Abstract
mRNA regulation is crucial for many aspects of metazoan development and physiology, including regulation of stem cells and synaptic plasticity. In the nematode germ line, RNA regulators control stem cell maintenance, the sperm/oocyte decision, and progression through meiosis. Of particular importance to this work are three GLD (germ-line development) regulatory proteins, each of which promotes entry into the meiotic cell cycle: GLD-1 is a STAR/Quaking translational repressor, GLD-2 is a cytoplasmic poly(A) polymerase, and GLD-3 is a homolog of Bicaudal-C. Here we report that the gld-1 mRNA is a direct target of the GLD-2 poly(A) polymerase: polyadenylation of gld-1 mRNA depends on GLD-2, the abundance of GLD-1 protein is dependent on GLD-2, and the gld-1 mRNA coimmunoprecipitates with both GLD-2 and GLD-3 proteins. We suggest that the GLD-2 poly(A) polymerase enhances entry into the meiotic cell cycle at least in part by activating GLD-1 expression. The importance of this conclusion is twofold. First, the activation of gld-1 mRNA by GLD-2 identifies a positive regulatory step that reinforces the decision to enter the meiotic cell cycle. Second, gld-1 mRNA is initially repressed by FBF (for fem-3 binding factor) to maintain stem cells but then becomes activated by the GLD-2 poly(A) polymerase once stem cells begin to make the transition into the meiotic cell cycle. Therefore, a molecular switch regulates gld-1 mRNA activity to accomplish the transition from mitosis to meiosis.
Keywords: cytoplasmic poly(A) polymerase, RNA regulation, mitosis/meiosis decision
Expression of mRNA is tightly regulated during metazoan development (1). One common mechanism of mRNA control relies on regulated polyadenylation. In the nucleus, poly(A) tails are added by a poly(A) polymerase (PAP) that acts on virtually all RNA polymerase II transcripts (2). However, in the cytoplasm, poly(A) tails are maintained or lengthened by a cytoplasmic PAP (cPAP) that acts specifically on a subset of mRNAs (3). These cPAPs, known as GLD-2 in metazoans (4), were discovered in Caenorhabditis elegans and Schizosaccharomyces pombe (4–6). A common biological function of GLD-2 has been inferred from null mutants in C. elegans (7) and molecular experiments in Xenopus (8, 9). In both cases, GLD-2 controls germ-line progression through meiosis. Furthermore, in C. elegans, GLD-2 controls the decision between mitosis and meiosis (7). In this work, we identify a direct target of GLD-2 in C. elegans.
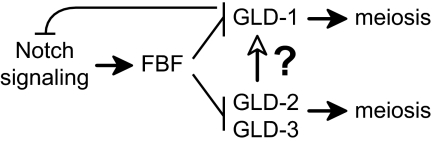
The mitosis/meiosis decision in C. elegans is controlled by Notch signaling and four broadly conserved RNA regulatory proteins (Fig. 1). Notch signaling and FBF (for fem-3 binding factor) are both required for maintenance of germ-line stem cells (10). FBF is an RNA-binding protein of the PUF (for Pumilio and FBF) family (11). Notch signaling activates transcription of the fbf-2 gene (12), and FBF represses gld-1 and gld-3 mRNAs (Fig. 1). Three gld genes (for germ-line development) promote entry into the meiotic cell cycle (7, 13). GLD-1 is a STAR RNA-binding protein and translational repressor (14, 15); GLD-2 is the catalytic subunit of a cPAP (4); and GLD-3 is a Bicaudal-C homolog that possesses five KH motifs and is predicted to bind RNA (4, 16). Like other cPAPs, nematode GLD-2 does not possess a recognizable RNA-binding domain (4). Instead, GLD-2 binds GLD-3, which stimulates its enzymatic activity in vitro (4). The GLD-2 and GLD-3 proteins appear to function together to promote entry into meiosis.
Fig. 1.
Regulatory circuit controlling the mitosis/meiosis decision. Both Notch signaling and FBF promote mitotic cell divisions, whereas the GLD proteins promote entry into the meiotic cell cycle. FBF negatively regulates both gld-1 and gld-3 mRNAs. GLD-1 and GLD-2/GLD-3 represent parallel branches of the regulatory circuit, because each can promote entry into meiosis in the absence of the other. This work tests the hypothesis that the gld-1 mRNA may be a direct target of the GLD-2 PAP (see text for details).
A key step in understanding how GLD-2 cPAP controls mRNAs is the identification of its direct targets. In this work, we present molecular data to demonstrate that the gld-1 mRNA is a direct target of GLD-2 cPAP. Consistent with our findings, genetic data suggest that gld-1 expression is controlled redundantly by GLD-2 and NOS-3 (17), which is a Nanos homolog (18). Identification of the gld-1 transcript as a direct GLD-2 target defines a mechanism for positive reinforcement in the circuitry controlling the mitosis/meiosis decision and suggests an attractive model for a regulatory switch from mitosis- to meiosis-promoting activity.
Results
GLD-2 Regulates gld-1 Poly(A) Tail Length.
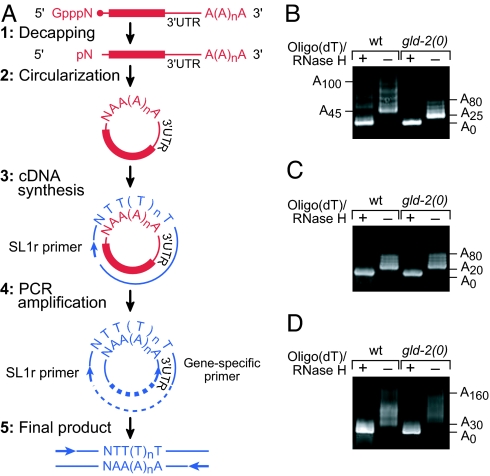
To identify target mRNAs of the GLD-2 PAP, we used a candidate gene approach. The gld-1 mRNA was a plausible candidate, because gld-1 and gld-2 both promote entry into meiosis (7). We first asked whether polyadenylation of gld-1 mRNA is dependent on GLD-2. Specifically, we compared the lengths of poly(A) tails on endogenous gld-1 mRNAs in wild-type animals and gld-2 null mutants by using a “circularization RT-PCR” (cRT-PCR) assay (Fig. 2A) (19). Total RNA was prepared from wild-type and gld-2(0) mutants and then decapped and ligated to generate circular RNAs. RT-PCR of these circular RNAs was then performed by using primers that flank the poly(A) tail: one primer was specific to sequences near the 3′ end of the mRNA of interest and the other primer was complementary to the trans-spliced leader present at the 5′ end of many C. elegans mRNAs (20). Poly(A) tail lengths were deduced from the lengths of PCR products and confirmed by sequencing of multiple cloned isolates. Our analysis focused on total mRNA prepared from larvae synchronized at the fourth larval stage (L4) because wild-type and gld-2(0) L4 larvae have morphologically similar germ lines (7). By contrast, adult gld-2(0) germ lines are severely abnormal (7) and unsuitable for comparison.
Fig. 2.
Poly(A) tail length of gld-1 mRNA is dependent on GLD-2. (A) Schematic of cRT-PCR (adapted from ref. 19). mRNAs are shown in red, and DNAs are shown in blue. (B) Gel analysis of poly(A) tail lengths of gld-1 mRNA in wild type and gld-2(0) mutants. As controls, samples were treated with RNase H in the presence of oligo(dT)12–18 to remove the poly(A) tail. The PCR products were resolved on a 5% gel. (C and D) Gel analyses of lag-1 mRNA (C) and tbg-1 mRNA (D), respectively.
The gld-1 poly(A) tail was shorter in gld-2(0) mutants than in wild type (Fig. 2B). In wild type, most gld-1 poly(A) tails were longer than ≈45 adenosine residues (A's), and they extended to ≈100 A's or longer, but in gld-2 mutants, most gld-1 poly(A) tails were shorter, averaging ≈25–30 A's. Treatment with oligo(dT)/RNase H before ligation and amplification showed that the deadenylated RNAs were of the same length in both wild type and gld-2(0). In contrast to gld-1 mRNA, the poly(A) tails of two control mRNAs, lag-1 and tbg-1, were of comparable length in wild type and gld-2(0) mutants (Fig. 2 C and D). LAG-1 and TBG-1 are ubiquitously expressed components of the Notch pathway and centrosomes, respectively (21, 22). Sequencing of the cloned cRT-PCR products confirmed our findings. We also compared gld-1 poly(A) tail lengths in wild-type and gld-2(0) adults and obtained similar results (data not shown). We conclude that the gld-1 poly(A) tail is shorter in animals lacking the GLD-2 PAP and that gld-1 mRNA may be a substrate for the GLD-2 enzyme.
GLD-2 Controls gld-1 Expression.
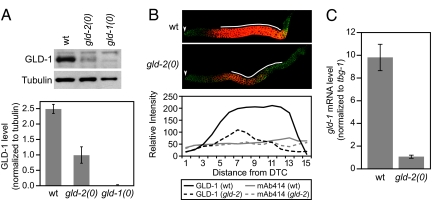
An extended poly(A) tail can enhance translation and increase mRNA stability (2). To investigate whether the abundance of GLD-1 protein was affected by GLD-2 activity, we compared GLD-1 protein levels in wild-type and gld-2(0) L4 larvae by Western blotting and immunohistochemistry (Fig. 3A and B). On immunoblots of whole-worm lysates, the amount of GLD-1 was 3-fold higher in wild type than in gld-2 mutants, using tubulin for normalization (Fig. 3A). By immunocytochemistry, more GLD-1 protein was observed in wild-type than in gld-2(0) germ lines, using a nuclear pore protein as an internal control (Fig. 3B). The difference in GLD-1 abundance was particularly dramatic in the region containing early stages of meiotic prophase nuclei. Previous work, which focused on adults instead of L4 larvae, suggested that GLD-1 levels were unaffected by gld-2(0) (17). However, adult gld-2(0) germ lines are defective in oogenesis (7), which complicates any comparison with wild type. We conclude that GLD-2 is critical for GLD-1 protein accumulation during the L4 stage of development.
Fig. 3.
GLD-2 regulates gld-1 expression. (A Upper) Western blot of proteins prepared from wild-type, gld-2(0), and gld-1(0) L4 larvae. (Lower) GLD-1 levels in three independent experiments after normalizing to level of tubulin. (B Upper) Germ lines dissected from wild-type and gld-2(0) L4 larvae and stained with anti-GLD-1 (red) and Mab-414 (green), which highlights nuclear pores. Germ lines were treated identically, and confocal images were taken with the same settings at the same magnification. The arrowheads indicate distal ends of germ lines. (Lower) Quantitation of protein abundance in wild type and gld-2(0) mutants. Solid black line, GLD-1 in wild type; dashed black line, GLD-1 in mutant; solid gray line, Mab-414 in wild type; dashed gray line, Mab-414 in mutant. x axis, the distance from the distal tip cell (DTC) in increments of 2 or 3 rows of germ cells. This profile is based on quantitation of the Upper images; similar profiles have been obtained in multiple independent experiments. (C) Real-time PCR analysis of gld-1 mRNA when normalized to tbg-1 mRNA.
To investigate whether the abundance of gld-1 mRNA was affected by GLD-2 activity, we compared gld-1 transcript levels in wild-type and gld-2(0) L4 larvae by quantitative RT-PCR (Fig. 3C). The gld-1 mRNA level was 9-fold lower in gld-2 mutants than in wild type when normalized to tbg-1 mRNA. We conclude that GLD-2 also affects gld-1 mRNA stability.
We next asked whether GLD-3 is critical for GLD-1 accumulation. To this end, we compared GLD-1 protein levels in L4 larvae from either wild type or gld-3(q730) mutants. In this case, no effect was observed on either Western blots or by immunocytochemistry (data not shown). We conclude that unlike GLD-2, GLD-3 is not crucial for activating gld-1 expression.
gld-1 mRNA Is Associated with GLD-2/GLD-3 PAP.
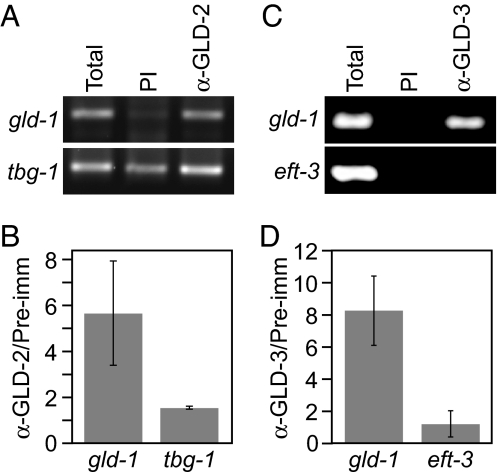
We next asked whether the GLD-2 and GLD-3 proteins interact physically with gld-1 mRNA in vivo. Specifically, we incubated extracts that had been prepared from wild-type animals with GLD-2-specific or GLD-3-specific antibodies, isolated the immunoprecipitate (IP), and used both RT-PCR and quantitative real-time PCR to assess the presence of gld-1 and control mRNAs (Fig. 4). By RT-PCR, gld-1 mRNA was enriched in the GLD-2 IP compared with a precipitate with preimmune serum (PI), whereas tbg-1 mRNA was present in both preimmune and GLD-2 IPs (Fig. 4A). By real-time PCR, gld-1 mRNA was enriched ≈6-fold in the anti-GLD-2 IP versus preimmune serum (Fig. 4B; range 3- to 8-fold in multiple experiments); tbg-1 mRNA was not enriched (Fig. 4B). The simplest explanation is that gld-1 mRNA associates specifically with GLD-2 protein in worm extracts.
Fig. 4.
GLD-2 and GLD-3 are physically associated with gld-1 mRNA. (A) Coimmunoprecipitation of gld-1 mRNA using GLD-2 antibody but not with preimmune serum (PI). (B) Real-time PCR analysis of gld-1 and tbg-1 mRNAs. (C) Coimmunoprecipitation of gld-1 mRNA using GLD-3 antibody but not with preimmune serum (PI). (D) Real-time PCR analysis of gld-1 and eft-3 mRNAs.
gld-1 mRNA was also enriched in IPs with GLD-3 antibody compared with preimmune serum. The negative control eft-3 mRNA was not detected in GLD-3 IP (Fig. 4C). eft-3 encodes a translation elongation factor 1α that is expressed in the germ line (23). By real-time PCR, gld-1 mRNA was enriched ≈8-fold in the anti-GLD-3 IP versus preimmune serum (Fig. 4D; range 6- to 10-fold in multiple experiments); eft-3 mRNA was not enriched (Fig. 4D). We conclude that gld-1 mRNA associates specifically with GLD-3 protein in worm extracts.
Discussion
GLD-2 PAP Activates Expression of its Target mRNA.
We have identified gld-1 mRNA as a direct target of the GLD-2 PAP. Importantly, GLD-2 activates gld-1 expression. In gld-2 null mutants, both gld-1 mRNA and GLD-1 protein are less abundant than in wild-type animals. We cannot exclude the formal possibility that GLD-2 controls transcription. However, GLD-2 is a cytoplasmic protein with PAP activity (4), and its Xenopus counterpart regulates cytoplasmic mRNAs (8, 9). Therefore, a more likely interpretation is that GLD-2 controls RNA stability and perhaps translation.
Both gld-1 mRNA and GLD-1 protein are reduced in gld-2 mutants, but the reduction is more severe for the mRNA than for the protein (9-fold versus 2.5-fold). This difference can be explained in various ways. The simplest explanation is that levels of mRNA and protein may be differentially subject to indirect effects of GLD-2 loss. For example, the GLD-1 protein, but not the RNA, may be stabilized in gld-2 mutants because of effects on some other regulator. A more intriguing idea is that GLD-2 is involved in translational repression in addition to its effect on mRNA stability. By this scenario, the less abundant gld-1 mRNA would be translationally more active in gld-2 mutants.
The ability of GLD-2 to activate expression of target mRNAs is similar to findings for its closely related Xenopus homolog: Xl-GLD-2 activates expression of cyclin B mRNA (8, 9). By contrast, the more distant homolog in Saccharomyces cerevisiae, known as TRF4, polyadenylates selected RNAs in the nucleus and targets them for destruction (24). We suggest that metazoan GLD-2 PAPs activate expression of their target mRNAs in the cytoplasm.
gld-1 Activation by GLD-2 Reinforces Decision to Enter Meiosis.
Entry into meiosis is controlled by two redundant pathways, which together provide the major control for the transition from mitosis to meiosis (7) (Fig. 1). In one branch is the GLD-1 translational repressor, which likely promotes entry into meiosis by down-regulating mRNAs required for mitosis. In the other branch is the GLD-2 translational activator, which likely promotes entry into meiosis by activating mRNAs required for meiosis. Also in the second branch is GLD-3, which appears to act with GLD-2 to generate a heterodimeric PAP (4, 13) (see below).
In single mutants lacking gld-1, gld-2, or gld-3, germ cells are capable of entering meiosis, although entry does not occur normally (7). Indeed, the switch from mitosis to meiosis appears to be delayed in gld-2 and gld-3 null mutants (13). A molecular understanding of the switch requires identification of direct targets of both the GLD-1 repressor and GLD-2 activator. Numerous direct targets of GLD-1 translational repression are known, but none to date control the mitosis/meiosis decision (15, 25). This work identifies gld-1 mRNA as a direct target of the GLD-2 activator. However, gld-1 mRNA cannot be the only GLD-2 target, because GLD-2 is able to promote entry into meiosis in the absence of GLD-1. Instead, we suggest that gld-1 activation by GLD-2 strengthens the switch from mitosis to meiosis, an idea that is consistent with the switch delay in gld-2 null mutants.
Do GLD-2 and GLD-3 Work Together to Activate gld-1 mRNA?
A remaining question is whether GLD-3 functions together with GLD-2 to polyadenylate gld-1 mRNA. Several lines of evidence support that idea. First, gld-2 and gld-3 null mutants are similar with respect to their effects on the decision between mitosis and meiosis. In gld-2 and gld-3 single mutants, the mitotic region is enlarged to an equivalent extent, suggesting that the two proteins play a similar role in the transition into meiosis (13). As double mutants with gld-1, they also have similar effects: both gld-1 gld-2 and gld-1 gld-3 double mutants have tumorous germ lines (7, 13). Second, the GLD-2 and GLD-3 proteins coimmunoprecipitate from worm extracts and therefore appear to be physically associated in vivo (13). Third, GLD-3 enhances GLD-2 enzymatic activity in vitro (4). These results, when taken together, support the model that GLD-2 and GLD-3 work together as a heterodimeric enzyme to promote meiosis. Consistent with that idea, gld-1 mRNA immunoprecipitates with antibodies specific to either GLD-2 or GLD-3 (this work). The simplest explanation is that GLD-2 and GLD-3 function together to polyadenylate the gld-1 transcript.
Although GLD-2 and GLD-3 are likely to work together to control entry into meiosis, they do not have equivalent roles in gld-1 mRNA activation. Whereas GLD-1 levels decreased in gld-2 null mutants, they remained normal in gld-3 null mutants. Molecular redundancy appears to be the rule in the circuitry controlling the mitosis/meiosis decision (7, 12). Moreover, GLD-2 binds additional RNA-binding proteins whose functions are not yet known (L. Wang and J.K., unpublished data). We suggest that GLD-2 can accomplish gld-1 polyadenylation with either GLD-3 or another RNA-binding protein (Protein X in Fig. 5Lower) that remains unknown.
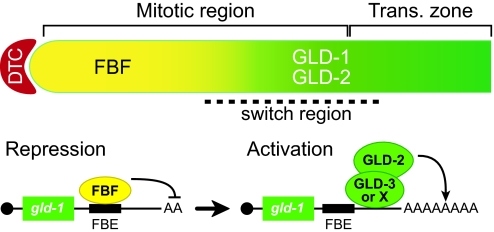
Fig. 5.
Model for molecular switch controlling gld-1 mRNA. (Upper) Diagram of distal germ line. DTC is the somatic distal tip cell that provides the niche for germ-line stem cells; the mitotic region spans the extent of any mitotically dividing germ cells; the transition zone includes nuclei in early stages of meiotic prophase; and the switch region contains nuclei transitioning from mitotic to meiotic cell cycle. Solid yellow, FBF promotes continued mitotic divisions; solid green, GLD proteins promote meiosis; gradient from yellow to green, transition from mitosis to meiosis. (Lower) FBF repression of gld-1 mRNA (Left), and GLD-2 activation of same mRNA (Right). FBE, FBF-binding element in the gld-1 3′ untranslated region. Figure is not to scale.
Dual Control of gld-1 mRNA Suggests Existence of a Molecular Switch.
The identification of gld-1 mRNA as a direct target of the GLD-2 PAP is of particular interest because gld-1 mRNA is also a direct target of repression by the PUF protein FBF (26). Fig. 5 presents a simple model for this dual control of gld-1 mRNA as germ cells move from the niche provided by the distal tip cell and progress proximally. As germ cells escape Notch signaling and FBF repression, they leave the mitotic cell cycle and enter meiosis. FBF promotes continued mitotic divisions, in part by repressing gld-1 mRNA (26). GLD-2 promotes entry into meiosis, in part by activating the same mRNA. The mechanism by which gld-1 mRNAs switch from FBF-dominated repression to a GLD-2-dominated activation is currently unknown.
One attractive possibility is that FBF and GLD-2 are components of a molecular switch. By this scenario, FBF would repress mRNAs in cells within the stem cell niche, which lack GLD-2, but FBF might also mark those same mRNAs for activation once cells had left the niche and GLD-2 became available. Consistent with that idea, FBF, which is encoded by two nearly identical genes, fbf-1 and fbf-2 (11), acts genetically in the GLD-2/GLD-3 branch of control: the germ line of gld-1; fbf-1 fbf-2 triple mutants is tumorous (26), an effect similar to that seen in gld-1; gld-2 and gld-1; gld-3 double mutants (7, 13). Regardless, we conclude that the GLD-2 PAP activates the same mRNA that FBF represses and that both controls are critical for the germ-line switch from mitosis to meiosis.
Methods
Nematode Strains and Methods.
All strains were maintained at 20°C as described (27). Strains included wild-type C. elegans (N2) as well as gld-2(q497), which is a nonsense mutation and putative null (4), and gld-3(q730), which is a deletion mutation and putative null (16). gld-2(q497) was balanced with hT2[qIs48]; gld-3(q730) was balanced with mIn1[mIs14 dpy-10(e128)].
cRT-PCR.
Total RNA from a mid-L4-staged population of animals was isolated by using TRIzol (Invitrogen, Carlsbad, CA). gld-2(q497) null animals were collected with a COPAS Biosort (Union Biometrica, Somerville, MA) from a population of gld-2(q497)/hT2[qIs48] animals, as described by the manufacturer. cRT-PCR was performed as described in ref. 19 with a minor modification: 4 μg of total RNA was used for the oligo(dT)/RNase H treatment, decapping, and circularization steps. SL1 reverse primer (SL1r) or SL2 reverse primer (SL2r) were used for reverse-transcriptase reaction with SuperScript III reverse transcriptase (Invitrogen). Nested PCRs were performed and analyzed by using 5% Criterion TBE gels (Bio-Rad, Hercules, CA). Five independent clones were obtained from each experiment and sequenced. The primer sequences are available from the authors upon request.
Western Blots and Immunocytochemistry.
Whole worm lysates prepared from mid- to late-L4 hermaphrodites were analyzed by both immunoblotting and immunohistochemistry. Anti-GLD-1 antibodies were used at a 1:750 dilution in Western blot analysis and at a 1:100 dilution in immunohistochemistry. Gonad dissections (28) and Western blots (29) were performed as described. The anti-α-tubulin antibody (Sigma, St. Louis, MO) was diluted 1:20,000, and Mab-414 (Berkeley Antibody, Richmond, CA) was diluted 1:400. Cy-3-labeled donkey anti-rabbit and FITC-labeled donkey anti-mouse (Jackson Laboratories, Bar Harbor, ME) were used as secondary antibodies (dilution 1:500) in immunohistochemistry. AP-conjugated donkey anti-rabbit (Pierce, Rockford, IL) and HRP-conjugated donkey anti-mouse (Jackson Laboratories) antibodies were used for Western blots (dilution 1:40,000). Confocal images were obtained on a Bio-Rad MRC1024 confocal microscope. Fluorescence was quantified by using National Institutes of Health ImageJ software as described (12).
Immunoprecipitations.
N2 worms were grown on standard NGM agar plates and collected by centrifugation, then washed several times in M9 buffer, with final wash in PBS with 1 μl/ml RNaseOUT (Invitrogen) and an EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN). Crude extract was then prepared by lysing the animals by using a French press at 19,000 psi two times. Samples were then centrifuged at 200 × g before being stored at −80°C and again at 16,000 × g immediately before use. Trisacryl-immobilized protein-A beads (Pierce) bound to anti-GLD-2 (rabbit α-GLD-2) (4) or preimmune controls were equilibrated with PBS and incubated overnight at 4°C with 500-μg extracts for each immunoprecipitation. Beads were subsequently washed five times with 0.5 ml of PBS. RNA was eluted from beads by extraction with TRIzol (Invitrogen) and solubilized in 20 μl of diethylpyrocarbonate water. Anti-GLD-3 (rb156) (16) or preimmune antibodies were first incubated for 1 h at 4°C with extracts supplemented with 1% Nonidet P-40 and subsequently captured on protein-A beads (Roche) for another hour before washing beads five times in 1× PBS/1% Nonidet P-40.
RT-PCR and Quantitative Real-Time PCR.
Reverse-transcriptase reactions were performed by using cDNA cloning primer (Integrated DNA Technologies, Inc., Coralville, IA) and SuperScript III reverse transcriptase (Invitrogen) with all coimmunoprecipitated samples or 5 μg of total input RNA. The reverse-transcription reaction (1 μl) was used for PCR with gene-specific primers for 30 cycles with an annealing temperature of 57°C (gld-1) and 53°C (tbg-1). RT-PCR products (40%) were analyzed with 2% agarose gel. We used real-time RT-PCR to quantify the mRNA enrichment in immunoprecipitated samples. Real-time RT-PCR was performed in 40 cycles of 95°C for 15 s and 60°C for 1 min by using the SmartCycler System (Cepheid, Sunnyvale, CA). The set of primers for each gene preferentially span an intron and sizes range from 100 to 150 bp, and gene-specific fluorogenic probes were obtained from Integrated DNA Technologies. The TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) was used according to the manufacturer's guidelines. The primer and probe sequences are available from the authors upon request.
Acknowledgments
We thank members of the J.K. and M.W. laboratories for comments on the manuscript, J. W. Pike for advice on using the Cepheid SmartCycler System, and E. B. Goodwin for the GLD-1 antibody. J.K. and M.W. are supported by the National Institutes of Health; J.K. is an Investigator of the Howard Hughes Medical Institute. C.R.E. is supported by the Max Planck Society.
Abbreviations
- GLD
germ-line development
- FBF
fem-3 binding factor
- PAP
poly(A) polymerase
- cPAP
cytoplasmic PAP
- IP
immunoprecipitate
- cRT-PCR
circularization RT-PCR
- L4
fourth larval stage.
Footnotes
The authors declare no conflict of interest.
References
- 1.Thompson B, Wickens M, Kimble J. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Woodbury, NY: Cold Spring Harbor Lab Press; 2007. in press. [Google Scholar]
- 2.Edmonds M. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 3.Richter JD. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Woodbury, NY: Cold Spring Harbor Lab Press; 2000. pp. 785–805. [Google Scholar]
- 4.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 5.Read RL, Martinho RG, Wang S-W, Carr AM, Norbury CJ. Proc Natl Acad Sci USA. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR III, Russell P. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- 7.Kadyk LC, Kimble J. Development (Cambridge, UK) 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- 8.Barnard DC, Ryan K, Manley JL, Richter JD. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. WormBook. 2005 Aug 15; doi: 10.1895/wormbook.1.13.1. www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 12.Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Eckmann CR, Crittenden SL, Suh N, Kimble J. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones AR, Francis R, Schedl T. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 15.Jan E, Motzny CK, Graves LE, Goodwin EB. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckmann CR, Kraemer B, Wickens M, Kimble J. Dev Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- 17.Hansen D, Wilson-Berry L, Dang T, Schedl T. Development (Cambridge, UK) 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- 18.Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- 19.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsen TW. Annu Rev Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 21.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. Development (Cambridge, UK) 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 22.Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maciejowski J, Ahn JH, Cipriani PG, Killian DJ, Chaudhary AL, Lee JI, Voutev R, Johnsen RC, Baillie DL, Gunsalus KC, et al. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson JT. Curr Biol. 2005;15:R635–R638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Lee M-H, Schedl T. Genes Dev. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 27.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee M-H, Hook B, Lamont LB, Wickens M, Kimble J. EMBO J. 2006;25:88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E, Lane D. Antibodies, A Laboratory Manual. Woodbury, NY: Cold Spring Harbor Lab Press; 1988. [Google Scholar]