Abstract
In comparison with CD4+ regulatory T cells, the generation and function of immunomodulatory CD8+T cells is less well defined. Here we describe the existence of regulatory anti-Kb-specific CD8+ T cells that are rendered tolerant during neonatal life via antigen contact exclusively on keratinocytes. These regulatory T cells maintain tolerance during adulthood as they prevent Kb-specific graft rejection by naïve CD8+ T cells. Third-party immune responses remain unaffected. Up-regulation of TGF-β1 and granzyme B in the regulatory CD8+ T cell population suggests the involvement of these molecules in common suppressive pathways shared with CD4+ regulatory T cells. In summary, CD8+ regulatory T cells can be induced extrathymically through antigen contact on neonatally accessible parenchymal cells and maintain tolerance throughout adult life.
Keywords: immune regulation, parenchymal cells, tolerance
The capacity of the immune system for specifically recognizing and eliminating an apparently limitless variety of foreign invaders by the enormous diversity of antigen-specific receptors on B and T lymphocytes has to be balanced by mechanisms preventing reactivity against self-antigens. Several tolerance mechanisms are operating in parallel under physiological conditions for silencing of T cells during their development in the thymus or in the periphery. Whereas in the past cell-intrinsic processes leading to deletion (1) or inactivation (2) of autoreactive T cells were regarded as the most important tolerance mechanisms, increased attention is given now to dominant forms of tolerance in which regulatory T (Treg) cells control autoreactive T cells that have escaped deletion in the thymus. These Treg cells are classified into naturally occurring Treg cells (3) and those that can be induced by suboptimal antigenic stimulation (4). Naturally occurring CD4+ CD25+ Treg cells are believed to be generated in the thymus, possibly by recognizing self-antigen on epithelial cells (5, 6). Such Treg cells have been shown to need for their survival in the periphery the presence of the respective self-antigen (7). Besides these most prominent CD4+ CD25+ Treg cells, CD8+ T cells have also been reported to be involved in the control of T cell responses (8–15). However, such CD8+ Treg cells are not well characterized yet. It is unknown, for example, whether naturally occurring CD8+ Treg cells are exclusively of thymic origin and represent a separate lineage as discussed for CD4+ CD25+ Treg cells (16).
The neonatal life is a unique developmental stage in which the peripheral T lymphocyte pool is evolving and in which newly generated T lymphocytes are extensively migrating through nonlymphoid tissues including the skin (17). We have previously shown that this tissue accessibility results in tolerance induction in neonatal mice when naïve T cells meet the respective self-antigen on parenchymal cells. CD8+ T cells with specificity for the MHC class I antigen Kb [Désiré T cell receptor (Des-TCR)] were rendered tolerant by recognition of the Kb antigen exclusively expressed on keratinocytes (2.4KerIV-Kb) (17). This type of tolerance induction was restricted to the neonatal life and was not found during adulthood. The observation was in agreement with the view that trafficking of naïve T cells through adult nonlymphoid tissue is restricted. Yet adult double-transgenic Des-TCR × 2.4KerIV-Kb mice are tolerant toward Kb-positive skin and tumor grafts. Therefore, the question was raised whether Treg cells are induced in the skin of neonatal double-transgenic Des-TCR × 2.4KerIV-Kb mice and maintain tolerance during adulthood when new thymic emigrants cannot reach the sessile Kb antigen in the skin.
Here we report that the observed tolerance in Des-TCR × 2.4KerIV-Kb mice is based on the induction of long-lived CD8+ Treg cells in the neonate. These CD8+ Treg cells prevent rejection of Kb-positive grafts by naïve anti-Kb CD8+ T cells independent of CD4+ T cells. Thus, naturally occurring CD8+ Treg cells can be induced extrathymically by parenchymal cells like keratinocytes and maintain tolerance throughout the adult life. This mechanism could assure tolerance against tissue-specific antigens that are not sufficiently highly expressed for cross-presentation by dendritic cells in the regional lymph nodes (18).
Results
Long-Lived Tolerant CD8+ T Cells Can Be Induced Neonatally in the Absence of CD4+ T Cells.
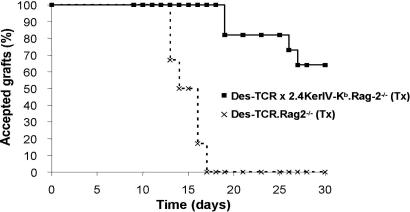
A dominant tolerance mechanism in double-transgenic Des-TCR × 2.4KerIV-Kb mice would require that T cells that had been rendered tolerant to Kb in the tolerance-sensitive period of the first 2 weeks of life (17) persist and interfere with the reactivity of naïve, Kb-specific T cells when confronted with a Kb-positive graft. To test whether neonatally generated tolerant CD8+ T cells are long-lived, Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice were thymectomized (Tx) at day 15 and tested for tolerance at 8 weeks of age by grafting C57BL/6 tail skin on their lateral thoracic wall. The majority of these mice accepted the Kb-positive skin graft, whereas all Tx, single-transgenic Des-TCR.Rag-2−/− mice rejected the transplants (Fig. 1). This finding was confirmed in a second test system in which 2 × 105 cells of the syngeneic tumor P815 transfected with Kb and B7 (P815.Kb.B7) was used as a Kb-positive graft (Table 1). The tolerance of the Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice was not due to deletion of the Kb-reactive T cells during antigen encounter on keratinocytes in the neonatal life as these mice showed no difference in absolute number and frequency of Des-TCR+ CD8+ T cells in comparison to the single-transgenic Tx Des-TCR.Rag-2−/− (5.0 ± 4.0 × 105 vs. 5.3 ± 4.2 × 105 cells per spleen) at the time of grafting. In addition, we could not detect any difference in the expression level of Des-TCR, CD8α, and CD8β on T cells isolated from both types of mice before or after tumor challenge (Fig. 5, which is published as supporting information on the PNAS web site, and data not shown). These results show that Des-TCR+ CD8+ T cells rendered tolerant during neonatal life are still present during adulthood. Furthermore, we can conclude that this CD8+ T cell tolerance can be induced in the absence of CD4+ T cells because the mice were Rag-2-deficient and therefore contained only Des-TCR+ CD8+ T cells.
Fig. 1.
Keratinocytes establish long-lived CD8+ T cell tolerance during the neonatal phase in the absence of CD4+ T cells. Shown is the acceptance of Kb-positive skin grafts on tolerant mice. Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− (n = 9; ■) and Tx Des-TCR.Rag2−/− (n = 6; ×) mice were grafted with C57BL/6 tail skin on their lateral thoracic wall. The transplants were monitored for 30 days. Results were confirmed in three independent experiments.
Table 1.
P815.Kb.B7 tumor growth in Kb-tolerant mice
| Group | Transgenic mice | Incidence of tumor growth (P815.Kb.B7) | Percentage |
|---|---|---|---|
| 1 | Rag-2−/− | 5/5 | 100 |
| 2 | Des-TCR.Rag-2−/− (Tx) | 1/23 | 4.3 |
| 3 | Des-TCR × 2.4KerIV-Kb.Rag-2−/− (Tx) | 28/36 | 77.8 |
A total of 2 × 105 P815.Kb.B7 tumor cells were inoculated s.c. into Rag-2−/−, Tx Des-TCR.Rag-2−/−, and Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice. Mice were assessed for tumor growth between days 10 and 18 after injection.
Neonatally Induced Tolerant CD8+ T Cells Prevent Graft Rejection in the Adult Mouse by Naïve CD8+ T Cells Bearing the Same TCR.
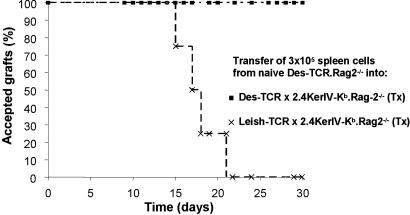
To investigate whether the CD8+ T cells in the tolerant animals could influence the reactivity of naïve Des-TCR+ CD8+ T cells we first determined the number of T cells from Des-TCR.Rag-2−/− animals that are needed to reject a Kb-positive skin graft. For the respective T cell transfer studies we used as a control group day-15 Tx 2.4KerIV-Kb.Rag-2−/− animals transgenic for an anti-Leishmania TCR (Leish-TCR) expressed on CD8+ T cells. Because this TCR is not cross-reactive with the Kb antigen Leish-TCR transgenic mice cannot respond to the Kb antigen. Transfer of 3 × 105 spleen cells of Des-TCR.Rag-2−/− mice containing 3 × 104 Des-TCR+ CD8+ T cells caused the rejection of C57BL/6 skin grafts within 21 days in all grafted mice (Fig. 2). In contrast, day-15 Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− tolerant mice that had received the same number of naïve Des-TCR+ CD8+ T cells accepted the C57BL/6 skin grafts. Again, these results were confirmed by monitoring the growth of the P815.Kb.B7 tumor in the respective animals (Table 2).
Fig. 2.
Regulation of naïve Des-TCR+ CD8+ T cells in tolerant mice. Acceptance of Kb-positive C57BL/6 skin grafts on Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− (n = 5; ■) but not on Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− (n = 4; ×) mice after i.v. transfer of 3 × 105 spleen cells from naïve Des-TCR.Rag2−/− mice. The transplants were followed up for 30 days.
Table 2.
P815.Kb.B7 tumor growth in tolerant mice after transfer of naïve Des-TCR+ CD8+ T cells
| Group | Transgenic mice | Incidence of tumor growth (P815.Kb.B7) | Percentage |
|---|---|---|---|
| 1 | Rag-2−/− (no transfer) | 5/5 | 100 |
| 2 | Leish-TCR × 2.4KerIV-Kb.Rag-2−/− (Tx) | 1/18 | 5.5 |
| 3 | Des-TCR × 2.4KerIV-Kb.Rag-2−/− (Tx) | 24/25 | 96 |
Growth of s.c. inoculated P815.Kb.B7 tumors in Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice but not in Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− mice after transfer of 3 × 105 spleen cells from naïve Des-TCR.Rag-2−/− mice. P815.Kb.B7 tumor cells were injected, and mice were assessed for tumor growth as described in Table 1.
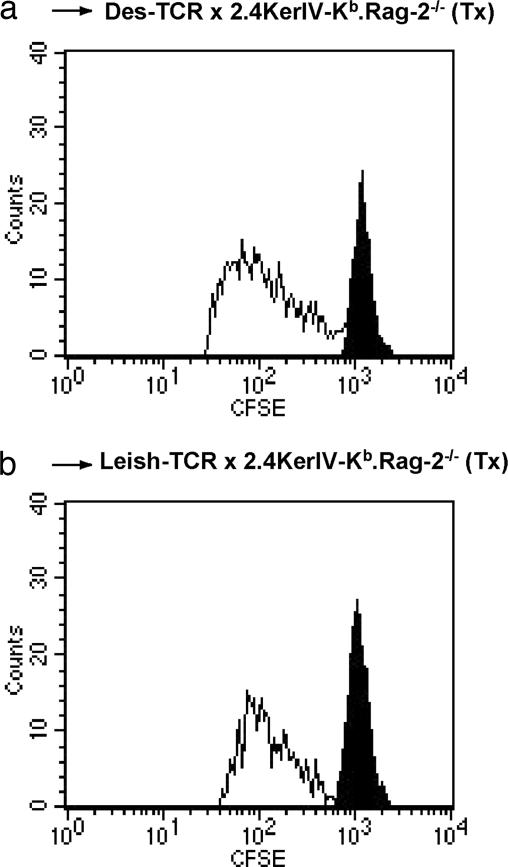
Lymphopenia-induced proliferation has been reported to expand and influence the reactivity of T cells transferred into a lymphopenic environment (19). To exclude the possibility that the difference in reactivity of the naïve Des-TCR+ CD8+ T cells transferred into the tolerant and nontolerant mice was due to differential lymphopenia-induced proliferation, spleen cells of Des-TCR.Rag-2−/− mice were carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled and transferred into the respective Tx mice. The same degree of expansion was observed in the Kb-tolerant and nontolerant mice (Fig. 3). Therefore, we conclude that Des-TCR+ CD8+ T cells from the double-transgenic, tolerant Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice do not inhibit lymphopenia-induced proliferation of naïve Des-TCR+ CD8+ T cells but regulate their capacity to reject Kb-positive grafts.
Fig. 3.
Tolerant T cells do not inhibit lymphopenia-induced division of naïve Des-TCR+ CD8+ T cells. A total of 3 × 105 CFSE-labeled spleen cells from naïve Des-TCR.Rag2−/− mice were transferred i.v. into Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice (a) and Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− mice (b). After 4 days, total lymph node cells pooled from two or three individual mice were analyzed by flow cytometry for CFSE-positive cells after gating on Des-TCR+ CD8+ T cells. The filled peak indicates the staining intensity of undivided cells. Representative data from three independent experiments are shown.
Third-Party Responses Are Not Affected by Kb-Specific Des-TCR+ CD8+ Treg Cells.
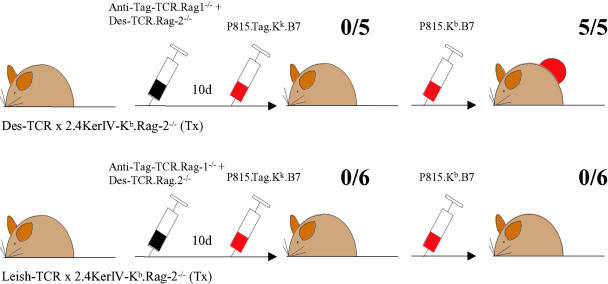
After having shown that the tolerant Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice contain a CD8+ Treg population that inhibits the function of naïve Des-TCR+ CD8+ T cells we investigated whether these CD8+ Treg cells interfere only with anti-Kb responses or have a nonspecific mode of action. A total of 3 × 105 spleen cells from TCR-transgenic Rag-1−/− mice (TCR 8) specific for the SV40 large T antigen (Tag) and from Des-TCR.Rag-2−/− mice were transferred into the above-mentioned nontolerant and tolerant animals. Both CD8+ T cell populations were of the H-2k haplotype. After T cell transfer all recipient mice received s.c. syngeneic P815 tumor cells expressing Tag, the H-2 restriction antigen Kk, and B7 (P815.Tag.Kk.B7). All recipient mice rejected the tumor graft independent of whether they were tolerant to the Kb antigen (Fig. 4). To learn whether a third-party CD8+ T cell response would override Kb tolerance, the same mice were inoculated with P815.Kb.B7 tumor cells 3 weeks later. The Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− mice rejected this second tumor graft. In contrast, tumor growth was observed in all Kb-tolerant Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice. These observations indicate that the Tag-specific response against the first tumor graft did not affect the regulation of the Kb-reactive CD8+ T cells. To confirm the Kb specificity of the Des-TCR+ CD8+ T cell regulation an in vivo cytotoxic T cell assay was performed as a second test system. Control and Kb-tolerant mice received 3 × 105 Tag-specific spleen cells from TCR 8 transgenic mice and were primed with dendritic cells (H-2k) loaded with Tag peptide. Spleen cells from CBA mice (H-2k) loaded with Tag peptide or untreated and differentially labeled with CFSE were used as target cells. Similar Tag-specific kill was observed in Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− mice and Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice (specific kill of 17.6 ± 12.6% vs. 30.6 ± 20.6%; P = 0.14). Thus, the regulation by Des-TCR+ CD8+ Treg cells is limited to Kb-specific responses.
Fig. 4.
Third-party immune reactivity remains intact within the Kb-tolerant mice. Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− and Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− mice received i.v. a mixture of naïve CD8 T cells from TCR-transgenic Rag1−/− mice reacting against Tag and from Des-TCR.Rag2−/− mice (3 × 105 spleen cells from each donor). All mice were challenged s.c. with a Tag-transfected P815 tumor (P815.Tag.Kk.B7). Then, after rejection of this tumor was seen in all animals, the P815.Kb.B7 tumor was inoculated into the same mice. Tumor growth was observed in five of five of the Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice but in none of the Tx Leish-TCR × 2.4KerIV-Kb.Rag-2−/− mice.
Des-TCR+ CD8+ Treg Cells Up-Regulate TGF-β1 and Granzyme B Expression.
To understand whether CD4+ CD25+ Treg cells and the tolerant Des-TCR+ CD8+ T cell population with regulatory capacity share marker molecules and possible regulatory pathways, gene expression of respective genes was analyzed. CD8+ T cells from Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice that had accepted the P815.Kb.B7 tumor and Tx Des-TCR.Rag-2−/− animals that had rejected the P815.Kb.B7 tumor were sorted and used for RNA isolation. Quantitative RT-PCR data for candidate gene expression in both cell types were obtained, and fold change values comparing the expression in tolerant vs. activated Des-TCR+ CD8+ T cells were calculated (Table 3). Gene products commonly associated with CD4+ Treg cells like FoxP3, GITR, CTLA-4, and PD-1 (16) were not significantly differentially expressed. Analyses of lymphokines associated with Treg cell functions revealed a prominent up-regulation of TGF-β1 expression whereas no differential signal was obtained for IL-4 and IL-10 (Table 3). Because granzyme B may also be involved in the function of CD4+ CD25+ Treg cells (20) we tested the differential expression of this gene and found a strong up-regulation in Des-TCR+ CD8+ Treg cells. When the experiment was repeated without a prior tumor challenge comparable results were observed, with the notable exception of TGF-β1. Granzyme B was still up-regulated in tolerant cells (4.8-fold). In contrast, TGF-β1 was not up-regulated, indicating induction of TGF-β1 in tolerant CD8+ T cells by the tumor. Taken together, this analysis suggests that CD4+ CD25+ Treg cells and Des-TCR+ CD8+ Treg cells share pathways in the regulation of T cell responses involving TGF-β1 and granzyme B.
Table 3.
Tolerant Des-TCR+ CD8+ T cells share elevated granzyme B levels with CD4+ CD25+ Treg cells
| GenBank accession no. | Gene symbol | Fold change (tolerant vs. activated Des-TCR+ CD8+ T cells) |
|---|---|---|
| NM_054039 | FoxP3 | 1.9 |
| NM_009400 | GITR | −1.3 |
| NM_009843 | CTLA-4 | 1.1 |
| X67914 | PD-1 | −1.2 |
| BC027514 | IL-4 | Not detected |
| NM_010548 | IL-10 | −0.6 |
| NM_011577 | TGF-β1 | 3.7 |
| M12302 | Granzyme B | 4.4 |
Tx Des-TCR × 2.4KerIV-Kb.Rag-2−/− and Tx Des-TCR.Rag-2−/− mice were analyzed for tolerance by s.c. inoculation of the P815.Kb.B7 tumors. Sorted CD8+ T cells from tolerant and nontolerant mice 14 days after tumor challenge were used for RNA extraction. The respective RT-PCR data (fold change mean of a minimum of three independent experiments) are depicted.
Discussion
It is well recognized that tissue-specific, self-reactive T cells can escape deletion in the thymus (4). There exist several possibilities of how these self-reactive T cells may be silenced in the periphery. T cells remain naïve if the expression of tissue antigens is too low to transmit a signal or if the respective tissue is inaccessible for these T cells. This phenomenon has been termed ignorance (21). Alternatively, autoreactive T cells may be controlled by CD4+ CD25+ Treg cells. Recent data show that the thymus expresses antigens that were formerly thought to be tissue-specific (22). The medullary thymic epithelial cells have been identified as the cell type responsible for promiscuous expression of a broad range of tissue-specific gene products and are believed to be involved in the induction CD4+ CD25+ Treg cells (5, 6, 23). Additionally, self-antigens that are sufficiently expressed in peripheral organs may be cross-presented to CD8+ T cells in the draining lymph nodes by dendritic cells (24). This cross-presentation may lead to tolerance based on deletion of the respective T cells in the absence of T cell help (18).
Here we report a dominant mechanism for the prevention of tissue damage by autoreactive T cells. In the neonatal phase T lymphocyte trafficking through nonlymphoid tissue, such as the skin, can result in the generation of CD8+ Treg cells that maintain tolerance to tissue antigens during adulthood. T cell tolerance seen in our transgenic model system is due to direct encounter of Kb on keratinocytes in the skin of the neonate. An antigen-specific contribution by the thymus to the observed tolerance was excluded because transfer of the thymus of 2.4KerIV-Kb mice did not lead to tolerance (17, 25). Moreover, replacement of the transgenic thymus by a wild-type thymus did not abrogate tolerance in the respective chimeras (25). In addition, cross-presentation in this system is excluded for two reasons. First, the Des-TCR can recognize only an intact Kb molecule presenting one of three possible peptides. It is not reacting with peptides of a degraded Kb molecule presented by another MHC class I antigen (26). Consequently, the cross-presentation pathway via dendritic cells is not relevant for Des-TCR recognition. Second, tolerance was abolished by antibody-mediated inhibition of T cell migration through the skin in the neonatal Des-TCR × 2.4KerIV-Kb mouse (17).
Des-TCR+ CD8+ T cells rendered tolerant during neonatal life were still found in adult mice that had been Tx at day 15, and they were capable of regulating the reactivity of naïve Des-TCR+ CD8+ T cells against Kb-positive skin and tumor grafts. Because continuous presence of the Kb antigen is essential for the maintenance of tolerance (27) we assume that the tolerant T cells can migrate through the skin of adult mice. This finding is in agreement with a recent description of a CD8+ Treg cell population that suppresses proliferation and IFNγ production by CD8+ T cells (28). The Des-TCR+ CD8+ Treg cells expressed normal levels of TCR, CD8α, and CD8β molecules and are therefore different from the described CD4−CD8− Treg cells and CD8low Treg cells that were not found in these mice (29, 30). The regulation of naïve Des-TCR+ CD8+ T cells occurred in the absence of CD4+ T cells. The CD4+ T cell-independent regulatory capacity of CD8+ Treg cells does not exclude the possibility that tolerance in 2.4KerIV-Kb mice with a polyclonal T cell repertoire is established by several mechanisms in parallel, including CD4+ T cells. CD4+ T cell-mediated regulation of Des-TCR+ CD8+ T cells has been shown in a transplantation tolerance model (31).
The Des-TCR+ CD8+ Treg cells did not interfere with the lymphopenia-induced proliferation (LIP)-induced expansion of naïve Des-TCR+ CD8+ T cells. The mechanism preventing rejection of Kb-positive grafts by nontolerant Des-TCR+ CD8+ T cells remains to be determined. However, because TGF-β1 was found to be up-regulated in the tolerant/regulatory Des-TCR+ CD8+ T cell population it is tempting to speculate that TGF-β1 acts directly on the Des-TCR+ CD8+ T cells. Such a mechanism has been suggested for the CD4+ CD25+ Treg cell-mediated control of activated CD8+ T cells in a model of autoimmune diabetes (32). Furthermore, the suppressive function of CD4+ CD25+ T cells has been reported to include the induction of cell death of effector T cells via granzyme B (20). This pathway could also be operative in the control of the transferred and expanding CD8+ T cells, because granzyme B is up-regulated in the regulatory Des-TCR+ CD8+ T cells. Further in vivo studies are needed to help to define the exact involvement of granzyme B in T cell regulation. Interestingly, granzyme B was constitutively expressed in the tolerant/regulatory CD8+ T cells, whereas TGF-β1 was induced only after challenge of the tolerant mice with the P815.Kb.B7 tumor. Induction of TGF-β1 instead of constitutive expression may help to avoid bystander suppression.
A difficulty in the elucidation of Treg cell function has been the lack of suitable markers to distinguish Treg cells from other T cell subsets. Presently, the main CD4+ Treg population is identified by the expression of CD25 and FoxP3 (33). But also other CD4+ Treg cells lacking these markers have been described (34–36). CD8+ Treg cells have been reported to express the CD122 antigen (12). However, this antigen does not seem to be a specific marker because it is also expressed on activated CD8+ T cells (12). Likewise, in the tolerance model described here the CD122 antigen was expressed in comparable amounts on tolerant Des-TCR+ CD8+ T cells and on activated Des-TCR+ CD8+ T cells (data not shown). Therefore, it is presently not possible to estimate the percentage of Des-TCR+ CD8+ Treg cells among the CD8+ T cell population in tolerant Des-TCR × 2.4KerIV-Kb.Rag-2−/− mice.
Recognition of antigen cross-presented by resting dendritic cells is presently regarded as the most prominent mechanism of peripheral T cell tolerance induction resulting in either T cell deletion (37) or the generation of Treg cells (14, 38). The system described here is unique in that it allows us to determine the contribution of parenchymal cells to peripheral CD8+ T cell tolerance induction independent of the involvement of dendritic cells. Because such a distinction is so far difficult in most tolerance models the role of parenchymal cells in peripheral T cell tolerance was possibly underestimated. The tolerance mechanism, namely direct recognition of antigen on parenchymal cells, may operate in parallel to cross-presentation, e.g., in the neonate when naïve T cells still have access to nonlymphoid tissues, such as the skin. Because cross-presentation in the draining lymph nodes requires high expression levels of the respective antigens (18), whereas tolerance can be induced directly by very small amounts of tissue antigen (17), this direct mechanism may ensure tolerance in situations where the presence of antigen is too low for cross-presentation.
In summary, our data identify a naturally occurring CD8+ Treg cell population that is induced by self-antigen expressed on parenchymal cells in neonatal mice. These CD8+ Treg cells maintain tolerance during adulthood by down-modulating effector functions of T cells with the same specificity independent of CD4+ T cells.
Materials and Methods
Mice.
Des-TCR mice (H-2Kk) are transgenic for a Kb-specific TCR (17). 2.4KerIV-Kb mice (H-2Kk or H-2Kd) express the Kb molecule under control of the 2.4KerIV promoter (17, 39). The Des-TCR and 2.4KerIV-Kb transgenic mice were crossed to a recombination-activating gene 2-deficient (Rag2−/−) background (H-2Kk) (17) and bred as single-transgenic or double-transgenic mice. Anti-Leish-TCR mice contain transgenic TCR α and β chains isolated from an H-2d-restricted CD4+ T cell clone reactive against an unknown Leishmania antigen (kindly provided by A. Gessner, Institute for Clinical Microbiology, Immunology, and Hygiene, University of Erlangen-Nürmberg, Erlangen, Germany) and were also maintained on the Rag2−/− background (H-2Kk). TCR 8 mice express a TCR transgene specific for the SV40 large T antigen (Tag) and were on the Rag1−/− background (H-2Kk) (40). C57BL/6 and CBA mice were obtained from Charles River Breeding Laboratories (Sulzfeld, Germany). All mice were kept under specific pathogen-free conditions at the animal facility of the German Cancer Research Center.
Antibodies and Flow Cytometry.
The CD3, CD8, CD25, and CD122 monoclonal antibodies (conjugated to various fluorochromes) were obtained from BD Biosciences (San Diego, CA). Flow cytometric analyses were done on a FACSCalibur with CellQuest software (BD Biosciences).
Thymectomy.
Up to 15-day-old mice were anesthetized with a mixture of ketamine (Fort Dodge Laboratories, Fort Dodge, IA) and xylazine (Abbott Laboratories, North Chicago, IL). A small incision of the upper thoracic region exposed the thymic lobes. The thymus was removed by the application of suction, and the wound was closed thereafter. Mice were kept under a heat lamp and observed after the procedure until they had regained consciousness. The complete extraction of both thymic lobes was controlled after the adult mice were killed.
Skin Grafting.
Full-thickness skin was obtained from the tails of donor mice. The graft beds were prepared on the lateral thoracic walls of the recipient mice. The procedure was performed under ketamine/xylazine anesthesia. Skin grafts were secured with suture clips and an adhesive bandage. The clips and bandages were removed on the seventh postoperative day, followed by daily inspection thereafter. Animal care was in accordance with the guidelines of the regulatory authorities.
Transfected Cell Lines.
The murine mastocytoma cell line P815 (H-2Kd) transfected with the genes encoding for the murine MHC class I molecule H-2Kb and the B7-1 gene plus the neomycin resistance gene has been described (17). P815 cells expressing Tag (SV40 large T antigen), the H-2 restriction antigen Kk, and B7-1 (P815.Tag.Kk.B7) were generated by transfection with the respective cDNAs. Selected clones were cultured in DMEM containing 10% heat-inactivated FCS, 10 mM Hepes, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.05 mM 2-mercaptoethanol, 100 units/ml penicillin, and 100 mg/ml streptomycin. For s.c. injection the cells were trypsinized (2 ml of 0.25% trypsin plus 2.5 mM EDTA for 5 min at room temperature) and washed twice with Dulbecco's PBS.
Cell Transfers.
Single-cell suspensions of spleens were prepared from donor mice of the indicated strains. For CFSE labeling, total spleen cells were incubated with 5 μM CFSE (Molecular Probes, Eugene, OR) in Dulbecco's PBS for 15 min at 37°C. Cells were washed twice with ice-cold Dulbecco's PBS and were finally resuspended in Dulbecco's PBS before injection. Cells were injected into the tail vein in a volume of 200 μl.
Cytotoxic T Lymphocyte-Mediated Cytotoxicity in Vivo.
Recipient mice were transferred with 3 × 105 naïve Tag-specific TCR 8 splenocytes i.v. and then immunized i.v. twice in a period of 6 days with 1–2 × 106 bone marrow-expanded dendritic cells (41) activated with 0.5 μM CpG-ODN 1668 and loaded with 10 μM Tag560–568 peptide. Seven days after the second immunization, the specific in vivo kill was quantified as previously described (42). Briefly, CBA spleen target cells (H-2k) from naïve donor mice were loaded with Tag560–568 peptide and a high CFSE concentration. Unloaded CBA splenocytes labeled with a low CFSE concentration were used as nonspecific target cells. The two target cell populations were mixed in equal ratios, and 20 × 106 cells in total were transferred i.v. into effector mice. Four hours later the specific cytotoxic T lymphocyte activity was quantified in spleens of effector mice. The ratio between CFSEhi and CFSElo cells recovered in nonimmunized Rag2−/− mice indicated 0% specific kill activity.
T Cell Isolation.
Single-cell suspensions of spleens were prepared, and total CD8+ T cells were purified by using CD8+-specific microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. To obtain increased purities (99.6% and higher), the MACS-isolated cells were stained with CD8+-specific antibodies and sorted on a FACSVantage (BD Biosciences).
Real-Time RT-PCR.
RNA was isolated with the RNeasy kit (Qiagen, Hilden, Germany) followed by cDNA synthesis using SuperScript II Reverse Transcriptase (Invitrogen, Karsruhe, Germany). Quantitative real-time RT-PCR was performed in a Gene Amp 5700 Sequence Detection System (Applied Biosystems, Darmstadt, Germany) by using a SYBR Green PCR kit (Applied Biosystems). Each sample was assayed in triplicate, and a 25-μl final reaction volume was used. Primer pairs were designed and synthesized according to Primer Express software (Applied Biosystems). A threshold was set in the linear part of the amplification curve, and the number of cycles needed to reach it was calculated for every gene. Melting curves established the purity of the amplified band. The relative mRNA frequencies were determined in relation to the housekeeping gene hypoxanthine phosphoribosyltransferase (HPRT). Briefly, we calculated for each sample the difference in the threshold cycles (CT) between the target gene and HPRT: ΔCTsample = (CTsample − CTHPRT). Relative mRNA frequencies were calculated as 2−ΔCT, and the experimental samples were normalized to the respective calibrator.
Supplementary Material
Acknowledgments
We thank Gorana Hollmann and Georg Pougialis for expert technical assistance and Birgit Vey for preparation of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 405) and by research funding from the Sixth European Commission–Research and Technology Development framework program through the Reprogramming the Immune System for the Establishment of Tolerance Integrated Project.
Abbreviations
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- TCR
T cell receptor
- Des-TCR
Désiré TCR
- Leish-TCR
Leishmania TCR
- Treg
regulatory T
- Tx
thymectomized.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kappler JW, Roehm N, Marrack P. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 2.Ramsdell F, Fowlkes BJ. Science. 1990;248:1342–1348. doi: 10.1126/science.1972593. [DOI] [PubMed] [Google Scholar]
- 3.Hori S, Nomura T, Sakaguchi S. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 4.Graca L, Chen TC, Le Moine A, Cobbold SP, Howie D, Waldmann H. Trends Immunol. 2005;26:130–135. doi: 10.1016/j.it.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Modigliani Y, Thomas-Vaslin V, Bandeira A, Coltey M, Le Douarin NM, Coutinho A, Salaun Z. Proc Natl Acad Sci USA. 1995;92:7555–7559. doi: 10.1073/pnas.92.16.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 7.Seddon B, Mason D. Immunol Today. 2000;21:95–99. doi: 10.1016/s0167-5699(99)01559-5. [DOI] [PubMed] [Google Scholar]
- 8.Chang CC, Ciubotariu R, Manavalan JS, Yuan J, Colovai AI, Piazza F, Lederman S, Colonna M, Cortesini R, Dalla-Favera R, Suciu-Foca N. Nat Immunol. 2002;3:237–243. doi: 10.1038/ni760. [DOI] [PubMed] [Google Scholar]
- 9.Jarvis LB, Matyszak MK, Duggleby RC, Goodall JC, Hall FC, Gaston JS. Eur J Immunol. 2005;35:2896–2908. doi: 10.1002/eji.200526162. [DOI] [PubMed] [Google Scholar]
- 10.Xystrakis E, Dejean AS, Bernard I, Druet P, Liblau R, Gonzalez-Dunia D, Saoudi A. Blood. 2004;104:3294–3301. doi: 10.1182/blood-2004-03-1214. [DOI] [PubMed] [Google Scholar]
- 11.Hu D, Ikizawa K, Lu L, Sanchirico ME, Shinohara ML, Cantor H. Nat Immunol. 2004;5:516–523. doi: 10.1038/ni1063. [DOI] [PubMed] [Google Scholar]
- 12.Rifa'I M, Kawamoto Y, Nakashima I, Suzuki H. J Exp Med. 2004;200:1123–1134. doi: 10.1084/jem.20040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James E, Scott D, Chai JG, Millrain M, Chandler P, Simpson E. Int Immunol. 2002;14:1333–1342. doi: 10.1093/intimm/dxf093. [DOI] [PubMed] [Google Scholar]
- 14.Faunce DE, Terajewicz A, Stein-Streilein J. J Immunol. 2004;172:1991–1995. doi: 10.4049/jimmunol.172.4.1991. [DOI] [PubMed] [Google Scholar]
- 15.Bienvenu B, Martin B, Auffray C, Cordier C, Becourt C, Lucas B. J Immunol. 2005;175:246–253. doi: 10.4049/jimmunol.175.1.246. [DOI] [PubMed] [Google Scholar]
- 16.Fontenot JD, Gavin MA, Rudensky AY. Nat Immunol. 2003;4:330–336. [PubMed] [Google Scholar]
- 17.Alferink J, Tafuri A, Vestweber D, Hallmann R, Hammerling GJ, Arnold B. Science. 1998;282:1338–1341. doi: 10.1126/science.282.5392.1338. [DOI] [PubMed] [Google Scholar]
- 18.Heath WR, Kurts C, Miller JF, Carbone FR. J Exp Med. 1998;187:1549–1553. doi: 10.1084/jem.187.10.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stockinger B, Kassiotis G, Bourgeopis C. Curr Opin Immunol. 2004;16:775–779. doi: 10.1016/j.coi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. J Immunol. 2005;174:1783–1786. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 21.Zinkernagel RM, Ehl S, Aichele P, Oehen S, Kundig T, Hengartner H. Immunol Rev. 1997;156:199–209. doi: 10.1111/j.1600-065x.1997.tb00969.x. [DOI] [PubMed] [Google Scholar]
- 22.Derbinski J, Schulte A, Kyewski B, Klein L. Nat Immunol. 2001;2:1032–1039. [Google Scholar]
- 23.Kyewski B, Klein L. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 24.Kurts C, Cannarile M, Klebba I, Brocker T. J Immunol. 2001;166:1439–1442. doi: 10.4049/jimmunol.166.3.1439. [DOI] [PubMed] [Google Scholar]
- 25.Arnold B. In: Autoimmune Liver Disease. Dienes HP, Leuschner U, Lohse AW, Manns MP, editors. Heidelberg: Springer; 2005. pp. 39–45. [Google Scholar]
- 26.Guimezanes A, Barrett-Wilt GA, Gulden-Thompson P, Shabanowitz J, Engelhard VH, Hunt DF, Schmitt-Verhulst AM. Eur J Immunol. 2001;31:421–432. doi: 10.1002/1521-4141(200102)31:2<421::aid-immu421>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Alferink J, Schittek B, Schonrich G, Hammerling GJ, Arnold B. Int Immunol. 1995;7:331–336. doi: 10.1093/intimm/7.2.331. [DOI] [PubMed] [Google Scholar]
- 28.Endharti AT, Rifa'I M, Shi Z, Fukuoka Y, Nakahara Y, Kawamoto Y, Takeda K, Isobe K, Suzuki H. J Immunol. 2005;175:7093–7097. doi: 10.4049/jimmunol.175.11.7093. [DOI] [PubMed] [Google Scholar]
- 29.Zhang ZX, Yang L, Young KJ, DuTemple B, Zhang L. Nat Med. 2000;6:782–789. doi: 10.1038/77513. [DOI] [PubMed] [Google Scholar]
- 30.Maile R, Pop SM, Tisch R, Collins E, Cairns BA, Frelinger JA. Eur J Immunol. 2006;36:397–410. doi: 10.1002/eji.200535064. [DOI] [PubMed] [Google Scholar]
- 31.Lin CY, Graca L, Cobbold SP, Waldmann H. Nat Immunol. 2002;3:1208–1213. doi: 10.1038/ni853. [DOI] [PubMed] [Google Scholar]
- 32.Green EA, Gorelik L, McGregor CM, Tran EH, Flavell RA. Proc Natl Acad Sci USA. 2003;100:10878–10883. doi: 10.1073/pnas.1834400100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakaguchi S. Nat Immunol. 2005;6:345–352. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 34.Groux H, O'Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MG. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 35.Buer J, Lanoue A, Franzke A, Garcia C, von Boehmer H, Sarukhan A. J Exp Med. 1998;187:177–183. doi: 10.1084/jem.187.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roncarolo MG, Bacchetta R, Bordignon C, Narula S, Levings MK. Immunol Rev. 2001;182:68–79. doi: 10.1034/j.1600-065x.2001.1820105.x. [DOI] [PubMed] [Google Scholar]
- 37.Probst HC, McCoy K, Okazaki T, Honjo T, van den Broek M. Nat Immunol. 2005;6:280–285. doi: 10.1038/ni1165. [DOI] [PubMed] [Google Scholar]
- 38.Dhodapkar MV, Steinman RM. Blood. 2002;100:174–177. doi: 10.1182/blood.v100.1.174. [DOI] [PubMed] [Google Scholar]
- 39.Schonrich G, Kalinke U, Momburg F, Malissen M, Schmitt-Verhulst AM, Malissen B, Hammerling GJ, Arnold B. Cell. 1991;65:293–304. doi: 10.1016/0092-8674(91)90163-s. [DOI] [PubMed] [Google Scholar]
- 40.Geiger T, Gooding L, Flavell R. Proc Natl Acad Sci USA. 1992;89:2985–2989. doi: 10.1073/pnas.89.7.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutz MB, Kukutsch N, Ogilvie AL. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 42.Garbi N, Arnold B, Gordon S, Hammerling GJ, Ganss R. J Immunol. 2004;172:5861–5869. doi: 10.4049/jimmunol.172.10.5861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.