). This donor plasmid is a minimal piggyBac vector with TR of 308 bp and 238 bp at the 5′ and 3′ ends, respectively.
Tol2 Transposon System.
For the Tol2 transposase expression construct, the Tol2 ORF was PCR-amplified from pBK-CMV-Tol2 (a gift from Vladimir Korzh, National University of Singapore, Singapore) by using the primers atcgggatccatgttcattggtcctttgg and cgattctagactactcaaagttgtaaaacc. The PCR fragment was then inserted into the BamHI/XbaI sites of pcDNA3.1Δneo. For the GAL4 DBD fusion the PCR fragment of GAL4 DBD plus linker (primer pair gatcgctagcaccATGACCCCCCCCAAGAAGAAGC and CCATggatccggatcggccgcggagcttgg) was cloned into the BamHI/NheI sites of pcDNA3.1Δneo. For the donor construct, the selection cassette was inserted blunt into the NotI site of the vector pGEM-T-easy-Tol2ends (a gift from Vladimir Korzh).
SB Transposon System.
The SB11 transposase expression construct (pCMV-SB11) was provided by Perry Hackett. The GAL4 DBD fusion was obtained by inserting the PCR fragment obtained by using the primers ATCGCGGCCGACCATGACCCCCCCCAAGAAG and CGATCGGCCGCGGAGCTTGGGGCCGCC into the EagI site of the vector pCMV-SB11. The donor plasmid was generated by inserting the selection cassette into the BglII/EcoRV sites of the vector pT2/HB (a gift from Perry Hackett).
Analysis of Transposon–Chromosomal Junction Mediated by GAL4-piggyBac in CHO Cells via Plasmid Rescue.
Individual clones were isolated and proliferated until confluence in a 100-mm plate. Genomic DNA was isolated by using a DNeasy Tissue kit according to the manufacturer's protocol (Qiagen, Valencia, CA). Five micrograms of genomic DNA was subjected to XhoI digestion followed by ligation. The ligation reactions were transformed into Escherichia coli DH10B cells. Plasmids rescued from transformants were subjected to DNA sequencing to retrieve the genomic sequence flanking the insertion site. The raw sequence is provided in Fig. 5, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Dr. Vladimir Korzh for the Tol2 donor plasmid (Tol2 ends), Dr. Stephen Beverley for the Mos1 (pELH6-0) plasmid, and Dr. Perry Hackett for the pCMV-SB11 and pT2/HB plasmids. We appreciate Dr. Ryuzo Yanagimachi's insightfulness and encouragement in the preparation of the manuscript.
Abbreviations
- SB
Sleeping Beauty
- DBD
DNA-binding domain
- TR
terminal repeat
- UAS
upstream activating sequence.
Footnotes
Conflict of interest statement: C.J.C., J.M.K., and S.M. are affiliated with Manoa Transgenics, Inc.
See Commentary on page 14981.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D, Sang H. Nat Biotechnol. 1998;16:1050–1053. doi: 10.1038/3497. [DOI] [PubMed] [Google Scholar]
- 4.Balu B, Shoue DA, Fraser M, Adams JH. Proc Natl Acad Sci USA. 2005;102:16391–16396. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessereau J, Wright A, Williams DC, Schuske K, Davis MW, Jorgensen EM. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 6.Pledger DW, Coates CJ. Insect Biochem Mol Biol. 2005;35:1199–1207. doi: 10.1016/j.ibmb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Kawakami K, Shima A, Kawakami N. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivics Z, Hackett P, Plasterk RH, Izsvák Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 9.Miskey C, Izsvak Z, Plasterk RH, Ivics Z. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser MJ, Ciszczon T, Elick T, Bauser C. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 11.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 13.Collier LS, Largaespada DA. Cancer Res. 2005;65:9607–9610. doi: 10.1158/0008-5472.CAN-05-3085. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 15.Balter M. Science. 2000;288:951–957. [Google Scholar]
- 16.Roberts JP. Scientist. 2005;19:20–24. [Google Scholar]
- 17.Hackett PB, Ekker SC, Largaespada DA, McIvor RS. In: Non-Viral Vectors for Gene Therapy (Advances in Genetics) 2nd Ed. Huang L, Wagner E, Hung M-C, editors. San Diego: Academic; 2005. pp. 189–232. [DOI] [PubMed] [Google Scholar]
- 18.Ivics Z, Izsvák Z. Methods Mol Biol. 2004;260:255–276. doi: 10.1385/1-59259-755-6:255. [DOI] [PubMed] [Google Scholar]
- 19.Izsvak Z, Ivics Z. Mol Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminski JM, Huber MR, Summers JB, Ward MB. FASEB J. 2002;16:1242–1247. doi: 10.1096/fj.02-0127hyp. [DOI] [PubMed] [Google Scholar]
- 22.Wilson MH, Kaminski JM, George AL., Jr FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 24.Lohe A, Hartl D. Mol Biol Evol. 1996;13:549–555. doi: 10.1093/oxfordjournals.molbev.a025615. [DOI] [PubMed] [Google Scholar]
- 25.Kawakami K, Noda T. Genetics. 2004;166:895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zayed H, Izsvak Z, Walisko O, Ivics Z. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 27.Maragathavally KJ, Kaminski JM, Coates CJ. FASEB J. 2006;20:1880–1882. doi: 10.1096/fj.05-5485fje. [DOI] [PubMed] [Google Scholar]
- 28.Fraser MJ, Cary L, Boonvisudhi K, Wang HG. Virology. 1995;211:397–407. doi: 10.1006/viro.1995.1422. [DOI] [PubMed] [Google Scholar]
- 29.Tosi LR, Beverley SM. Nucleic Acids Res. 2000;28:784–790. doi: 10.1093/nar/28.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izsvak Z, Ivics Z, Hackett PB. Mol Gen Genet. 1995;247:312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- 31.Koga A. Adv Biophys. 2004;38:161–180. [PubMed] [Google Scholar]
- 32.Ziegler K, Bui T, Frisque RJ, Grandinetti A, Nerurkar VR. J Virol Methods. 2004;122:123–127. doi: 10.1016/j.jviromet.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Harrell RA, Handler AM, Beam T, Hennessy K, Fraser MJ., Jr Insect Mol Biol. 2005;14:17–30. doi: 10.1111/j.1365-2583.2004.00525.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.