Abstract
Chromosomes condense during mitotic entry to facilitate their segregation. Condensation is typically assayed in fixed preparations, limiting analysis of contributing factors. Here, we describe a quantitative method to monitor condensation kinetics in living cells expressing GFP fused to a core histone. We demonstrate the utility of this method by using it to analyze the molecular requirements for the condensation of holocentric chromosomes during the first division of the Caenorhabditis elegans embryo. In control embryos, the fluorescence intensity distribution for nuclear GFP:histone changes during two distinct time intervals separated by a plateau phase. During the first interval, primary condensation converts diffuse chromatin into discrete linear chromosomes. After the plateau, secondary condensation compacts the curvilinear chromosomes to form shorter bar-shaped structures. We quantitatively compared the consequences on this characteristic profile of depleting the condensin complex, the mitosis-specific histone H3 kinase Aurora B, the centromeric histone CENP-A, and CENP-C, a conserved protein required for kinetochore assembly. Both condensin and CENP-A play critical but distinct roles in primary condensation. In contrast, depletion of CENP-C slows but does not prevent primary condensation. Finally, Aurora B inhibition has no effect on primary condensation, but slightly delays secondary condensation. These results provide insights into the process of condensation, help resolve apparent contradictions from prior studies, and indicate that CENP-A chromatin has an intrinsic role in the condensation of holocentric chromosomes that is independent of its requirement for kinetochore assembly.
Keywords: Aurora B, CENP-A, condensin, kinetochore, C. elegans
As cells enter mitosis, replicated chromosomes, consisting of two identical DNA molecules called sister chromatids, are compacted to facilitate segregation by the mitotic spindle. The packaging of each chromosome into a folded rod-shaped structure, often thousands of times shorter than the DNA molecule itself, is called condensation (1–4). Condensation resolves not only different chromosomes, but also the two sister chromatids, which form morphologically distinct rods that remain connected along one side by sister chromatid cohesion.
Two related protein complexes, condensins I and II, together with the topoisomerase family of DNA decatenating enzymes, are critical for proper condensation (1–4). Inhibition of condensin blocks the ability of mitotic Xenopus extracts to reorganize sperm chromatin into linear rod-shaped chromosomes (5, 6), suggesting an essential role in chromosome compaction and resolution. However, inhibiting condensin in Drosophila (7, 8), Caenorhabditis elegans (9), and vertebrate cells (10, 11) delays, but does not prevent, compaction. The compacted chromosomes that form after condensin inhibition are sensitive to hypotonic fixation conditions and exhibit defects during alignment and segregation that suggest compromised structural integrity (1).
Concurrent with condensation, kinetochores assemble on centromeric chromatin to provide a chromosomal attachment site for spindle microtubules. Interactions between kinetochores and spindle microtubules play a central role in chromosome alignment and segregation (12, 13). Two distinct chromosome architectures are prevalent among metazoans: monocentric, in which kinetochore assembly is restricted to a localized region of each chromatid, and holocentric, in which diffuse kinetochores assemble along the entire chromatid length (14). Both monocentric and holocentric chromosomes assemble kinetochores on chromatin containing the histone H3 variant CENP-A (15–18). After condensation in vertebrates (monocentric), CENP-A-containing chromatin is positioned on the poleward surface of each sister chromatid in a localized chromosomal region called the primary constriction. Holocentric chromosomes lack a primary constriction. Instead, a stripe of centromeric chromatin runs along the entire length of each mitotic chromatid.
Analysis of mitotic chromosome remodeling has been limited, with few exceptions (19–21), by the methods available to monitor this dynamic process. Here, we describe a simple but powerful quantitative analysis method that can be used to monitor the kinetics of chromosome condensation in time-lapse sequences of cells expressing GFP fused to a core histone. We demonstrate the utility of the method by combining it with RNA interference-mediated depletion in the C. elegans embryo to analyze the molecular requirements for the condensation of holocentric chromosomes.
Results
A Method to Quantitatively Monitor Chromosome Condensation.
Our goal was to develop an analysis method to extract quantitative kinetic information on the progress of chromosome condensation from time-lapse sequences. We decided to use the C. elegans embryo as a model system because of its stereotypical first mitotic division. Analyzing the consequences of depleting essential chromosome components is also straight-forward in this system because RNAi can be used to generate oocytes reproducibly >95% depleted of targeted proteins that can be monitored as they progress through their first mitotic division after fertilization (22). During this division, mitotic prophase initiates when the oocyte and sperm-derived pronuclei are on opposite sides of the embryo and continues as the pronuclei migrate toward each other. After the pronuclei meet, the nuclear envelopes become permeable [nuclear envelope breakdown (NEBD)], and the condensed chromosomes interact with spindle microtubules to align and segregate. These dynamic events can be monitored in embryos coexpressing GFP:histone H2b and GFP:γ-tubulin (18). The latter fusion protein marks the centrosomes, has no nuclear signal, and serves as an independent timer of cell cycle progression.
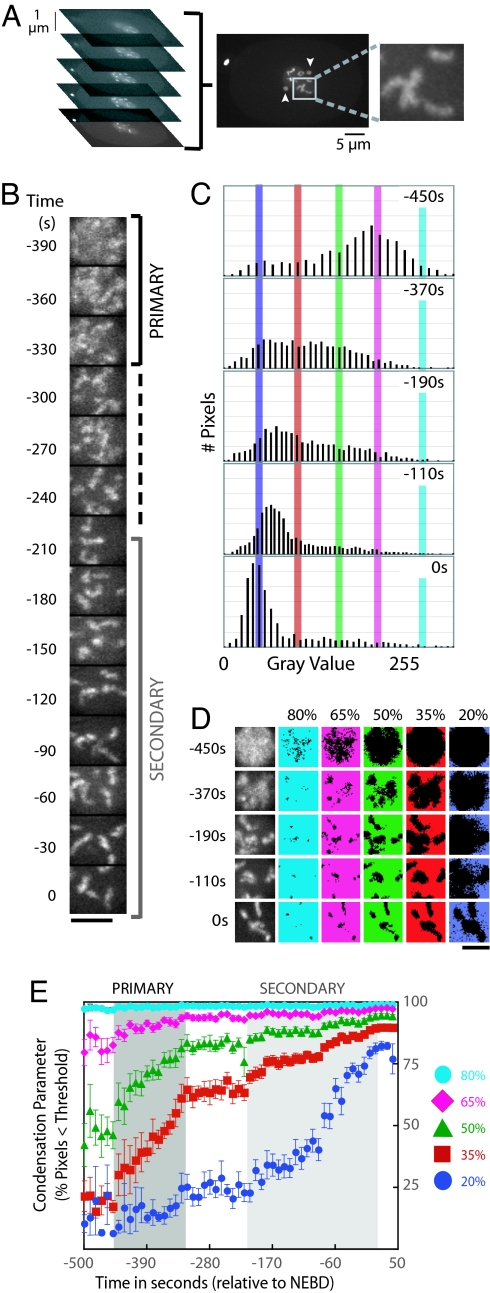
We imaged the GFP:histone by using spinning disk confocal optics to collect time-lapse z-series containing the nucleus. Maximum intensity projections were generated for each time point, and the largest square region that fit within the sperm pronucleus was cut out for further analysis (Fig. 1A) Condensation was analyzed in the sperm pronucleus to facilitate comparison between control embryos and embryos depleted of proteins by RNAi. After fertilization, the sperm chromatin decondenses, is replicated, and then condenses during prophase of the first mitotic division. In contrast, the chromosomes in the oocyte pronucleus complete two rounds of meiotic segregation before entering the S-phase that precedes the first mitotic division. Failure of meiotic segregation when proteins required for chromosome structure are depleted can result in defects in the oocyte chromatin that preclude analysis of their mitotic condensation. The chromatin in the sperm-derived pronucleus does not inherit meiotic defects because the meiotic divisions that produce the sperm occur at a developmental stage before RNAi initiation. The images of the chromatin in the sperm pronucleus were individually scaled, setting the minimum intensity to 0 and the maximum to 255 (Fig. 1B). Individual scaling ensures that the shape of the fluorescence intensity distribution at each time point is independent of fluctuations due to photobleaching or changes in illumination intensity.
Fig. 1.
A quantitative method to monitor chromosome condensation in living cells. (A) Embryos coexpressing GFP-histone and GFP-γ-tubulin were imaged by using spinning disk confocal optics. A five-plane z-series was collected every 10 s, and a maximum intensity projection was generated for each time point. The largest square region that fit within the sperm pronucleus was cut out for further analysis. The change in centrosomal γ-tubulin (arrowheads) serves as a chromatin-independent marker of cell cycle progression. (B) Representative images of a single nucleus after scaling (setting the minimum pixel intensity in each image to 0 and the maximum to 255). Times are with respect to NEBD. The time intervals corresponding to primary and secondary condensation are labeled (the pause is indicated by a dashed line). (Scale bar: 5 μm.) (C) Examples of the fluorescence intensity distribution for GFP:histone in individual nuclei at different time points. Vertical bars mark the thresholds used to measure the progressive change in the shape of the fluorescence intensity distribution that accompanies condensation (cyan, 80% of the image maximum; pink, 65%; green, 50%; red, 35%; blue, 20%). (D) Scaled images of nuclear GFP-histone (left column) were partitioned by using five different thresholds. In each row, the same image is repeated with the pixels below the indicated threshold in color. (Scale bar: 5 μm.) (E) Kinetic plot of the percentage of pixels below each threshold (the condensation parameter) as a function of time. The average values of the condensation parameters for each threshold were measured from 12 sequences time-aligned with respect to NEBD (error bars = SE). The intervals corresponding to primary (between ≈−450 and −325 s; dark gray) and secondary (between −200 and 0 s; light gray) condensation are indicated.
Chromosome condensation is accompanied by a progressive change in the shape of the fluorescence intensity distribution of the nuclear GFP:histone signal (Fig. 1C). Before condensation, the signal is relatively homogeneous, and, after scaling, the majority of pixels are distributed around a central peak at ≈65% of the image maximum (scaled intensity = 166; Fig. 1C, −450 s). As the chromatin condenses, the GFP:histone fluorescence concentrates in a smaller area of the image at the expense of signal elsewhere in the nucleus, progressively increasing the percentage of pixels further away from the image maximum (Fig. 1C). To quantify this shift in the shape of the fluorescence intensity distribution, we monitored the flux of pixels across a grating of thresholds set at 80%, 65%, 50%, 35%, and 20% of the maximum intensity of the image (scaled intensities: 204, 166, 127, 89, and 51, respectively; Fig. 1 C–E). Kinetic profiles to compare control and specifically perturbed embryos were generated by plotting the percentage of pixels below each threshold (the condensation parameter) as a function of time. Times were calculated relative to NEBD, defined as the time point when diffusion out of the nucleus resulted in equilibration of the free nuclear GFP:histone signal with the cytoplasm. For every condition, the condensation parameters measured from time-aligned sequences of 10–20 different embryos were averaged and plotted. Averaging minimizes the contribution of spatial fluctuations to emphasize the global physical changes in chromosome structure that reproducibly change the shape of the fluorescence intensity distribution.
When condensation initiated, the intensity of the majority of pixels was already less than the high threshold values (80% and 65%). Consequently, the kinetic profiles for these thresholds saturated early in the time course (Fig. 1 D and E; cyan and pink traces). In contrast, the percentage of pixels less than the 50% and 35% thresholds progressively increased throughout condensation. The percentage of pixels with intensities less than the 20% threshold was nearly constant during the early stages of condensation but changed dramatically during the later stages of compaction. Two advantages of an intensity distribution-based method over approaches based on segmenting the nucleus to estimate chromosome volume are as follows: (i) the ability to monitor early compaction before the establishment of defined chromosomes on which accurate measurements of length and diameter can be made and (ii) sensitive detection of changes late in condensation, accompanied by relatively small changes in chromosome volume, from relatively low z-resolution image stacks. The latter is true because small increases in the peak chromosomal signal generate a differential that pushes the pixels in the remainder of the nucleus below progressively lower thresholds. The analysis method is simple and robust and can be used to analyze data acquired using wide-field (Fig. 3, which is published as supporting information on the PNAS web site) as well as confocal optics.
Chromosome Condensation Is Temporally Biphasic in C. elegans.
Quantitative analysis revealed that condensation is temporally biphasic in control C. elegans embryos. Changes in the shape of the fluorescence intensity distribution were confined to two distinct time intervals. Condensation was first detected ≈450 s before NEBD (Fig. 1 B and E). Over the subsequent 125-s interval, a consistent increase in the condensation parameters was observed for thresholds between 35% and 65%. We term this initial phase “primary condensation.” Qualitatively, primary condensation corresponds to the organization of diffuse chromatin into distinct linear chromosomes (Fig. 1B). Primary condensation was followed by an ≈125-s pause during which there was no statistically significant change in the percentage of pixels falling below any threshold (Fig. 1E), indicating that the shape of the fluorescence intensity distribution for the nuclear GFP:histone signal did not change. Distinct linear chromosomes with a high degree of curvature were observed throughout this plateau (Fig. 1B). After the pause, a second shift in the shape of the intensity distribution, which we term “secondary condensation,” was detected over the 200-s interval immediately preceding NEBD. Secondary condensation, which corresponds to a change from elongated, highly curved chromosomes to compact, bar-shaped chromosomes, was characterized by a consistent increase in the condensation parameters for thresholds between 20% and 50% (Fig. 1 B and E). Thus, our analysis method revealed that condensation occurs in two temporally distinct phases: a primary phase (lasting ≈125 s), when diffuse chromatin is compacted to form distinct chromosomes, and a subsequent secondary phase (lasting ≈200 s) initiated after a pause, when curved linear chromosomes are further compacted to generate bar-shaped mitotic chromosomes.
The Condensin Subunit SMC-4 Is Required for Primary Condensation.
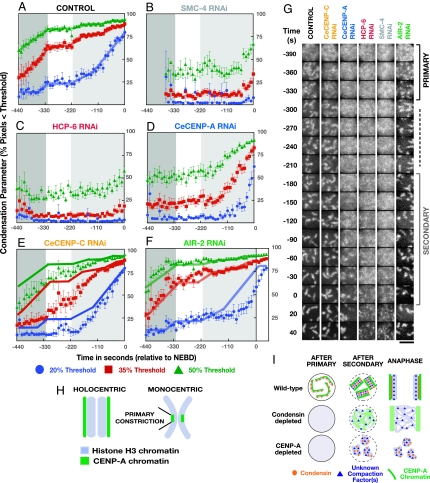
The related condensin I and II complexes play critical roles in chromosomes condensation (2). The core of both condensin complexes consists of the same two essential structural maintenance of chromosomes (SMC) family ATPase subunits. In C. elegans, condensin II contributes to condensation, whereas a condensin I-like complex is thought to have been adapted for sex chromosome dosage compensation (23). To examine the role of condensin, we depleted SMC-4, which is predicted to abolish condensin function. We used the three most informative thresholds (20%, 35%, and 50%) for this analysis. In SMC-4-depleted embryos, no condensation was detected coincident with the primary and plateau phases in WT (Fig. 2B). However, ≈60 s before NEBD, when secondary condensation is well underway in controls, the percentage of pixels falling below the 50% and 35% thresholds began to increase. Visual inspection revealed that this increase was due to a precipitous clumping of chromatin. Distinct chromosomes were not observed at any stage (Fig. 2G and Movie 1, which is published as supporting information on the PNAS web site). Thus, consistent with prior work (9–11), our results indicate that condensin has a critical role in the timely compaction of chromatin during prophase. Although some compaction occurs in condensin-depleted embryos, the chromatin aggregates into a tangled meshwork, and distinct chromosomes are not observed.
Fig. 2.
Kinetic analysis of chromosome condensation in embryos depleted of conserved chromosomal proteins. Shown are plots of the average value of the condensation parameters vs. time for three thresholds (A–F) (green, 50%; red, 35%; blue, 20%) and images from representative time-lapse sequences (G). The time intervals when primary (dark gray) and secondary (light gray) condensation occurs in control embryos are marked on all graphs for reference. Condensation kinetics in control embryos (A; n = 12) and embryos depleted of SMC-4 (B; n = 18), HCP-6 (C; n = 12), CeCENP-A (D; n = 15), CeCENP-C (E; n = 10), and AIR-2 (F; n = 12). In E and F, control traces (solid lines) are superimposed to facilitate comparison. (Scale bar in G: 5 μm.) (H) Schematic comparing the organization of holocentric chromosomes to the region of the primary constriction of monocentric chromosomes. (I) Speculative model for the formation of mitotic C. elegans chromosomes. In WT, condensin (orange circles) and chromatin containing CENP-A (green) are both required for primary condensation. We speculate that unknown compaction factor(s) (blue triangles) drive secondary condensation. In condensin-depleted embryos, primary condensation fails, but compaction into a disorganized meshwork still occurs. CENP-A-containing chromatin is present and functions to attach the disorganized chromatin meshwork to the mitotic spindle (9). In CENP-A-depleted embryos, primary condensation fails. The chromatin ultimately compacts into discrete masses because of the action of condensin and other unknown factors but is unable to assemble kinetochores that can attach to spindle microtubules (18).
The Condensation of Holocentric Chromosomes Is Highly Aberrant After Depletion of CENP-A.
In the region of the primary constriction of mitotic monocentric chromosomes, histone H3 containing “inner-centromeric chromatin” is sandwiched between chromatin containing the H3 variant CENP-A that forms the structural base for the kinetochore. Analysis of the role of CENP-A chromatin in the assembly of mitotic chromosomes is limited by the fact that the primary constriction is only a small proportion of total chromatin. Characterizing the bulk properties of chromatin in holocentrics, where an architecture analogous to the primary constriction extends along the entire length of each chromosome (Fig. 2H), may therefore be useful to investigate the structural properties of CENP-A chromatin. Consistent with a prior qualitative observation (24), we found that depletion of CeCENP-A dramatically altered condensation kinetics. In CeCENP-A-depleted embryos, as in SMC-4 depleted embryos, no condensation was detected coincident with the primary or plateau phases in controls. However, coincident with secondary condensation in controls, the chromatin in CeCENP-A-depleted embryos abruptly compacted (Fig. 2D). The condensation defect in CeCENP-A-depleted embryos was kinetically distinct from that in condensin-depleted embryos; condensation initiated earlier and progressed farther, reaching a WT extent of compaction before NEBD. In addition to this kinetic difference, visual inspection revealed spatially distinct chromosomes by the end of prophase, in contrast to the disorganized chromatin meshwork in condensin-depleted embryos (Fig. 2G and Movie 1). All CeCENP-A-depleted embryos exhibited a kinetochore-null phenotype (18), and quantitative immunoblotting confirmed >97% depletion (data not shown), indicating that the less severe condensation defect is not due to poor protein depletion. We conclude that, in C. elegans, centromeric chromatin is required for the timely compaction and resolution of chromosomes in early prophase. However, the differences in compaction kinetics and final chromosome morphology between CeCENP-A and condensin-depleted embryos indicate that condensin-mediated compaction occurs in the absence of centromeric chromatin.
The Role of CENP-A Chromatin in the Condensation of Holocentric Chromosomes Is Independent of Its Role in Directing Kinetochore Assembly.
To determine whether the role of CENP-A in condensation is linked to its role in kinetochore assembly, we characterized embryos depleted of the kinetochore structural component CENP-C. Depletion of CeCENP-C blocks the recruitment of all known kinetochore components except for CeCENP-A (18, 25). Condensation in CeCENP-C-depleted embryos was qualitatively similar to controls, in that curved linear chromosomes, and subsequently shorter bar-shaped chromosomes, are formed (Fig. 2G). However, the rate of primary condensation was slowed relative to controls, and condensation continued coincident with the plateau phase in WT (Fig. 2E). The extent of chromosome condensation in CeCENP-C-depleted embryos quantitatively resembled that in comparable control embryos ≈110 s before NEBD, roughly when the chromatin in CeCENP-A-depleted embryos first begins to compact (compare Fig. 2 D and E). The conditions used for this experiment result in >95% depletion of CeCENP-C (26), and a clear kinetochore-null phenotype was evident in all embryos filmed.
The fact that depletion of CeCENP-C slows primary condensation is interesting in light of previous work demonstrating a role for CeCENP-C in sister kinetochore resolution, which normally occurs after primary condensation is complete (ref. 27 and our unpublished data). These results suggest either that successful resolution of sister kinetochores depends on the timely completion of primary condensation, or that depletion of CeCENP-C inhibits an upstream process required for both a normal rate of primary condensation and sister kinetochore resolution. Further work will be needed to distinguish between these possibilities. More importantly, the effect of depleting CeCENP-C on chromosome condensation is clearly much less severe than depleting CeCENP-A. We therefore conclude that CeCENP-A-containing chromatin has a critical role in the condensation of holocentric chromosomes that is independent of its requirement for kinetochore assembly.
HCP-6 Depletion Results in a Condensation Defect Kinetically Identical to Depletion of SMC-4.
HCP-6 is the C. elegans homolog of CAP-D3, a non-SMC subunit of condensin II (24, 28, 29). In contrast to the SMC subunits SMC-4 and MIX-1 (9, 24), the accumulation of HCP-6 on metaphase chromosomes requires CENP-A (24, 29). However, during meiotic prophase, when condensin activity restructures the recombined bivalent chromosomes, HCP-6 localizes to chromosomes independently of CENP-A (24). Whether HCP-6 requires CENP-A to target to chromosomes during mitotic prophase is not clear. The signals obtained with antibodies to condensin subunits during prophase are weak. In addition, because it is difficult to determine whether proteins are present on chromosomes before they form distinct units, the condensation delay in CENP-A-depleted embryos complicates analysis of HCP-6 targeting. In light of our finding that the condensation defect in SMC-4-depleted embryos is more severe than that in CENP-A-depleted embryos, there are two possibilities for how HCP-6 contributes to mitotic chromosome formation: (i) HCP-6 is an integral subunit of condensin that targets to prophase chromosomes in the absence of CENP-A and contributes to their condensation, or (ii) HCP-6 is a specific adaptor that targets a subset of condensin to chromosomes in a CENP-A-dependent fashion during prophase, possibly accounting for the condensation defect in CENP-A-depleted embryos. To distinguish between these possibilities, we analyzed HCP-6-depleted embryos to determine whether condensation resembled that after depletion of SMC-4 or CeCENP-A. Kinetic analysis revealed that the condensation profile after depletion of HCP-6 is identical to that after depletion of SMC-4 (Fig. 2 B and C), and more severe than that resulting from depletion of CeCENP-A. This result suggests that, during mitotic prophase, HCP-6 is an integral subunit of condensin that targets to chromosomes independently of CENP-A.
Depletion of Aurora B Delays Secondary Condensation.
During mitotic entry, the mitotic kinase Aurora B phosphorylates histone H3 (30) and promotes removal of cohesin and remodeling complexes from chromatin (31, 32). However, the role of Aurora B in chromosome condensation is less clear. In budding yeast, vertebrate cells, and Xenopus extracts, Aurora B does not have a significant role in recruiting condensin to the chromosome arms or in prophase condensation (31–34). However, in human cells, Aurora B is required to recruit condensin to the region of the primary constriction and for the proper morphology of this chromosomal region (34). In C. elegans, where the entire chromosome has an architecture similar to that of the primary constriction in vertebrate cells, previous work has shown that the two SMC subunits of condensin fail to localize to metaphase chromosomes in embryos depleted of Aurora B (9, 35). Nevertheless, qualitative inspection did not suggest a major condensation defect (9, 18, 35).
To better understand its role in the condensation of holocentric chromosomes, we used our assay to characterize Aurora-B-depleted embryos. Consistent with prior work, primary condensation proceeded with normal kinetics (Fig. 2F). However, Aurora B-depleted embryos exhibited a delay in secondary condensation. In particular, the pronounced increase in the percentage of pixels below the 20% threshold, a hallmark of secondary condensation, was delayed by ≈65 s relative to controls.
The relatively mild condensation defect in Aurora-B-depleted embryos seemed at odds with a role for Aurora-B in targeting condensin to chromosomes. To reexamine this issue, we performed immunofluorescence using antibodies against SMC-4 in fixed embryos and used spinning disk confocal microscopy to examine the localization of a GFP fusion with F55C5.4, the C. elegans homolog of the hCAP-G2 subunit of condensin (36), in living embryos. By both methods, condensin localized to chromosomes beginning in prophase and persisting through metaphase in control as well as Aurora-B-depleted embryos (Fig. 4 and Movie 2, which are published as supporting information on the PNAS web site). We conclude that condensin targets to holocentric C. elegans chromosomes in Aurora-B-depleted embryos, consistent with their relatively normal condensation.
Discussion
A Quantitative Live Imaging Assay for Chromosome Condensation.
Live cell imaging, in combination with specific functional perturbations, is a powerful approach for the mechanistic dissection of cellular processes (37, 38). However, extracting meaningful kinetic measurements remains challenging, and methods to quantify large-scale changes in cellular architecture visible in live imaging data would enhance the utility of this approach. Here, we describe a simple, robust strategy to monitor the redistribution of nuclear GFP-histone signal during chromosome condensation. The principle underlying this method could also prove useful to analyze other dynamic processes, such as redistribution of the Golgi apparatus during the cell cycle or rearrangement of cortical components during assembly of the cytokinetic furrow.
Chromosome Condensation Is Temporally Biphasic in C. elegans.
Our analysis revealed that C. elegans chromosomes condense with biphasic kinetics, suggesting that compaction occurs in at least two discrete steps. Primary condensation converts diffuse chromatin into discrete linear chromosomes, and secondary condensation further compacts these chromosomes to shorter bar-shaped structures. Consistent with this idea, a recent study using EM and immunofluorescence to characterize prophase in fixed vertebrate cells reported two prominent classes of cells: (i) “middle prophase” cells containing well defined chromosomes ≈0.4–0.5 μm in diameter, and (ii) “late prophase” cells containing shorter chromosomes ≈0.8–1.0 μm in diameter (39). These two classes likely correspond to cells that have completed primary and secondary condensation, respectively, suggesting that biphasic kinetics will be a conserved aspect of condensation not limited to organisms with holocentric chromosomes.
Condensin Is Required to Form Distinct Chromosomes During the First Embryonic Division of the C. elegans Embryo.
Primary condensation fails in condensin-depleted C. elegans embryos, and, although some late compaction occurs, discrete chromosomes of normal structure are never observed. An early role for condensin is consistent with previous studies showing that compaction is delayed in the absence of condensin (9–11), but inconsistent with the proposal that condensin is only required in late prophase, when it is first observed to concentrate along the chromatid axis (39). The fact that individual chromosomes are not observed in depleted embryos suggests that condensin has a critical role in coupling the untangling of interphase chromatin to compaction in this system. This result is in contrast to recent studies in vertebrate cultured cells in which individual compacted chromosomes with reduced structural integrity were able to form after condensin depletion (10, 11) but is similar to results previously reported after depletion of condensin in Xenopus extracts (5). One explanation for these differences could be the nature of the remodeling in distinct experimental systems. In Xenopus extracts and the C. elegans embryo, the ability of sperm chromatin to be remodeled into mitotic chromosomes is being assayed. In contrast, in vertebrate somatic cells, the ability of previously formed chromosomes to continue undergoing cycles of condensation and decondensation is examined. It is possible that that these two processes have differing requirements for condensin function. For example, the chromosomes in vertebrate somatic cells could possess a scaffold that provides them with a structural memory to direct their compaction in the absence of condensin (1). Such a scaffold could be lacking when chromosomes reform from sperm chromatin.
Centromeric Chromatin Has a Critical Role in the Condensation of Holocentric Chromosomes.
The amplification of the structural role of centromeric chromatin in holocentric chromosomes allowed us to use our method to examine the role of CENP-A chromatin in condensation. Like condensin, CeCENP-A is required for primary condensation; chromatin compaction is delayed in CeCENP-A-depleted embryos, and well formed linear chromosomes are not observed at any point. However, the defect resulting from CeCENP-A depletion is markedly less severe than that after depletion of condensin; compaction initiates earlier, and spatially distinct chromatin masses, although abnormal in morphology, are formed. Interestingly, depletion of CeCENP-C, which abolishes the targeting of all known kinetochore components except CeCENP-A, resulted in only minor condensation defects. The comparison between the two depletion phenotypes strongly argues that CeCENP-A-containing chromatin has an intrinsic role in condensation that is independent of its role in kinetochore assembly.
How could CeCENP-A-containing chromatin, which is only a small fraction of total chromatin (≈5% based on our unpublished results), exert such a dramatic effect on primary condensation? One possibility is that centromeric chromatin locally concentrates condensin to initiate condensation. Alternatively, CENP-A-containing chromatin may have an organizational role that, in cooperation with condensin, helps structure the chromosome and ensure timely compaction (Fig. 2I). An enticing hypothesis is that the contribution of CENP-A-containing chromatin to condensation may serve to ensure its final placement on opposing surfaces of sister chromatids, a necessary condition for attachment of sister kinetochores to opposite spindle poles. Centromeric chromatin-mediated condensation may also provide structural integrity at the base of the kinetochore, allowing the efficient translation of forces generated by kinetochore–spindle interactions into chromosome movement.
Methods
Live Imaging.
Embryos were imaged by using a spinning disk confocal (McBain Instruments, Los Angles, CA) mounted on a Nikon TE2000e inverted microscope (Nikon Instruments, Melville, NY). Images were acquired by using a 60 × 1.4 N.A. Plan Apo objective lens with ×1.5 auxiliary magnification using an Orca ER CCD camera (Hamamatsu Photonics, Bridgewater, NJ) with 2 × 2 binning. Acquisition parameters, shutters, and focus were controlled by MetaMorph software (Universal Imaging, Downingtown, PA). The condensation parameter was measured by using three custom macros (available upon request).
RNAi.
L4 hermaphrodites of the C. elegans strain TH32 (expressing GFP-histone H2B and GFP-γ-tubulin) were injected with dsRNAs (Table 1, which is published as supporting information on the PNAS web site), incubated for 48 h at 20°C, and dissected to obtain recently fertilized depleted embryos. All depletions were confirmed by immunofluorescence and/or immunoblotting as described (26).
Supplementary Material
Acknowledgments
We thank R. Green, I. Cheeseman, R. Gassmann, and A. Maddox for helpful comments. P.S.M. is the Fayez Sarofim Fellow of the Damon Runyon Cancer Research Foundation (DRG-1808-04). K.O. is a Pew Scholar in the Biomedical Sciences. A.D. is the Connie and Bob Lurie Scholar of the Damon Runyon Cancer Research Foundation (DRS 38-04). This work was supported in part by National Institutes of Health Grant R01GM074215-01 and the Human Frontiers Science Program (to A.D.). K.O. and A.D. received additional support from the Ludwig Institute for Cancer Research.
Abbreviations
- NEBD
nuclear envelope breakdown
- SMC
structural maintenance of chromosomes.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gassmann R, Vagnarelli P, Hudson D, Earnshaw WC. Exp Cell Res. 2004;296:35–42. doi: 10.1016/j.yexcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Hirano T. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 3.Huang CE, Milutinovich M, Koshland D. Philos Trans R Soc London B Biol Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasmyth K, Haering CH. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 5.Hirano T, Kobayashi R, Hirano M. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 6.Hirano T, Mitchison TJ. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 7.Coelho PA, Queiroz-Machado J, Sunkel CE. J Cell Sci. 2003;116:4763–4776. doi: 10.1242/jcs.00799. [DOI] [PubMed] [Google Scholar]
- 8.Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MM, Sunkel CE. Curr Biol. 2001;11:295–307. doi: 10.1016/s0960-9822(01)00096-3. [DOI] [PubMed] [Google Scholar]
- 9.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. J Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- 11.Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Dev Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 12.Kline-Smith SL, Sandall S, Desai A. Curr Opin Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiato H, DeLuca J, Salmon ED, Earnshaw WC. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 14.Maddox PS, Oegema K, Desai A, Cheeseman IM. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 15.Blower MD, Karpen GH. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 17.Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura H, Cook PR. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manders EM, Kimura H, Cook PR. J Cell Biol. 1999;144:813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swedlow JR, Sedat JW, Agard DA. Cell. 1993;73:97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- 22.Oegema K, Hyman AA. WormBook. 2006 Jan 19; doi: 10.1895/wormbook.1.72.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagstrom KA, Meyer BJ. Nat Rev Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 24.Chan RC, Severson AF, Meyer BJ. J Cell Biol. 2004;167:613–625. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore LL, Roth MB. J Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore LL, Stanvitch G, Roth MB, Rosen D. Mol Cell Biol. 2005;25:2583–2592. doi: 10.1128/MCB.25.7.2583-2592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirano T. Curr Biol. 2005;15:R265–R275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 29.Stear JH, Roth MB. Genes Dev. 2002;16:1498–1508. doi: 10.1101/gad.989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, et al. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 31.Losada A, Hirano M, Hirano T. Genes Dev. 2002;16:3004–3016. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacCallum DE, Losada A, Kobayashi R, Hirano T. Mol Biol Cell. 2002;13:25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavoie BD, Hogan E, Koshland D. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono T, Fang Y, Spector DL, Hirano T. Mol Biol Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. Curr Biol. 2002;12:798–812. doi: 10.1016/s0960-9822(02)00820-5. [DOI] [PubMed] [Google Scholar]
- 36.Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 37.Rieder CL, Khodjakov A. Science. 2003;300:91–96. doi: 10.1126/science.1082177. [DOI] [PubMed] [Google Scholar]
- 38.Tsien RY. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. J Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.