Abstract
Nonspecific protein–DNA interactions are inherently dynamic and involve both diffusion of the protein along the DNA and hopping of the protein from one DNA molecule or segment to another. Understanding how gene regulatory proteins interact nonspecifically with DNA in terms of both structure and dynamics is challenging because the experimental observables are an ensemble average of many rapidly exchanging states. By using a variety of NMR spectroscopic techniques, including relaxation analysis, paramagnetic relaxation enhancement, and residual dipolar couplings, we have characterized structural and kinetic aspects of the interaction of the HoxD9 homeodomain with a nonspecific, 24-bp DNA duplex in a system in which the protein is not constrained to any particular site. The data reveal that HoxD9 binds to nonspecific DNA with the same binding mode and orientation as that observed in the specific complex. The mobility, however, of Arg side-chains contacting the DNA is increased in the nonspecific complex relative to the specific one. The kinetics of intermolecular translocation between two different nonspecific DNA molecules have also been analyzed and reveal that at high DNA concentrations (such as those present in vivo) direct transfer from one nonspecific complex to another nonspecific DNA molecule occurs without going through the intermediary of free protein. This finding provides a simple mechanism for accelerating the target search in vivo for the specific site in a sea of nonspecific sites by permitting more effective sampling of available DNA sites as the protein jumps from one segment to another.
Keywords: dynamics, nonspecific binding, protein–DNA interactions, structure
The process whereby transcription factors are able to efficiently and rapidly locate their specific DNA target sequence in the presence of an enormous background of nonspecific DNA sites has been the subject of considerable interest in molecular biophysics. Nonspecific DNA binding is thought to play a critical role in increasing the efficiency with which a transcription factor locates its specific target site, either through one-dimensional diffusion along the DNA or by intersegment transfer from one location on the DNA to another (1, 2). Because the typical equilibrium dissociation constant (KD) for nonspecific DNA binding is much smaller than the DNA concentration in the nucleus [≈100 mg/ml corresponding to ≈150 mM on a base pair basis (3)], any excess of a given gene-regulatory protein will be bound to nonspecific DNA rather than remain free in solution (4). Understanding how proteins are bound nonspecifically to DNA and how they diffuse on the DNA in their search for a specific target sequence is therefore important.
Although some crystallographic and NMR structural studies of nonspecific protein–DNA complexes have been reported, in each case the system was constrained such that the protein could only be located at a single site on the DNA oligonucleotide used (5–9). Although such studies provide structural information at atomic resolution, the biological relevance of the observed binding modes remains questionable because it is hard to exclude the possibility that these structures may represent partially specific complexes or be heavily influenced by either crystal packing forces or the use of cross-linking agents to fix the complex at a single location. Structural characterization of nonspecific protein–DNA interactions without constraining the protein to a particular site on the DNA is therefore essential but challenging given that the experimental observables represent an ensemble average of various rapidly exchanging states in which the protein is located at multiple sites on the DNA.
By using a variety of NMR spectroscopic techniques, in this paper we characterize the structural and kinetic features of a nonspecific protein–DNA complex in which the protein can both slide and hop on the DNA. The model system involves homeodomain–DNA interactions that have been extensively characterized by biophysical and biochemical means (10). The sequence specificities of homeodomains are modest and the affinities for specific sites are ≈200-fold greater than those for nonspecific sites (11–13). In previous work on the specific HoxD9 homeodomain–DNA complex, we were able to directly detect very low population transient intermediates stochastically bound to nonspecific sites by means of intermolecular paramagnetic relaxation enhancement (PRE) measurements (11). Although some general structural features of the nonspecific complexes could be deduced, the information was very limited because of the high population (>99%) of the specific complex. Here, we disrupted the specific target site in the original DNA sequence and analyzed the protein bound to two nonspecific 24-bp DNA duplexes. Because these oligonucleotides are much longer than the length of DNA covered by homeodomain binding and because neither specific nor semispecific recognition sequences are present, the protein can freely diffuse along the DNA.
Results and Discussion
NMR of the Homeodomain Bound to Nonspecific DNA.
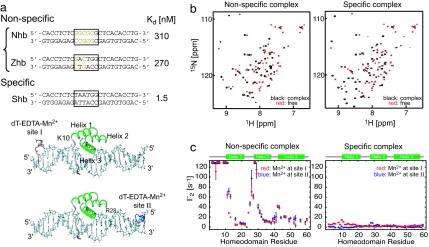
Two nonspecific complexes of the HoxD9 homeodomain with the DNA duplexes, Nhb and Zhb, containing five and two mutations, respectively, within the specific 6-bp homeodomain-binding motif (Shb duplex) were studied (Fig. 1a). At 100 mM NaCl, the KD values for the two nonspecific complexes, determined by fluorescence anisotropy, are 330 and 270 nM, respectively, compared with a value of 1.5 nM for the specific complex (11), fully consistent with the values reported previously for other homeodomains (12, 13). The 1H-15N heteronuclear single quantum correlation (HSQC) spectra of the nonspecific complexes of the HoxD9 homeodomain bound to the Nhb (Fig. 1b) and Zhb (data not shown) oligonucleotides are very similar, well resolved, and significantly different from those of either the specific complex or the free protein (Fig. 1b). Only a single 1H-15N cross-peak per residue was observed for the nonspecific complexes, despite the presence of many potential sites for nonspecific binding. 1HN-Γ2 PRE data were recorded for the nonspecific complex with the Nhb DNA duplex and the specific complex with the Shb DNA duplex under identical experimental conditions at a salt concentration of 20 mM NaCl with oligonucleotides derivatized with dT-EDTA at two sites (I and II) located at each end of the duplex (Fig. 1a). The PRE profiles for the specific and nonspecific complexes were completely different (Fig. 1c). For the specific complex, the 1HN-Γ2 values were small, with maximum values <20 s−1 and consistent with the Mn2+-1HN distances in the structure of the specific complex (modeled from the 2.4-Å resolution crystal structure of the highly homologous Antennapedia homeodomain–DNA complex) (14) with a PRE Q factor (15) of ≈0.32. In contrast, the 1HN-Γ2 values for the nonspecific complex were very large, with several cross-peaks, located at the N terminus and at the N-terminal ends of helices 1 and 2, broadened beyond the limits of detection (Γ2 > 100 s−1). The PRE profiles for the nonspecific complex were inconsistent with the structure of the specific complex with a PRE Q factor of ≈0.83. Equally importantly, the PRE profiles arising from Mn2+ at sites I and II are very similar to each other in the nonspecific complex [as we previously reported for the nonspecific HMGB-1A/DNA complex (16)]. These observations can be accounted for by nonspecific protein–DNA interactions in which HoxD9 can be located anywhere on the 24-bp Nhb duplex with an almost uniform distribution in both directions. In addition, translocation between sites is fast on both the chemical shift and PRE time scales.
Fig. 1.
Specific and nonspecific HoxD9–DNA complexes. (a) DNA duplexes of 24 bp were used in the current study. Shb contains a single 6-bp homeodomain-binding motif (boxed area), which is disrupted by 2- and 5-bp mutations (orange) for the nonspecific Zhb and Nhb DNA duplexes, respectively. KD values (11) at 100 mM NaCl are listed. (b) 1H-15N HSQC spectra of the uniformly 2H,15N-labeled HoxD9 homeodomain free (red) and bound (black) to the Nhb nonspecific (Left) and Shb specific (Right) DNA duplexes. For the specific complex, all cross-peaks are observed; for the nonspecific complexes (Nhb and Zhb), Tyr-8 is not observed, and Trp-48 and Asn-51 are significantly broadened. (c) Intermolecular 1HN-Γ2 PREs arising from the dT-EDTA-Mn2+ measured on uniformly 2H,15N-labeled HoxD9 homeodomain bound to the Nhb nonspecific (Left) and Shb-specific (Right) DNA duplexes at 20 mM NaCl. Two PRE data sets arising from dT-EDTA-Mn2+ at sites I (red) and II (blue) are plotted for the two complexes. The amide protons broadened beyond detection because of very large PRE are indicated by asterisks.
Kinetics of Intermolecular Translocation.
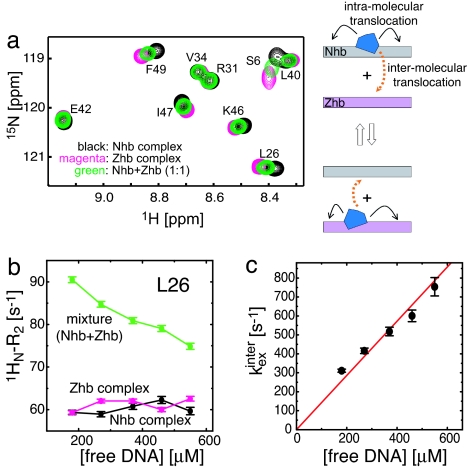
To investigate the kinetics of intermolecular translocation of the HoxD9 homeodomain between two nonspecific DNA duplexes, we used the mixture approach described previously for the analysis of translocation between two specific sites (17). We used a 1:1 mixture of two nonspecific complexes comprising the 24-bp Nhb and Zhb DNA duplexes (Fig. 1a). The positions of the 1H-15N cross-peaks in the mixture of the two complexes are located at the weighted average of the cross-peak positions for the individual complexes, which immediately yields the populations of the two complexes in the mixture (Fig. 2a). Thus, at 20 mM NaCl, the intermolecular translocation process whereby a HoxD9 homeodomain is transferred from one nonspecific DNA molecule to another is in fast exchange on the chemical shift time scale. In contrast, translocation between specific sites under the same experimental conditions is in the slow exchange regime (17). Because z-exchange spectroscopy is not applicable in the fast-exchange regime, we measured exchange contributions to 1H transverse relaxation rates (R2) to quantitatively analyze the kinetics of intermolecular translocations between nonspecific duplexes. 1HN-R2 rates were measured for the two individual nonspecific complexes and their 1:1 mixture by using Lorentzian line-shape fitting of cross-peaks in the 1H acquisition dimension of two-dimensional 1H-15N HSQC spectra. For residues with significant chemical shift differences between the two nonspecific complexes, the R2 relaxation rates measured for the 1:1 mixture are larger than those for the individual nonspecific complexes (Fig. 2b). The increase in R2 is due to the additional exchange contribution arising from the intermolecular translocation process. The overall R2 relaxation rate for the mixture is given by R2mixture = pNhbR2Nhb + pZhbR2Zhb + Rexinter, where pNhb and pZhb are the populations of the Nhb and Zhb nonspecific complexes, respectively; R2Nhb and R2Zhb are the corresponding transverse relaxation rates for the individual states; and Rexinter is the exchange contribution arising from intermolecular translocation of HoxD9 between the Nhb and Zhb DNA duplexes. Note that exchange contributions arising from intramolecular translocation (which are independent of the concentration of free DNA) are included in the R2Nhb and R2Zhb terms and that the chemical shift differences for HoxD9 bound to each potential nonspecific site are expected to be just as large as the difference in the observed chemical shifts between the two nonspecific complexes, which represent ensemble averages of all states present. Because the measured R2Nhb and R2Zhb rates appear to be independent of free DNA concentration, whereas the R2mixture rate clearly decreases as a function of free DNA concentration, one can deduce that the rates for intramolecular translocation are in all likelihood much faster than those for intermolecular translocations (Fig. 2b). The two-state exchange process (with no refocusing pulses involved) can be described by the McConnell equations (18), giving Rexinter as (19)
and
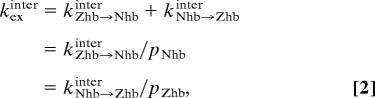 |
where kZhb→Nhbinter and kNhb→Zhbinter are the protein translocation rate constants, and ∣δNhb − δZhb∣ is the chemical shift difference between the two nonspecific complexes measured in hertz. Because the populations and chemical shift differences can be obtained directly from the 1H-15N HSQC spectra of the 1:1 mixture and the individual complexes, respectively, the determination of the overall intermolecular translocation rate, kexinter, is straightforward. kexinter was measured as a function of free DNA concentration while keeping the molar ratio of Nhb and Zhb DNA duplexes constant at 1:1 and was found to be proportional to the free DNA concentration (Fig. 2c). If the principal mechanism for intermolecular translocation were to involve spontaneous dissociation and subsequent reassociation, kexinter should be independent of the free DNA concentration (17) because the rate-limiting step at high DNA concentration is the unimolecular dissociation process, which is independent of free DNA concentration. The DNA concentration dependence of kexinter suggests that the protein is directly transferred from a nonspecific complex to a free nonspecific DNA molecule without going through the intermediary of free protein. This direct transfer mechanism is a second-order reaction with rate constants of 1.5 × 106 M−1·s−1 for the transfer from Nhb to Zhb and 1.3 × 106 M−1·s−1 for the reverse process. These values are ≈25 times larger than that for the transfer between specific sequences under similar conditions (17).
Fig. 2.
Kinetics of intermolecular translocation between nonspecific DNA duplexes. (a) Overlay of three 1H-15N HSQC spectra measured on nonspecific complexes comprising Nhb (black) and Zhb (magenta) duplexes and their 1:1 mixture (green). (b) Amide 1HN-R2 rates for Leu-26 as a function of free DNA concentration measured by Lorentzian line-shape-fitting in the 1H dimension of the 1H-15N HSQC spectra. HoxD9 was 2H,15N-labeled, yielding a pure Lorentzian line shape without 3JHNHα splitting. In processing, a 5-Hz, line-broadening exponential window function was applied in the 1H-dimension and then subtracted from the fitted value of R2. (c) Overall apparent intermolecular translocation rate kexinter plotted as function of [DNA]free{[Nhb + Zhb]free for the mixture with [Nhb]:[Zhb] = 1:1}. The value of kexinter was obtained by fitting all of the data for residues with ∣δNhb − δZhb∣ > 20 Hz (at a 1H frequency of 500 MHz) simultaneously, minimizing a χ2 function defined as Σ(Rex,calcinter − Rex,obsinter)2/σ2, where σ is the experimental error and Rex,calcinter is given by Eq. 1). Values of Rex,obsinter were obtained from the experimental values of R2Nhb, R2Zhb, and R2mixture.
Binding Mode of the Homeodomain Diffusing on Nonspecific DNA.
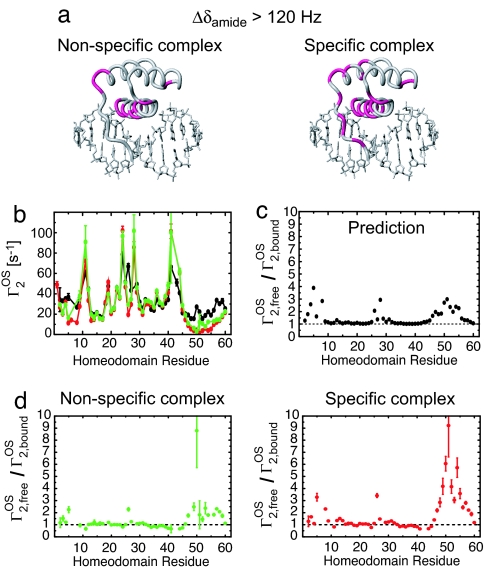
The binding interface used by HoxD9 to interact nonspecifically with DNA was analyzed by using chemical shift perturbation and PRE arising from paramagnetic cosolute molecules. Large 1HN/15N chemical shift perturbations are observed for residues in helix 3 and the loop between helices 2 and 3 in the nonspecific complex (Fig. 3a), implying that these residues are located at the binding interface just as in the specific complex. PRE arising from paramagnetic cosolute molecules (20, 21) provides more quantitative information on the binding interface (22). We used a neutral compound that chelates paramagnetic metal Gd3+, 3 mM Gd–diethylenetriamine pentaacetic acid–bismethylamide (Gd-DTPA-BMA), as a cosolute and measured 1HN-Γ2 PREs arising from the cosolute (referred to as Γ2OS) on the nonspecific and specific complexes and the free protein (Fig. 3b). Because the PRE correlation time is dominated by translational diffusion of the cosolute (20), the magnitude of Γ2OS is almost independent of the size of the observed macromolecule. Amino acid residues at the binding interface are less exposed to solvent in the complex and hence should exhibit smaller Γ2OS rates upon DNA binding. Residues at the interface can be readily identified as those with a Γ2,freeOS/Γ2,boundOS ratio significantly larger than 1. The same regions of the homeodomain in both nonspecific and specific complexes exhibit large Γ2,freeOS/Γ2,boundOS ratios (Fig. 3d). These data indicate that the HoxD9 homeodomain makes use of the identical binding interface for both nonspecific and specific DNA interactions.
Fig. 3.
The HoxD9 homeodomain binds to nonspecific DNA by using the same interface as that in the specific complex. (a) Residues exhibiting a large (>120 Hz at a 1H frequency of 600 MHz) 1HN/15N chemical shift perturbation, Δδamide [defined as (ΔδH2 + ΔδN2)1/2 in Hz], upon complex formation are colored in lilac on the structure of the specific complex. (b) 1HN-Γ2 PREs (Γ2OS) arising from 3 mM paramagnetic cosolute Gd-DTPA-BMA. Data measured on the nonspecific complex, specific complex and free protein are shown in green, red, and black, respectively. (c) Predicted profile of Γ2,freeOS/Γ2,boundOS for the specific complex. The ratio was predicted from the structures by using a grid-based approach (20, 21). The radius of the Gd–DTPA-BMA molecule was set to 3.5 Å. The correlation time for the PRE was assumed to be dominated by translational diffusion of the paramagnetic cosolute molecule and virtually identical for the free and bound states of the protein. (d) Experimental Γ2,freeOS/Γ2,boundOS ratios for the nonspecific and specific complexes. The binding interface can be identified from regions with Γ2,freeOS/Γ2,boundOS > 1.
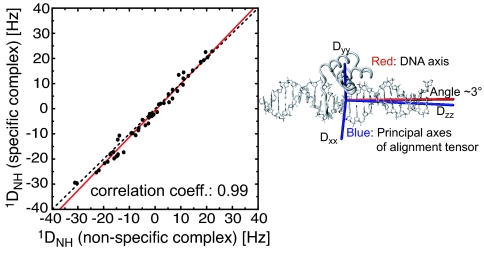
To ascertain the binding orientation of HoxD9 relative to DNA in nonspecific complexes, we analyzed 1DNH residual dipolar couplings (RDCs) induced by phage Pf1 (23). One might expect that the RDC data for the nonspecific complex should be very different from those for the specific complex because of the presence of multiple states in the nonspecific complex. However, the results were surprising: 1DNH data measured on the nonspecific and specific complexes are almost the same, with a correlation coefficient of 0.99, a slope of 1.06, and a pairwise rms difference of 2.3 Hz (Fig. 4). The agreement between observed RDCs for the nonspecific and specific complexes and those calculated from the crystal structure of the Antennapedia homoedomain–DNA complex are in excellent agreement with the RDC R factors (24) of 15.2% and 13.7%, respectively. Thus, the backbone structure of HoxD9 in the specific and nonspecific complexes is effectively identical to that of the Antennapedia homeodomain within the errors of the crystal coordinates. The values for the magnitude of the principal component of the alignment tensor, DaNH, are −18.2 and −19.4 Hz for the nonspecific and specific complexes, respectively; and both complexes are essentially axially symmetric, with rhombicities of 0.055 and 0.020, respectively. The principal axis, Dzz, of the alignment tensor is close to parallel to the long axis of the DNA (Fig. 4), with an angle between the two axes of 3° for the specific complex and 6° for the nonspecific complex. The small 3° difference in the direction of Dzz for the specific and nonspecific complexes is within the experimental uncertainty of ≈5° (25). To account for the RDC data for the nonspecific complex, the following must hold: (i) for each nonspecific site, the binding orientation of HoxD9 with respect to the long axis of the DNA is essentially identical to that for the specific complex (note that, because of axial symmetry, the RDCs are invariant to rotations about the principal axis of the alignment tensor and a 180° rotation about an axis perpendicular to the principal axis); (ii) end-effects that could potentially contribute alternative binding modes are insignificant; and (iii) negative charges on the 24-bp DNA dominate Pf1-induced alignment such that the alignment tensor is minimally sensitive to protein location along the DNA.
Fig. 4.
Correlation between Pf1-induced 1DNH RDCs measured on the nonspecific (Nhb) and specific (Shb) HoxD9–DNA complexes. The red line is the linear regression (slope of 1.06). The data extend from residues 2–60 and include both the N-terminal arm (residues 1–9) and the core domain. The Pf1 concentration was 12 mg/ml, and the buffer contained 10 mM Tris·HCl (pH 6.8) and 20 mM NaCl. The quadrupole splittings of the solvent 2H resonance were 9.7 and 9.4 Hz for the nonspecific and specific complexes, respectively.
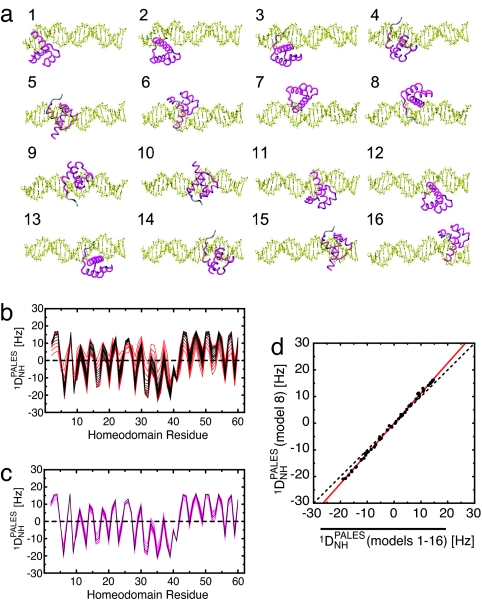
We examined the latter hypothesis by using a computational approach. The homeodomain covers a 9-bp region in the specific complex. Hence, a nonspecific 24-bp DNA duplex provides 32 [(24 − 9 + 1) × 2] nonspecific binding sites. To represent the nonspecific complexes, we generated 16 structure models [derived from the crystal structure of the specific Antennapedia homedodomain–DNA complex (14)] located at each possible site with the same binding orientation relative to the DNA and the same binding surface on HoxD9 (Fig. 5a). Note that the other 16 states with the opposite protein orientation are also represented with these 16 structures (e.g., the opposite orientation at the binding site of model 1 corresponds to model 16). The RDCs for the individual states were predicted on the basis of the three-dimensional shapes and charge distributions using the program PALES (26). The RDC values predicted for the specific complex were highly correlated with those observed experimentally, with a correlation coefficient of 0.95 (see the supporting information, which is published on the PNAS web site), confirming the reliability of the PALES calculation. Fig. 5b shows the 1DNH profiles predicted for the individual 16 models. The profiles are similar to each other despite the different locations of the protein on the DNA (Fig. 5b), and the ensemble averages of the RDCs for consecutive multiple states are in excellent agreement with those predicted for the specific complex (Fig. 5 c and d). Even the small deviation from unity in the slope of the correlation between the RDCs for the nonspecific and specific complexes is reproduced by the PALES calculations. These theoretical results together with the experimental RDC data suggest that the binding orientation of the homeodomain with respect to the long axis of nonspecific DNA is identical regardless of base sequence and is the same as that in the specific complex.
Fig. 5.
Prediction of Pf1-induced 1DNH RDCs for nonspecific complexes based on three-dimensional shapes and charge distribution using the program PALES (26). (a) Sixteen structure models for nonspecific complexes along a 24-bp DNA duplex with HoxD9 binding to DNA in the same binding mode but at different locations. Model 8 corresponds to the HoxD9–specific complex. (b) Predicted 1DNH RDCs for each residue. Results for models 1–16 are plotted (black, models 4–13; red, models 1–3 and 14–16). (c) 1DNH RDCs predicted for the specific complex (equivalent to model 8, black) and the average of the 1DNH RDCs predicted for 10 contiguous models (magenta; 7 lines correspond to averaging for models 1–10, 2–11, 3–12, 4–13, 5–14, 6–15, and 7–16). (d) Correlation between predicted 1DNH RDCs for the specific complex (model 8) and the average 1DNH values obtained for all 16 complexes. The red line is the linear regression (slope of 1.13). Further details are given in the supporting information.
Based on the RDC, PRE, and chemical-shift perturbation data, we conclude that the homeodomain diffuses on nonspecific DNA while retaining the same binding mode as that in the specific complex. This conclusion had been the subject of much speculation for some time, because additional homeodomain molecules bound to nonspecific sites were found in crystals of specific complexes (27, 28). Although the nonspecific binding mode observed in these crystals was very similar to that of the specific complex, the crystallographic data do not permit one to exclude the possibility that the observed nonspecific binding mode is adopted only for particular DNA sequences that are semispecific or only in the presence of crystal packing forces. The present study provides direct and unambiguous evidence that the homeodomain does indeed adopt essentially the same binding mode for nonspecific and specific protein–DNA interactions in solution. In the case of the Lac repressor headpiece dimer, on the other hand, sequence-specific DNA-binding is coupled to the formation of a helix at the dimer interface, and the structures of specific and nonspecific complexes are rather different (8). Our data suggest that a homeodomain protein searches the DNA by using the same binding mode until it hits a “sticky spot” (i.e., the specific target site) for which intermolecular protein–base hydrogen bonding interactions are optimal. This phenomenon might be directly related to the relatively weak sequence specificity of homeodomain proteins with a ratio of nonspecific to specific equilibrium dissociation constants of only ≈200 (11–13).
Arg Side-Chain Dynamics in the Nonspecific HoxD9–DNA Complex.
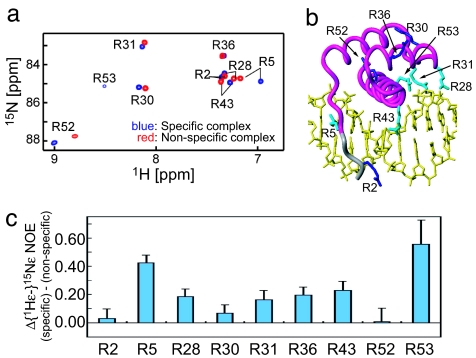
In the specific complex, five Arg side-chains are in contact with DNA (14). To investigate the mobility of these five Arg side-chains (Fig. 6b), we compared {1Hε-}15Nε heteronuclear NOEs for the Arg guanidino group in the nonspecific and specific DNA complexes. Because the {1H-}15N heteronuclear NOE is governed by motions in the picosecond-to-nanosecond time scale, the heteronuclear NOE data on the nonspecific complex should reflect dynamics within the individual states rather than the translocation process itself, which is much slower. The values of the {1Hε-}15Nε NOEs are smaller for the nonspecific than the specific complex (Fig. 6c; see also the supporting information); therefore, the Arg side-chains contacting the DNA are more mobile in the nonspecific complex than the specific one.
Fig. 6.
Dynamics of Arg side-chains. (a) 1Hε-15Nε correlation spectrum for Arg side-chains in the nonspecific complex (red) and specific (blue) HoxD9–DNA complexes. (b) Locations of Arg residues in HoxD9. Arg residues in contact with DNA (14) appear cyan. (c) Difference between {1Hε-}15Nε heteronuclear NOEs measured for nonspecific and specific complexes.
Arg-5, located in the N-terminal arm, is the only Arg that makes a base-specific contact in the specific complex (14). The sizeable reduction (≈0.4 units) in the {1Hε-}15Nε heteronuclear NOE for Arg-5 in the nonspecific complex (+0.01) relative to the specific one (+0.43) can be directly attributed to the removal of the base-specific hydrogen bond between the guanidino group of Arg-5 and the O2 atom of a thymine in the minor groove. The loss of this hydrogen bond, however, results in a much smaller increase in backbone mobility for the N-terminal arm because (i) the difference in backbone {1H-}15N heteronuclear NOEs between the nonspecific and specific complexes in this region is <0.1 units, and (ii) the RDC data for this region in the nonspecific complex are fully consistent with the structure in the specific complex (see Fig. 4).
Concluding Remarks.
We have characterized the structural and kinetic features of a nonspecific homeodomain–DNA complex in a truly dynamic system with rapid exchange between a large array of nonspecific sites. Relaxation analysis revealed that intermolecular translocation from one DNA molecule to another occurs via direct transfer. At the high DNA concentrations found in the nucleus, intermolecular translocation affords an important mechanism of transfer for DNA-binding proteins, accelerating the target search process for the specific site among a sea of nonspecific sites by permitting more effective sampling of available DNA sites as the protein jumps from one segment to another. Comparison of RDC and PRE data measured for nonspecific and specific protein–DNA complexes demonstrates that the HoxD9 homeodomain diffuses rapidly along nonspecific DNA by using essentially the same the binding mode and interface as that used for the specific complex. However, heteronuclear {1Hε-}15Nε NOE data indicate that the Arg side-chains in contact with DNA are more mobile in the nonspecific complex than the specific one, indicative of “looser” intermolecular interactions. The information provided by the present work is fundamental for understanding how a homeodomain-class gene-regulatory protein diffuses and hops along the DNA in search of specific target sequences.
Materials and Methods
Sample Preparation.
HoxD9 homeodomain uniformly labeled with 13C,15N, 2H,15N, and unlabeled 24-bp DNA duplexes were prepared as described (11). For complex formation, the protein and DNA were mixed at a ratio of 1:2 (DNA-excess) to ensure that all protein was bound to DNA in the NMR samples. For NMR analysis, the protein–DNA complexes were dissolved in 10 mM Tris·HCl, pH6.8/20 mM NaCl/7% 2H2O. For the free protein, 140 mM Na2SO4 was added to the buffer to increase solubility.
NMR Spectroscopy.
Data were recorded on Bruker DMX-500, DRX-600, and DMX-800 spectrometers (Billerica, MA) equipped with cryogenic triple resonance z-gradient probes. All NMR measurements were performed at 35°C, with the exception of experiments used to determine exchange rates that were carried out at 30°C. 1H, 13C, and 15N resonances for the free and DNA-bound proteins were assigned by using three-dimensional, double- and triple-resonance experiments (29). PRE 1HN-Γ2 data were acquired at 600 MHz by using the two-dimensional 1H-15N correlation-based experiment as described (15). For PRE arising from EDTA-Mn2+ conjugated to DNA (Fig. 1c), the diamagnetic control was the Ca2+-chelated state. For PRE arising from paramagnetic cosolute molecule (Fig. 3b), samples with and without 3 mM Gd-DTPA-BMP (20) were used. A stock solution of 0.5 M Gd-DTPA-BMA (product name, OmniScan) was purchased from Nycomed (Princeton, NJ). 1DNH RDCs induced with 12 mg/ml Pf1 phage were measured at 750 MHz by using a two-dimensional 1H-15N in-phase/anti-phase HSQC experiment (30). Arg {1Hε}-15Nε NOEs were measured at 600 MHz by using the water flip-back scheme (31) with a sample pH of 5.8 for better sensitivity. Arg 1Hε/15Nε resonances were assigned by using 13Cγ, 13Cδ resonances correlated with 15Nε in the HNCACB experiment and NOEs to 1Hε observed in the 15N-separated NOE spectrum.
Prediction of RDCs.
RDCs were predicted from the three-dimensional charge distribution and shape by using the software PALES, which approximates the electrostatic interaction between a solute and an ordered phage particle as that between the solute surface charges and the electric field of the phage (26). The solute was treated as a particle in the external field of the liquid crystal, and its electrostatic potential was obtained by solving the nonlinear three-dimensional Poisson–Boltzmann equation. The ionic strength was set to the experimental value of 20 mM NaCl. The Pf1 phage particle was represented by an infinite cylinder with a uniform surface charge density of −0.475 e/nm2 and a cylinder radius of 3.35 nm. The liquid crystal order parameter was set to 0.9, and the phage concentration was 12 mg/ml. The protein was represented by the charges of its ionizable residues, and their protonation state was calculated by using the Henderson–Hasselbach equation. Charges were distributed evenly over the heavy atoms involved. In the case of DNA, a charge of −0.5 e was assigned to each of the phosphate oxygen atoms.
Supplementary Material
Acknowledgments
We thank Dr. Denis Torchia for useful discussion. This work was supported by funds from the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (National Institutes of Health) and in part by the AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C.). M.Z. was supported by a Deutsche Forschungsgemeinschaft Emmy Noether Grant ZW 71/1-5.
Abbreviations
- HSQC
heteronuclear single quantum correlation
- PRE
paramagnetic relaxation enhancement
- RDC
residual dipolar coupling
- Gd-DTPA-BMA
guadolinium complex and diethylenetriamine pentaacetic acid-bismethylamide.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.von Hippel PH, Berg OG. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 2.Halford SE, Marko JF. Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewin B. Genes VII. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 4.Bruinsma RF. Phys A. 2002;313:211–237. [Google Scholar]
- 5.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gewirth DT, Sigler PB. Nat Struct Biol. 1995;2:386–394. doi: 10.1038/nsb0595-386. [DOI] [PubMed] [Google Scholar]
- 7.Viadiu H, Aggarwal AK. Mol Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 8.Kalodimos CG, Biris N, Bonvin AMJJ, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee A, Santos WL, Verdine GL. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 10.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wüthrich K. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 11.Iwahara J, Clore GM. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 12.Affolter M, Percival-Smith A, Müller M, Lepin W, Gehring WJ. Proc Natl Acad Sci USA. 1990;87:4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seimiya M, Kurosawa Y. FEBS Lett. 1996;398:279–284. doi: 10.1016/s0014-5793(96)01246-x. [DOI] [PubMed] [Google Scholar]
- 14.Fraenkel E, Pabo CO. Nat Struct Biol. 1998;5:692–697. doi: 10.1038/1382. [DOI] [PubMed] [Google Scholar]
- 15.Iwahara J, Schwieters CD, Clore GM. J Am Chem Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 16.Iwahara J, Schwieters CD, Clore GM. J Am Chem Soc. 2004;126:12800–12808. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]
- 17.Iwahara J, Clore GM. J Am Chem Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 18.McConnell HM. J Chem Phys. 1958;28:430–431. [Google Scholar]
- 19.Reuben J, Fiat D. J Chem Phys. 1969;55:4918–4927. [Google Scholar]
- 20.Pintacuda G, Otting G. J Am Chem Soc. 2001;124:372–373. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- 21.Hernández G, Teng C-L, Bryant RG, LeMaster DM. J Am Chem Soc. 2002;124:4463–4472. doi: 10.1021/ja017340k. [DOI] [PubMed] [Google Scholar]
- 22.Sakakura M, Noba S, Luchette PA, Shimada I, Prosser RS. J Am Chem Soc. 2005;127:5826–5832. doi: 10.1021/ja047825j. [DOI] [PubMed] [Google Scholar]
- 23.Clore GM, Starich MR, Gronenborn AM. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 24.Clore GM, Garrett DS. J Am Chem Soc. 1999;121:9008–9012. [Google Scholar]
- 25.Zweckstetter M, Bax A. J Biomol NMR. 2002;23:127–137. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
- 26.Zweckstetter M, Hummer G, Bax A. Biophys J. 2004;86:3444–3460. doi: 10.1529/biophysj.103.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 28.Aishima J, Wolberger C. Proteins. 2003;51:544–551. doi: 10.1002/prot.10375. [DOI] [PubMed] [Google Scholar]
- 29.Clore GM, Gronenborn AM. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 30.Ottiger M, Delaglio F, Bax A. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 31.Grzesiek S, Bax A. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.