Abstract
Enzyme replacement therapy (ERT) is available for several lysosomal storage diseases. Except for Gaucher disease, for which an enzyme with exposed mannosyl residues targets mannose receptors (MR) on macrophages, ERT targets primarily the mannose 6-phosphate receptor (MPR). Most recombinant lysosomal enzymes contain oligosaccharides with both terminal mannosyl and mannose 6-phosphate residues. Effective MPR-mediated delivery may be compromised by rapid clearance of infused enzyme by the MR on fixed tissue macrophages, especially Kupffer cells. To evaluate the impact of this obstacle to ERT, we introduced the MR-null mutation onto the mucopolysaccharidosis type VII (MPS VII) background and produced doubly deficient MR−/− MPS VII mice. The availability of both MR+/+ and MR−/− mice allowed us to study the effects of eliminating the MR on MR- and MPR-mediated plasma clearance and tissue distribution of infused phosphorylated (P) and nonphosphorylated (NP) forms of human β-glucuronidase (GUS). In MR+/+ MPS VII mice, the MR clearance system predominated at doses up to 6.4 mg/kg P-GUS. Genetically eliminating the MR slowed plasma clearance of both P- and NP-GUS and enhanced the effectiveness of P-GUS in clearing storage in kidney, bone, and retina. Saturating the MR clearance system by high doses of enzyme also improved targeting to MPR-containing tissues such as muscle, kidney, heart, and hepatocytes. Although ablating the MR clearance system genetically is not practical clinically, blocking the MR-mediated clearance system with high doses of enzyme is feasible. This approach delivers a larger fraction of enzyme to MPR-expressing tissues, thus enhancing the effectiveness of MPR-targeted ERT.
Keywords: β-glucuronidase deficiency, immune tolerance, lysosomal storage disease
The mucopolysacchridoses (MPS) are a group of lysosomal storage diseases (LSDs) in which a deficiency of one or more lysosomal enzymes interferes with the degradation of glycosaminoglycans (GAGs), resulting in their storage in the lysosome. Accumulated GAG storage in lysosomes results in progressive cellular and organ dysfunction (1). In recent years, advances have been made in the treatment of several LSDs (2–5). In these diseases, lysosomal storage can be partially or completely reversed in many target tissues by i.v. infusion of the missing lysosomal enzyme. Traditionally, it was assumed that lysosomal enzymes would be delivered to target tissues so long as the enzymes contained the mannose 6-phosphate (M6P) recognition marker (6). However, other factors can influence the delivery of lysosomal enzymes to target tissues. Among these is the presence of nonphosphorylated oligosaccharides on the enzymes and the extent to which these have been processed from high mannose to complex-type oligosaccharides (7). The latter determines the number of exposed mannose residues on the enzyme, which can target the enzyme preferentially to the mannose receptor (MR). In addition, the distribution of the MR and the mannose 6-phosphate receptor (MPR) and their relative abundance in target tissues affect cells targeted by infused enzyme (6).
The murine model of MPS VII has been used successfully to study many different therapies for lysosomal storage diseases, including enzyme replacement therapy (ERT; ref. 8). We have modified this model by making it immunologically tolerant to human β-glucuronidase (hGUS; ref. 9). This reduces the risk of complications from an immune response when hGUS is infused repeatedly in ERT.
Recently, another useful murine model has been developed in which the MR has been knocked out (10). We bred this strain on to the B6 MPS VII/E540ATg background to produce a model that is mouse GUS-deficient, hGUS-tolerant, and MR-deficient (MPS VII/MR−/−). In the current studies, we used both the MPS VII/E540ATg MR+/+ (referred to later as MR+/+) and the MPS VII/E540ATg MR−/− (referred to later as MR−/−) mice to study the individual roles of the MPR and the MR in enzyme therapy.
Results
Effect of the MR on Clearance of Low-Dose Phosphorylated (P)-GUS vs. Nonphosphorylated (NP)-GUS.
We compared the plasma clearance of infused P-GUS with that of NP-GUS (both at 0.2 mg/kg) in MR+/+ and MR−/− MPS VII mice (Fig. 1A). Both were cleared at approximately the same initial rate (t1/2 of ≈5 min) in MR+/+ mice. In contrast, the clearance of infused P-GUS in MR−/− mice was significantly prolonged (t1/2 of ≈9 min). Markedly slower still was the clearance of NP-GUS in the MR−/− mice (t1/2 of ≈4 h). This experiment shows the contributions of both the MR and MPR in clearance of recombinant hGUS in the MPS VII mouse. Both systems contribute to the rapid clearance of P-GUS in the MR+/+ mouse. The prolonged circulation of infused NP-GUS in the MR−/− mouse reflects the fact that neither receptor-mediated clearance system is operative. The more rapid clearance of P- than NP-GUS in the MR−/− mice shows the isolated effect of MPR-mediated clearance.
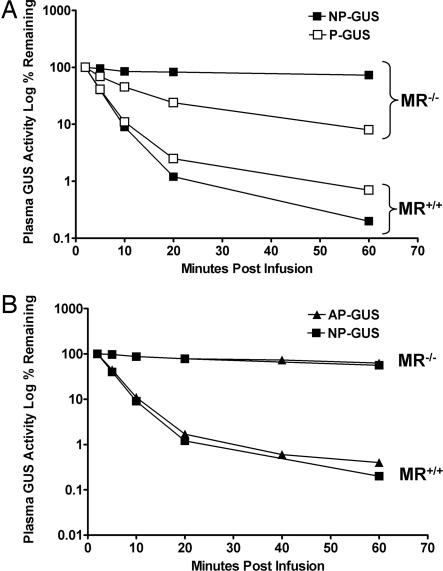
Fig. 1.
Plasma clearance of P- and NP-GUS (A) or AP- and NP-GUS (B) in MR−/− and MR+/+ mice. (A) P- (□) and NP-GUS (■) were infused at a dose of 0.2 mg/kg and plasma GUS levels measured over the next hour. Note that the NP-GUS in the MR+/+ mice was very rapidly cleared (AUC = 411 units/h per ml) in contrast to the much slower clearance in MR−/− mice (AUC = 4,703 units/h per ml). P-GUS is also rapidly cleared in the MR+/+ mice (AUC = 473 units/h per ml) but shows an intermediate rate of clearance in MR−/− mice (AUC = 1,524 units/h per ml), reflecting the M6P-mediated clearance in the absence of MR. (B) Clearance rates of AP-GUS (▴) (AUC = 4,395 units/h per ml) and NP-GUS (■) (AUC = 4,703 units/h per ml) appear nearly equivalent in MR−/− mice.
The dominant role of the MR in the clearance of both NP- and P-GUS is evident from Fig. 1A, as there was no difference in the areas under the curve (AUCs) for both enzymes in MR+/+ mice over the first 60 min (see legend to Fig. 1A). Thus, at this dose, we could not demonstrate the impact of the MPR on enzyme clearance in MR+/+ mice. The AUCs were increased for both P- and NP-GUS in the MR−/− mice compared with those for MR+/+ mice. However, the AUC for P-GUS was increased only 4-fold, whereas that of NP-GUS was increased 10-fold. This difference indicates that, in the absence of the MR, the role of the MPR-mediated clearance can be readily demonstrated.
Sands et al. (6) showed that NP-GUS, produced in a baculovirus/insect cell expression system, contains oligosaccharides with terminal mannose residues that interact with the MR but not the MPR. To verify that more rapid clearance of P- than NP-GUS in the MR−/− mice was MPR-mediated, we dephosphorylated P-GUS with placental alkaline phosphatase (AP) and compared its clearance with that of NP-GUS. Fig. 1B shows that NP- and AP-GUS (both lacking M6P) showed similarly slowed clearance (t1/2 of ≈4 h) in the MR−/− mice and similarly rapid clearance in the MR+/+ mice, indicating they are equivalent ligands for the MR-mediated clearance system. In addition, this experiment shows that the more rapid clearance of P-GUS in the MR−/− mouse depends on the M6P recognition marker, which was destroyed by AP (11–13).
Effect of Increasing Dose on Clearance of P-GUS in the Presence or Absence of the MR.
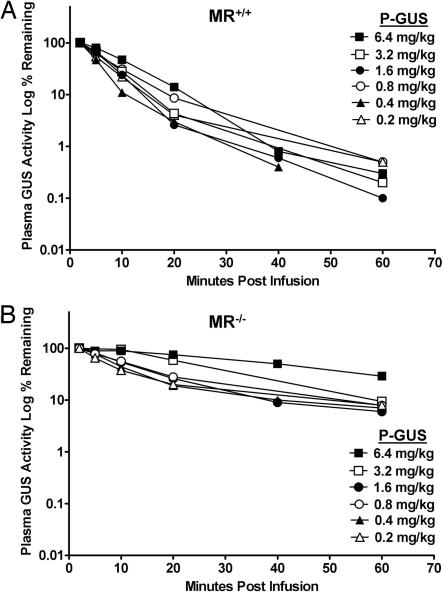
Even though P-GUS contains phosphorylated oligosaccharides, it also contains high-mannose oligosaccharides with exposed terminal mannoses (14). To determine what dose of P-GUS is required to saturate the MR vs. the MPR, we did an enzyme saturation curve at doses from 0.2 to 6.4 mg/kg P-GUS in MR+/+ (Fig. 2A) and MR−/− (Fig. 2B) mice. In MR+/+ mice, it was difficult to see saturation <6.4 mg/kg, at which the t1/2 of clearance was prolonged to ≈10 min. Saturation of MPR-mediated clearance by P-GUS in MR−/− mice was evident at a lower dose. (Clearance t1/2 was 23 min at 3.2 mg/kg and 42 min at 6.4 mg/kg.) This corresponds to 4- and 3.4-fold increases in AUC over the first 60 min for P-GUS at 3.2 and 6.4 mg/kg, respectively, in MR−/− mice (Table 1).
Fig. 2.
Effect of increasing dose of P-GUS on plasma clearance in MR+/+ (A) and MR−/− (B) mice. P-GUS was infused at doses of 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 mg/kg, and plasma GUS levels measured over the next hour. At the two highest doses in MR−/− mice (B), clearance was slower than at lower doses and much slower than in all doses in MR+/+ mice (A), reflecting the rapid clearance by the MR in MR+/+ mice and the saturation of the MPR with the higher doses in MR−/− mice.
Table 1.
Calculated AUC for plasma clearance of P-GUS in MR+/+ and MR−/− mice infused with increasing doses of P-GUS
| Dose, mg/kg |
||||||
|---|---|---|---|---|---|---|
| 0.2 | 0.4 | 0.8 | 1.6 | 3.2 | 6.4 | |
| MR+/+ | 641 | 474 | 852 | 646 | 740 | 1,052 |
| MR−/− | 1,351 | 1,347 | 1,740 | 1,513 | 2,918 | 3,589 |
AUC calculated from data in Fig. 2.
Effect of Increasing Dose of P-GUS on Its Tissue Distribution in the Presence or Absence of the MR.
To address this question, we infused P-GUS at doses of 0.4, 1.6, or 6.4 mg/kg into MR+/+ and MR−/− mice. In all mice, increasing the dose led to increased enzyme delivery (Table 2). In MR−/− mice, spleen, which normally depends largely on MR-mediated delivery, received only 20% of the enzyme seen in MR+/+ mice at the highest doses. However, liver showed only slightly less enzyme in the MR−/− mice at all doses.
Table 2.
Tissue GUS levels 24 h after infusion of different doses of P-GUS in MR+/+ and MR−/− mice
| Tissue* | Dose, mg/kg | Tissue level, units/mg |
||
|---|---|---|---|---|
| MR+/+ | MR−/− | |||
| Brain | 0.4 | 0.10 ± 0.05 | 0.12 ± 0.04 | 1.20 |
| 1.6 | 0.14 ± 0.04 | 0.20 ± 0.06 | 1.43 | |
| 6.4 | 0.34 ± 0.17 | 0.70 ± 0.10 | 2.06 | |
| Liver | 0.4 | 87.6 ± 12.4 | 72.5 ± 12.1 | 0.83 |
| 1.6 | 381 ± 20.3 | 324 ± 15.5 | 0.85 | |
| 6.4 | 1,577 ± 211 | 1,290 ± 181 | 0.82 | |
| Spleen | 0.4 | 19.9 ± 2.2 | 12.4 ± 4.0 | 0.62 |
| 1.6 | 145 ± 15 | 41.7 ± 9.0 | 0.29 | |
| 6.4 | 890 ± 298 | 198 ± 18 | 0.22 | |
| Heart | 0.4 | 2.9 ± 1.2 | 4.0 ± 1.7 | 1.38 |
| 1.6 | 6.8 ± 2.9 | 7.6 ± 0.7 | 1.12 | |
| 6.4 | 27.5 ± 6.3 | 63.5 ± 31 | 2.30 | |
| Kidney | 0.4 | 3.1 ± 1.5 | 3.3 ± 1 | 1.06 |
| 1.6 | 8.2 ± 3.2 | 11.0 ± 1.9 | 1.34 | |
| 6.4 | 19.9 ± 3.9 | 64.5 ± 18 | 3.2 | |
| Lung | 0.4 | 4.0 ± 1.4 | 4.9 ± 0.9 | 1.2 |
| 1.6 | 10.2 ± 5 | 11.2 ± 2.2 | 1.1 | |
| 6.4 | 33.2 ± 15 | 41.7 ± 9.1 | 1.3 | |
| Muscle | 0.4 | 0.57 ± 0.19 | 0.85 ± 0.47 | 1.49 |
| 1.6 | 1.02 ± 0.54 | 1.35 ± 0.65 | 1.32 | |
| 6.4 | 2.70 ± 1.77 | 17.2 ± 4.2 | 6.37 | |
| Bone (including marrow) | 0.4 | 8.4 ± 6.1 | 6.6 ± 2.9 | 0.79 |
| 1.6 | 35.0 ± 24 | 41.2 ± 32.1 | 1.17 | |
| 6.4 | 198 ± 137 | 196 ± 133 | 0.99 | |
| Plasma (units/ml) | 0.4 | 1.41 ± 0.51 | 203 ± 69 | 144 |
| 1.6 | 7.75 ± 4.25 | 890 ± 24 | 115 | |
| 6.4 | 36.8 ± 26.5 | 4,169 ± 734 | 113 | |
*Tissues were collected after perfusion to remove enzyme in blood.
We then compared the fraction of total infused enzyme delivered to liver in MR+/+ and MR−/− mice. After 1 h, 99% of the infused enzyme had been cleared in the MR+/+ and 94% in the MR−/− mice. The fraction of administered enzyme in liver was 41.8 ± 0.2% in MR+/+ mice and 38.6 ± 3.6% in MR−/− mice (Table 3). The surprisingly large amount of GUS still delivered to liver in the MR−/− mice suggests that nearly 40% of the total MPR accessible to infused P-GUS enzyme is contained in the liver.
Table 3.
Liver accumulation 1 h after infusion of MR−/− and MR+/+ mice with 1 mg/kg P-GUS
| Genotype | P-GUS, units infused* | Total P-GUS, units in liver* | Percent of infused enzyme in liver* |
|---|---|---|---|
| MR−/− | 120,900 ± 3,100 | 46,576 ± 3,189 | 38.5% ± 3.6 |
| MR+/+ | 107,467 ± 13,940 | 44,907 ± 5,736 | 41.8% ± 0.2 |
*Mean ± SD.
In other tissues that express both the MR and MPR (such as lung and bone), there is also a dose-dependent increase in enzyme activity (Table 2). However, the amount of enzyme in these tissues is not greatly different between MR−/− and MR+/+ mice regardless of dose. By contrast, heart, kidney, and muscle, which normally express the MPR but little or no MR, showed a greater increase with increasing dose in MR−/− than in MR+/+ mice. The increased activity with increasing dose likely reflects increased accessibility of MPR in these tissues, because P-GUS initially saturates the MPR in liver and persists much longer in the circulation of MR−/− mice (see Fig. 1 and Table 1). Fig. 5, which is published as supporting information on the PNAS web site, shows histochemical localization of GUS activity in MR+/+ and MR−/− mice treated with increasing enzyme doses.
Comparison of Effectiveness of P-GUS in Reducing Lysosomal Storage in MR+/+ and MR−/− Mice.
MR−/− and MR+/+ mice were infused i.v. with 1 mg/kg P-GUS weekly for 3 wk and killed 7 days later for histopathology, histochemistry, and measurement of residual enzyme levels in each organ. We have used this protocol previously to demonstrate differences in effectiveness between P- and NP-GUS and between GUS and GUS-glycosylation-independent lysosomal targeting in MR+/+ mice (6, 15). This dose is also comparable to that used in many human ERT trials (2–5).
Untreated MR−/− and MR+/+ MPS VII mice were morphologically indistinguishable (Fig. 3) and had lysosomal storage identical to that described in MPS VII/B6 mice (16). In enzyme-treated mice, the liver (Fig. 3), spleen, bone marrow, and renal tubular epithelial cells (not shown) had similar moderate to marked reduction in lysosomal storage in both MR+/+ and MR−/− mice. The brain and meninges had no reduction in storage in either group. However, the visceral epithelial cells of the glomeruli, the retinal pigment epithelium, and the osteoblasts lining the rib cortical bone all showed less lysosomal distention in MR−/− than in MR+/+ mice (Fig. 3).
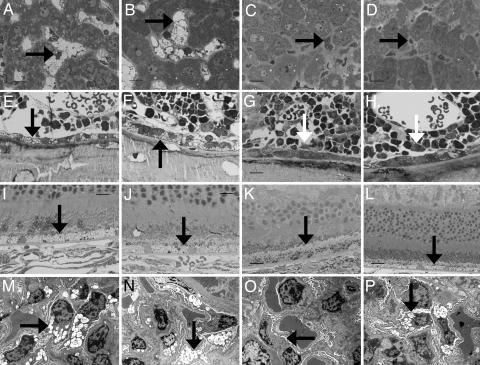
Fig. 3.
Impact of three doses of 1 mg/kg P-GUS on lysosomal storage of P-GUS in liver, bone, retina, and kidney in MR−/− and MR+/+ mice. (A–D) Compared with an untreated MR−/− mouse (A) and an untreated MR+/+ mouse (B), both MR−/− (C) and MR+/+ (D) treated mice had essentially complete clearance of lysosomal storage in the Kupffer cells in the liver (arrow) 1 wk after the last enzyme treatment. (E and F) The bone of untreated MR−/− (E) and MR+/+ (F) mice had lysosomal storage in osteoblasts (arrow) and marrow sinus-lining cells. (G and H) Treated MR−/− (G) and MR+/+ (H) mice had similar marked reduction in storage in the sinus-lining cells. However, only treated MR−/− mice (G) had a marked reduction in storage in the osteoblasts (arrow). Osteoblasts in treated MR+/+ mice (H) had only a slight reduction in storage (arrow). (I and J) Untreated MR−/− (I) and MR+/+ (J) mice had abundant lysosomal storage in retinal pigment epithelial cells (arrow). (K and L) In treated MR−/− mice (K), the retinal pigment epithelial cells had reduced storage. Treated MR+/+ mice (L) showed minimal or no reduction in storage in the retinal pigment epithelial cells. (M and N) Untreated MR−/− (M) and MR+/+ (N) mice both had marked lysosomal distention in the renal visceral epithelial cells (podocytes; arrow). (O and P) After ERT with P-GUS, MR−/− mice (O) showed a marked reduction in lysosomal storage in these cells, but treated MR+/+ mice (P) showed much less treatment effect. (Scale bar: A–L, toluidine blue, 20 μm; M–P, uranyl acetate-lead citrate, 10 μm.)
Table 4 shows the residual enzyme content in tissues from the above experiment, i.e., 7 days after the third infusion. MR−/− livers showed slightly more enzyme than MR+/+ livers, whereas the opposite was true in livers 24 h after infusion where MR−/− mice had ≈83% of the amount present in MR+/+ mice (Table 2). These results are not contradictory. The results at 24 h reflect the loss of MR-mediated uptake by Kupffer cells in MR−/− mice, partially compensated by increased MPR-mediated delivery to hepatocytes (6). The 7-day reversal could be explained by the more rapid turnover in Kupffer cells than hepatocytes in MR+/+ mice, which leads to a lower total amount of residual enzyme in MR+/+ liver at 7 days. The spleen, which normally expresses MR abundantly on macrophages, shows 50% less residual enzyme in MR−/− than in MR+/+ mice. On the other hand, in the heart, there is 60% more GUS in MR−/− mice, and in skeletal muscle, there is a smaller increase.
Table 4.
Tissue levels of GUS 7 days after third weekly infusion of P-GUS in MR−/− and MR+/+ mice
| Tissue | MR−/−, units/mg protein |
MR+/+, units/mg protein |
||||||
|---|---|---|---|---|---|---|---|---|
| Noninfused |
Plue enzyme* |
Noninfused |
Plus enzyme* |
|||||
| Triplicate mice | Mean ± SD | Triplicate mice | Mean ± SD | Triplicate mice | Mean ± SD | Triplicate mice | Mean ± SD | |
| 0.06 | 0.11 | 0.06 | 0.07 | |||||
| Brain | 0.08 | 0.06 ± 0.01 | 0.12 | 0.10 ± 0.02 | 0.05 | 0.05 ± 0.004 | 0.07 | 0.07 ± 0.00 |
| 0.05 | 0.08 | 0.05 | 0.07 | |||||
| 0.30 | 88.14 | 0.34 | 79.20 | |||||
| Liver | 0.31 | 0.29 ± 0.02 | 70.73 | 84.1 ± 9.7 | 0.29 | 0.30 ± 0.03 | 74.78 | 70.9 ± 8.7 |
| 0.27 | 93.54 | 0.26 | 58.95 | |||||
| 0.16 | 10.27 | 0.08 | 15.84 | |||||
| Spleen | 0.27 | 0.23 ± 0.05 | 6.96 | 7.54 ± 2.03 | 0.12 | 0.13 ± 0.05 | 17.25 | 15.61 ± 1.44 |
| 0.27 | 5.40 | 0.19 | 13.74 | |||||
| 0.20 | 1.35 | 0.07 | 0.77 | |||||
| Heart | 0.21 | 0.17 ± 0.05 | 1.22 | 1.25 ± 0.07 | 0.05 | 0.06 ± 0.009 | 0.79 | 0.78 ± 0.009 |
| 0.09 | 1.18 | 0.07 | 0.77 | |||||
| 0.08 | 1.80 | 0.07 | 0.91 | |||||
| Kidney | 0.07 | 0.10 ± 0.03 | 0.85 | 1.32 ± 0.48 | 0.04 | 0.05 ± 0.02 | 0.62 | 2.93 ± 3.07 |
| 0.14 | 1.31 | 0.03 | 7.28 | |||||
| 0.13 | 2.57 | 0.04 | 1.82 | |||||
| Lung | 0.13 | 0.10 ± 0.04 | 1.76 | 1.91 ± 0.49 | 0.07 | 0.05 ± 0.01 | 1.90 | 1.71 ± 0.12 |
| 0.04 | 1.39 | 0.04 | 1.59 | |||||
| 0.04 | 0.38 | 0.03 | 0.26 | |||||
| Muscle | 0.02 | 0.03 ± 0.009 | 0.28 | 0.31 ± 0.05 | 0.02 | 0.02 ± 0.005 | 0.32 | 0.25 ± 0.06 |
| 0.04 | 0.27 | 0.02 | 0.18 | |||||
| 0.07 | 3.59 | 0.04 | 3.09 | |||||
| Bone + Marrow | 0.05 | 0.05 ± 0.02 | 3.50 | 3.25 ± 0.42 | 0.01 | 0.03 ± 0.01 | 2.55 | 2.71 ± 0.26 |
| 0.03 | 2.65 | 0.03 | 2.51 | |||||
*MR−/− and MR+/+ mice received 1 mg/kg P-GUS weekly for 3 wk.
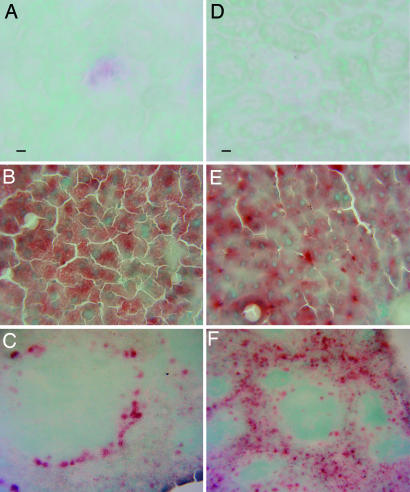
Fig. 4 presents the histochemical staining for GUS 1 wk after the last enzyme infusion. The kidney of MR−/− mice showed a small amount of histochemical staining in the glomeruli, but MR+/+ mice showed no staining in kidney. In liver, Kupffer cells still contained more enzyme than hepatocytes in MR+/+ mice, whereas MR−/− mice had more diffuse activity, especially in hepatocytes. The spleen in MR+/+ mice showed diffuse red pulp GUS activity, whereas staining in MR−/− is limited to red pulp perifollicular cells.
Fig. 4.
Histochemical staining for GUS performed 1 wk after the last 1 mg/kg enzyme infusion in MR−/− (A–C) and MR+/+ (D–F) mice. The kidney of the MR−/− mice (A) showed a small amount of histochemical staining in the glomeruli, but MR+/+ mice (D) showed no enzyme activity in kidney. MR−/− mice (B) had diffuse enzyme activity in liver, especially in hepatocytes, but MR+/+ mice (E) contained enzyme in Kupffer cells and hepatocytes. MR−/− mice (C) showed reduced enzyme in spleen overall but intense staining in the perifollicular cells, whereas MR+/+ mice (F) showed more GUS activity diffusely distributed in red pulp (also see spleen data in Table 4). (Scale bar: 6-bromo-2-hydroxy-β-glucuronide, 20 μM.)
Discussion
MR-null mice have no obvious phenotype other than elevated serum lysosomal enzymes related to slower clearance of serum glycoproteins, including the acid hydrolases (10). When crossed on an MPS VII background, the MR null allele had no additional effect on the phenotype of the MPS VII mouse. The doubly deficient mouse allowed comparison of the MR- and MPR-dependent components of the clearance of infused GUS. At doses of enzyme up to 6.4 mg/kg, the MR clearance system predominated in MR+/+ mice. Even at 6.4 mg/kg, the MR-dependent clearance system was barely saturated. The t1/2 of clearance of P-GUS was 5 min at 0.2 mg/kg, 7 min at 3.2 mg/kg, and 9 min at 6.4 mg/kg. Eliminating the MR component greatly prolonged clearance of P-GUS and showed that the MPR-mediated clearance system saturated at lower doses (t1/2 of 10 min for MR+/+ mice and 42 min for MR−/− mice at 6.4 mg/kg).
We had hypothesized that elimination of the MR clearance system would shift a large fraction of enzyme normally targeted to liver to MPR-expressing parenchymal cells in other organs. In fact, ablation of the MR clearance system reduced the amount of enzyme targeted to liver far less than we expected. The fraction of enzyme found in liver 1 h after injection of 1 mg/kg was 38.6% in the MR−/− mouse and 41.8% in the MR+/+ mouse. Increasing the enzyme dosage did not alter the relative proportion of enzyme delivered to liver. The ratio of enzyme activity in liver in MR−/− mice was 0.82–0.85 that in MR+/+ mice 24 h after infusion of up to 6.4 mg/kg (Table 2). What was altered substantially was the distribution of enzyme in liver, which was shifted predominantly to hepatocytes in MR−/− mice. These experiments indicate that a large fraction of total MPR available to infused P-GUS resides in liver, which gets nearly the same fraction of enzyme even in the absence of the MR. Kupffer cells in MR−/− mice still received some enzyme, because they also express the MPR.
While completely eliminating the MR clearance system (i.e., MR−/− vs. MR+/+) reduced uptake by liver by <18%, it still enhanced the effectiveness of P-GUS in clearing storage in kidney, bone, and retina at 1 mg/kg, the dose used in many conventional ERT trials. These observations suggest that one could achieve wider distribution and enhanced clearance of storage by MPR-targeted enzyme if one could block the MR clearance system pharmacologically with an inhibitor. In fact, Ioannou et al. (7) reported wider distribution of infused α-galactosidase after preinfusion of yeast mannans, which bind the MR and act as competitive inhibitors (17). Yeast mannans are too toxic and immunogenic for clinical use as inhibitors. However, most recombinant enzymes produced in CHO cells are themselves competitive ligands for the MR, because they contain oligosaccharides with exposed mannose residues as well as phosphorylated oligosaccharides. Using a high enough dose of recombinant enzyme might be a nontoxic way to saturate the MR clearance system and enhance MPR-mediated enzyme delivery. Favorable effects in overcoming the blood–brain barrier to ERT in certain parts of the CNS with high doses of infused GUS may have been partially attributable to saturation of the MR clearance system (18–21).
Another interesting question is the mechanism of eventual clearance of NP-GUS from plasma in the MR−/− mouse. Although the clearance of NP-GUS is greatly slowed (t1/2 of 4 h), >90% of the enzyme is cleared in 24 h (data not shown). Possibly, this very slow clearance is mediated by nonspecific fluid-phase pinocytosis or by adsorptive endocytosis after binding of the enzyme to cell surface proteoglycans, like heparan sulfate. We previously demonstrated that the chimeric enzyme containing the highly basic peptide, TAT, binds cell surface proteoglycans and is taken up by adsorptive endocytosis (22).
Du et al. (23) recently reported related studies in which they found that lysosomal acid lipase (LAL) prepared in Pichia pastoris, which normally targets the MR in MR+/+ mice, is taken up by hepatocytes in the LAL−/−/MR−/− doubly deficient mouse (but not in the LAL−/−/MR+/+ mouse) after i.p. injection. These observations led them (23) to propose an uptake mechanism in hepatocytes, independent of the classical MR and MPR, which remains to be identified. This system was observable only if the rapid clearance by the MR was eliminated.
Another striking and unexpected observation was the staining activity in perifollicular cells in the spleens of P-GUS-treated MR−/− mice (Figs 4 and 5). This staining was even more intense after infusion of NP-GUS (data not shown). The greater intensity in MR−/− mice could be attributable to the much longer exposure to circulating enzyme (t1/2 of 4 h). Nonetheless, the preferential localization in these cells indicates a selective uptake system that does not rely on the classical MR or the MPR. The nature of this uptake system remains to be established. These cells could be enriched in cell surface proteoglycans that mediate adsorptive endocytosis of the bound enzyme. Another possible candidate for mediating this uptake would be the recently discovered fourth member of the MR family, Endo180, which is reported to be expressed on fibroblasts, endothelial cells, and macrophages (24).
Materials and Methods
MPS VII/E540ATg mice are deficient in mouse GUS activity but produce a low level of inactive hGUS protein that confers immunologic tolerance to infused hGUS (9). These mice were bred with an MR-deficient mouse strain (10) to produce the MPS VII/MR−/− mouse that is mouse GUS-deficient, hGUS-tolerant, and MR-deficient. Mice were screened for all three markers (MPS VII, the human GUS E540A transgene, and MR) by PCR primers by using tail DNA to confirm their genotype before use in the experiments described. PCR primers and conditions were as described (9, 10).
Recombinant phosphorylated hGUS (P-GUS) was produced in CHO cells, and enzyme secreted into conditioned medium was collected and purified as described (18). This enzyme contains the M6P recognition marker and exhibits a Kuptake value of 1.5 nM, as measured in human MPS VII fibroblasts. Two nonphosphorylated forms of recombinant hGUS were produced, one (NP-GUS) in a baculovirus/SF 21 insect cell expression system (6) and the second (AP-GUS) by treatment of P-GUS with recombinant human placental AP (12). Both NP- and AP-GUS lack the M6P recognition marker and exhibit essentially no uptake by human fibroblasts. Production and characterization of AP-GUS are described in Supporting Text, which is published as supporting information on the PNAS web site.
For plasma clearance experiments, weight-based enzyme doses were determined immediately before each injection. Aliquots of P- or NP-GUS were infused in 125 μl of 10 mM Tris, pH 7.5/150 mM NaCl/1 mM β-glycerophosphate into tail veins of MPS VII/MR+/+ or MPS VII/MR−/− mice. Timed 50-μl blood samples obtained by orbital bleed using heparinized hematocrit tubes were centrifuged and plasma assayed for GUS activity on 4-methyl umbelliferyl β-d-glucuronide (25).
Tissue distribution of GUS was determined after enzyme infusion. For tissue distribution at 24 h, mice were infused with 0.4, 1.6, and 6.4 mg/kg GUS in triplicate. Twenty-four hours after infusion, mice were killed and perfused with 25 ml of cold 10 mM Tris, pH 7.2/140 mM NaCl. Tissues were then harvested and frozen in liquid nitrogen. Tissues were thawed and homogenized by Polytron and sonication in 10–30 volumes of cold 10 mM Tris, pH 7.2/140 mM NaCl/1 mM PMSF. Aliquots of total homogenate were assayed for GUS as described above and for protein (26). Tissues were also obtained for histochemical analysis.
To determine the fraction of infused enzyme delivered to liver, MR+/+ (n = 3) and MR−/− (n = 2) mice received 1 mg/kg i.v. P-GUS. Plasma was collected 1 h later, and mice were killed after perfusion, as above. The livers were homogenized, and the fraction of infused GUS in liver was calculated.
To determine tissue GUS levels and the effectiveness of the clearance of MPS storage after multiple infusions, three mice each of both MR+/+ and MR−/− strains were infused with 1 mg/kg GUS 1× weekly for 3 wk. Seven days after the last infusion, mice were killed, tissues for GUS assay were frozen in liquid nitrogen, and the samples were processed for histochemical analysis for GUS activity and histological analysis of lysosomal storage (6, 18).
Supplementary Material
Acknowledgments
We thank Dr. Michel Nussensweig (The Rockefeller University, New York, NY) for generously providing breeding pairs of MR−/− mice; Colleen Isele and Timothy Becker for excellent technical assistance in management of the mouse colonies and animal husbandry; and Drs. Emil Kakkis, Jung-San Huang, and Mark Sands for helpful critical comments on the manuscript. This work was supported by National Institutes of Health Grant GM34182 (to W.S.S.).
Abbreviations
- MR
mannose receptor
- MPR
mannose 6-phosphate receptor
- M6P
mannose 6-phosphate
- ERT
enzyme replacement therapy
- MPS
mucopolysaccharidosis
- GUS
β-glucuronidase
- hGUS
human GUS
- P-GUS
phosphorylated GUS
- NP-GUS
nonphosphorylated GUS
- AUC
area under the curve
- AP
alkaline phosphatase.
Footnotes
The authors declare no conflict of interest.
References
- 1.Neufeld EF, Muenzer J. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. New York: McGraw-Hill; 2001. pp. 3421–3451. [Google Scholar]
- 2.Desnick RJ. J Inherit Metab Dis. 2004;27:385–410. doi: 10.1023/B:BOLI.0000031101.12838.c6. [DOI] [PubMed] [Google Scholar]
- 3.Brady RO, Barton NW. Lipids. 1996;31(Suppl):S137–S139. doi: 10.1007/BF02637065. [DOI] [PubMed] [Google Scholar]
- 4.Kakkis ED, Muenzer J, Tiller GE, Waber L, Belmont J, Passage M, Izykowski B, Phillips J, Doroshow R, Walot I, et al. N Engl J Med. 2001;344:182–188. doi: 10.1056/NEJM200101183440304. [DOI] [PubMed] [Google Scholar]
- 5.Harmatz P, Giugliani R, Schwartz I, Guffon N, Teles EL, Miranda MC, Wraith JE, Beck M, Arash L, Scarpa M, et al. J Pediatr. 2006;148:533–539. doi: 10.1016/j.jpeds.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Sands MS, Vogler CA, Ohlemiller KK, Roberts MS, Grubb JH, Levy B, Sly WS. J Biol Chem. 2001;276:43160–43165. doi: 10.1074/jbc.M107778200. [DOI] [PubMed] [Google Scholar]
- 7.Ioannou YA, Zeidner KM, Gordon RE, Desnick RJ. Am J Hum Genet. 2001;68:14–25. doi: 10.1086/316953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogler C, Barker J, Sands MS, Levy B, Galvin N, Sly WS. Pediatr Dev Pathol. 2001;4:421–433. doi: 10.1007/s10024001-0079-1. [DOI] [PubMed] [Google Scholar]
- 9.Sly WS, Vogler C, Grubb JH, Zhou M, Jiang J, Zhou XY, Tomatsu S, Bi Y, Snella EM. Proc Natl Acad Sci USA. 2001;98:2205–2210. doi: 10.1073/pnas.051623698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SJ, Evers S, Roeder D, Parlow AF, Risteli J, Risteli L, Lee YC, Feizi T, Langen H, Nussenzweig MC. Science. 2002;295:1898–1901. doi: 10.1126/science.1069540. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan A, Achord DT, Sly WS. Proc Natl Acad Sci USA. 1977;74:2026–2030. doi: 10.1073/pnas.74.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talkad V, Sly WS. J Biol Chem. 1983;258:7345–7351. [PubMed] [Google Scholar]
- 13.Natowicz MR, Chi MM, Lowry OH, Sly WS. Proc Natl Acad Sci USA. 1979;76:4322–4326. doi: 10.1073/pnas.76.9.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natowicz M, Baenziger JU, Sly WS. J Biol Chem. 1982;257:4412–4420. [PubMed] [Google Scholar]
- 15.LeBowitz JH, Grubb JH, Maga JA, Schmiel DH, Vogler C, Sly WS. Proc Natl Acad Sci USA. 2004;101:3083–3088. doi: 10.1073/pnas.0308728100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogler C, Birkenmeier EH, Sly WS, Levy B, Pegors C, Kyle JW, Beamer WG. Am J Pathol. 1990;136:207–217. [PMC free article] [PubMed] [Google Scholar]
- 17.Achord DT, Brot FE, Bell CE, Sly WS. Cell. 1978;15:269–278. doi: 10.1016/0092-8674(78)90102-2. [DOI] [PubMed] [Google Scholar]
- 18.Vogler C, Levy B, Grubb JH, Galvin N, Tan Y, Kakkis E, Pavloff N, Sly WS. Proc Natl Acad Sci USA. 2005;102:14777–14782. doi: 10.1073/pnas.0506892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunder U, Kaartinen V, Valtonen P, Vaananen E, Kosma VM, Heisterkamp N, Groffen J, Mononen I. FASEB J. 2000;14:361–367. doi: 10.1096/fasebj.14.2.361. [DOI] [PubMed] [Google Scholar]
- 20.Roces DP, Lullmann-Rauch R, Peng J, Balducci C, Andersson C, Tollersrud O, Fogh J, Orlacchio A, Beccari T, Saftig P, et al. Hum Mol Genet. 2004;13:1979–1988. doi: 10.1093/hmg/ddh220. [DOI] [PubMed] [Google Scholar]
- 21.Matzner U, Herbst E, Hedayati KK, Lullmann-Rauch R, Wessig C, Schroder S, Eistrup C, Moller C, Fogh J, Gieselmann V. Hum Mol Genet. 2005;14:1139–1152. doi: 10.1093/hmg/ddi126. [DOI] [PubMed] [Google Scholar]
- 22.Orii KO, Grubb JH, Vogler C, Levy B, Tan Y, Markova K, Davidson BL, Mao Q, Orii T, Kondo N, et al. Mol Ther. 2005;12:345–352. doi: 10.1016/j.ymthe.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du H, Levine M, Ganesa C, Witte DP, Cole ES, Grabowski GA. Am J Hum Genet. 2005;77:1061–1074. doi: 10.1086/498652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isacke CM, van der GP, Hunter T, Trowbridge IS. Mol Cell Biol. 1990;10:2606–2618. doi: 10.1128/mcb.10.6.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser JH, Sly WS. J Lab Clin Med. 1973;82:969–977. [PubMed] [Google Scholar]
- 26.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.