Abstract
The nucleolus is the largest subnuclear structure and is plurifunctional in nature. Here, we demonstrate that nucleolar localization of a key herpesvirus regulatory protein is essential for its role in virus mRNA nuclear export. The herpesvirus saimiri ORF57 protein is a nucleocytoplasmic shuttle protein that is conserved in all herpesviruses and orchestrates the nuclear export of viral intronless mRNAs. We demonstrate that expression of the ORF57 protein induces nucleolar redistribution of human TREX (transcription/export) proteins that are involved in mRNA nuclear export. Moreover, we describe a previously unidentified nucleolar localization signal within ORF57 that is composed of two distinct nuclear localization signals. Intriguingly, point mutations that ablate ORF57 nucleolar localization lead to a failure of ORF57-mediated viral mRNA nuclear export. Furthermore, nucleolar retargeting of the ORF57 mutant was achieved by the incorporation of the HIV-1 Rev nucleolar localization signal, and analysis demonstrated that this modification was sufficient to restore viral mRNA nuclear export. This finding represents a unique and fundamental role for the nucleolus in nuclear export of viral mRNA.
Keywords: mRNA export, nucleolus, virus
The eukaryotic cell nucleus is a highly organized environment containing distinct and often dynamic compartments (1). Of these, the nucleolus is the most prominent, and, for many years, its exclusive role was thought to be the site of ribosomal RNA transcription, processing, and assembly into the ribosome subunits (2). Recent studies, however, suggest that it has additional nonclassical roles in many aspects of cell biology, including cell cycle regulation, viral replication, tumorigenesis, and cellular stress responses (3–5). This plurifunctional nature of the nucleolus has been highlighted by extensive proteomic analysis of human nucleoli (6, 7). To date, the nucleolar proteome database archives 728 nucleolar proteins, and functional classification of these proteins reinforces the multiple roles of the nucleolus (8). Furthermore, there is constant dynamic trafficking of nucleolar proteins, and this concomitant dynamic nature of nucleolar structure may provide regulation of nonclassical nucleolar functions.
Interestingly, an increasing number of key proteins from both RNA and DNA viruses have been shown to localize to the nucleolus. These proteins include those encoded by viruses such as coronaviruses, influenza, HIV-1, adenoviruses, and herpesviruses (9). Therefore, virus–nucleolar interactions are likely to have important implications in the life cycle of many viruses. However, at present, the precise functional role of these virus–nucleolar colocalizations has not been determined. One such key viral protein that traffics to the nucleolus is the herpesvirus saimiri (HVS) ORF57 protein. HVS is the prototype γ-2 herpesvirus, or rhadinovirus (10), which has become an important family of viruses since the identification of the first human γ-2 herpesvirus, the oncogenic Kaposi's sarcoma-associated herpesvirus (11, 12). ORF57 encodes an essential multifunctional transregulatory protein that is functionally conserved in all herpesvirus subfamilies (13). We have characterized the role of the ORF57 protein in the herpesvirus life cycle and demonstrated that transactivation of late viral genes by ORF57 occurs independently of target gene promoter sequences and appears to be mediated at a posttranscriptional level (14). Further analysis has demonstrated that the ORF57 protein has the ability to bind viral RNA and shuttle between the nucleus and cytoplasm and is required for efficient cytoplasmic accumulation of virus mRNA (15). More recently, we have shown that ORF57 interacts with two distinct cellular pathways involved in cellular nuclear import and export (16, 17). We have demonstrated that the ORF57 protein interacts with importin-α through a classical nuclear localization signal (NLS) within ORF57, allowing it to shuttle into the nucleus (16). In addition, we have demonstrated that ORF57 interacts with a cellular protein, Aly/Ref, and utilizes this interaction to gain access to the TAP (transporter associated with antigen processing)-mediated nuclear export pathway (17). Aly also associates with members of the human THO complex to form the recently described hTREX (human TREX, human transcription/export) complex, which has been shown to be essential for cellular mRNA nuclear export (18). These findings show that ORF57 is a nucleocytoplasmic shuttle protein that plays a pivotal role in mediating the nuclear export of viral transcripts.
Intriguingly, we demonstrate that expression of ORF57 results in the redistribution of cellular mRNA nuclear export factors, such as hTREX components, into the nucleolus. This finding suggests that the nucleolus plays a critical role in ORF57-mediated viral mRNA export and possibly has a role in herpesvirus RNA processing. The data presented here confirm this hypothesis and demonstrate a key role for the nucleolus in viral mRNA nuclear export.
Results
ORF57 Localizes to the Nucleolus.
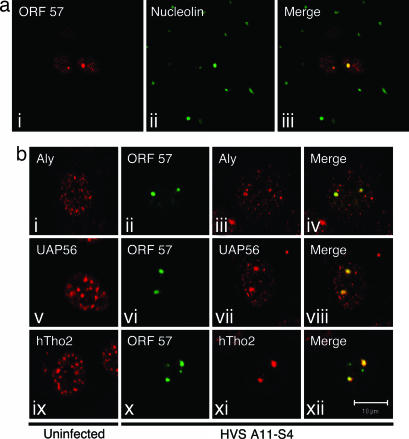
We have previously shown that ORF57 localizes to subnuclear structures during infection (19). To investigate this observation in more detail, a time-course experiment of HVS-infected cells was performed. Owl monkey kidney (OMK) cells grown on glass coverslips were infected with the HVS A11-S4 strain at a multiplicity of infection of 1 and cultured at 37°C for 6, 9, 12, 15, 18, 21, and 24 h. ORF57 expression was first detected around the 12-h mark, and, after 18 h, ORF57 expression was abundant in a large percentage of the cells. In addition to a speckled nuclear pattern of localization that was evident from 12 h, we observed defined nuclear substructure staining (reminiscent of the nucleolus) for ORF57 in many of the infected cells from 16 h onward (Fig. 1a i). To investigate whether ORF57 was localizing to the nucleolus, OMK cells 18 h postinfection were costained for ORF57 and nucleolin (Fig. 1a ii). Results clearly show that ORF57 and nucleolin colocalize, confirming that ORF57 does traffic to the nucleolus (Fig. 1a iii).
Fig. 1.
Redistribution of TREX components by ORF57 into the nucleolus. (a) ORF57 localizes to the nucleolus during HVS infection. OMK cells were cultured on glass coverslips until fully confluent and then infected with HVS A11-S4 strain and incubated at 37°C for 18 h. Cells were fixed, and coimmunofluorescence was carried out by using an ORF57-specific monoclonal antibody (i) and a nucleolin-specific polyclonal antibody (ii). A merged image is shown in iii. (b) Components of the hTREX complex are redistributed to the nucleolus and colocalize with ORF57 in HVS-infected cells. OMK cells were cultured on glass coverslips until fully confluent and then infected with the HVS A11-S4 strain and incubated at 37°C for 18 h. Cells were fixed, and coimmunofluorescence was carried out by using an ORF57-specific monoclonal antibody (ii, vi, and x) and Aly-, UAP56-, and hTho2-specific polyclonal antibodies (iii, vii, and xi). (i, v, and ix) Uninfected OMK cells served as negative controls.
HVS Infection Results in a Redistribution of Key mRNA Export Proteins to the Nucleolus.
The major functional role attributed to ORF57 (and its homologous herpesvirus proteins) is the nuclear export of intronless viral mRNA. Cooper et al. (19) showed that, during a HVS infection or after transfection of cells with CMV-ORF57, the splicing factor SC-35 displayed an altered pattern of localization. Therefore, we were interested in determining whether ORF57 localization to the nucleolus during infection had any effect on the distribution of key cellular mRNA export proteins. The hTREX complex has only recently been described in metazoan cells, but early data suggest that it is a crucial exponent of nuclear mRNA export in the cell (18, 20). Indirect immunofluorescence on uninfected OMK cells using polyclonal antibodies to several members of the hTREX complex revealed a speckled pattern of staining that is consistent with the findings of others (18) (Fig. 1b i, v, and ix). However, double indirect immunofluorescence labeling of A11-S4 HVS-infected OMK cells with ORF57 and hTREX antibodies revealed a drastic alteration in the localization for each of the mRNA export proteins. In contrast to the speckled pattern of localization seen in uninfected OMK cells, the hTREX components were almost exclusively redistributed to the nucleolus (Fig. 1b iii, vii, and xi), where they colocalized with ORF57 (Fig. 1b, iv, viii, and xii). This finding is unique in its demonstration that the hTREX complex is relocalized to the nucleolus and indicates a possible role for the nucleolus in viral mRNA nuclear export.
Characterization of a Previously Unidentified Nucleolar Localization Signal (NoLS) Within ORF57.
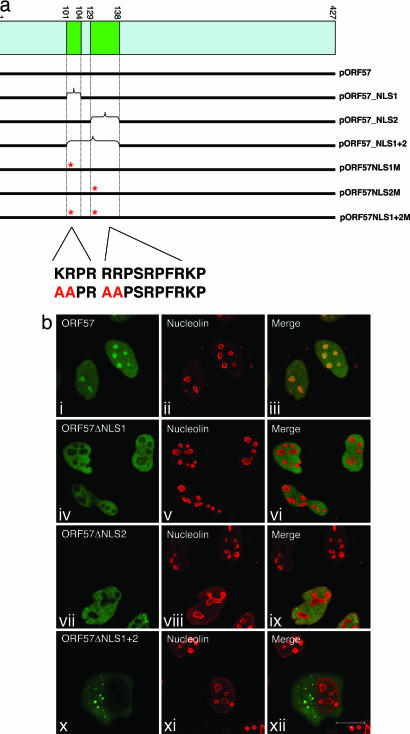
To delineate a minimal NoLS within ORF57, a deletion series of ORF57–GFP fusion constructs was engineered (Fig. 2a). Characterization of these deletion mutants by transient transfection of HeLa cells and immunofluorescence microscopy identified two distinct NLSs positioned toward the N terminus of the protein. To assess whether these NLSs were contributing to the observed nucleolar localization phenotype, each NLS was deleted in the context of the full-length ORF57 protein. Interestingly, analysis of these deletion mutants revealed that a loss of either NLS was sufficient to prevent ORF57 from localizing to the nucleolus; however, the nuclear localization of ORF57 remained unperturbed, confirming that both NLSs were functional (Fig. 2b iv–ix). As one would predict, removal of both NLSs resulted in an accumulation of ORF57 in the cytoplasm (Fig. 2b x–xii). To discount any artifacts that may arise from the use of deletion mutants, we next generated site-directed point mutants for either or both NLSs. The first two basic residues in each ORF57 NLS were converted to alanines, and the phenotype of these mutants was compared with their deletion mutant counterparts. Immunofluorescence data for the point mutants matched those of the deletion mutants exactly, demonstrating that both NLSs were required for nucleolar targeting (see Figs. 7 and 8, which are published as supporting information on the PNAS web site). These data describe a previously uncharacterized NoLS composed of two distinct NLSs that function in tandem to localize ORF57 to the nucleolus.
Fig. 2.
Characterization of a previously unidentified NoLS within ORF57. (a) Schematic representation of the ORF57 deletion and point mutants. Deletion analysis identified two distinct nuclear localization sequences within ORF57. These sequences were deleted from ORF57–GFP constructs and subsequently rendered nonfunctional by the alanine substitution mediated by site-directed mutagenesis. (b) HeLa cells were cultured on glass coverslips to 80% confluency and transfected with the respective ORF57–GFP constructs. Twenty-four hours after transfection, cells were fixed, and immunofluorescence was carried out by using a nucleolin-specific polyclonal antibody as a marker of the nucleolus.
Nucleolar Trafficking of ORF57 Is Essential for Viral mRNA Nuclear Export.
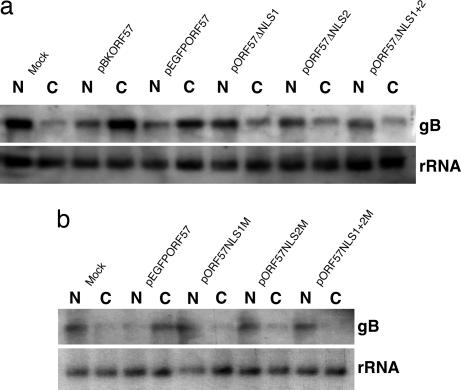
The core function of ORF57 is the nuclear export of intronless viral mRNA. Having shown earlier that several key cellular mRNA export factors are redistributed to the nucleolus, where they colocalize with ORF57, one intriguing possibility is that the nucleolar localization of ORF57 may be important for its role in viral mRNA nuclear export. To test this hypothesis, viral mRNA export assays were performed. 293T cells were transfected with a vector expressing glycoprotein B (gB, an intronless late structural gene from HVS), in addition to mock control, CMV-ORF57, and ORF57–GFP, and the ORF57–GFP deletion and point mutants described above. Twenty-four hours after transfection, total RNA was extracted from the nuclear and cytoplasmic cell fractions. RNA from each fraction was then analyzed by Northern blot using a gB-specific radiolabeled probe. As observed in Fig. 3a, cells that were transfected with the gB vector alone retain the vast majority of gB mRNA in the nuclear RNA fraction (Fig. 3a, lanes 1 and 2). Upon cotransfection of gB and CMV-ORF57 or ORF57–GFP, there is a shift in gB mRNA localization from the nuclear fraction to predominantly the cytoplasmic fraction, symptomatic of ORF57-mediated viral mRNA nuclear export (Fig. 3a, lanes 3–6). However, when gB is cotransfected with ORF57ΔNLS1, ORF57ΔNLS2, or the double ORF57ΔNLS1+2, the majority of gB mRNA is no longer found in the cytoplasmic fraction; instead, it is retained in the nuclear pool, indicative of a failure in ORF57-mediated viral mRNA nuclear export (Fig. 3a, lanes 7–12). This experiment was also performed by using the ORF57 point mutants, and data shown in Fig. 3b clearly illustrate the same loss of viral mRNA nuclear export phenotype depicted in Fig. 3a.
Fig. 3.
ORF57 mutants that lack a NoLS can no longer export viral mRNA. (a) 293T cells were cultured in 35-mm plates to 80% confluency and transfected with the respective ORF57–GFP deletion constructs plus pBK-gB. Cells were incubated at 37°C for 24 h, and total RNA was extracted from nuclear and cytoplasmic fractions by using a PARIS kit. RNA was separated on a denaturing agarose gel, and Northern blotting was carried out by using a gB-specific radiolabeled probe. A probe to the 18S subunit of ribosomal RNA was used as a loading control. (b) The experiment outlined in a was repeated by using the ORF57–GFP point mutants to confirm that these mutants possessed the mRNA export defect.
ORF57 NoLS Mutants Still Bind Viral mRNA and Interact with Cellular mRNA Export Machinery with Similar Affinities to WT Protein.
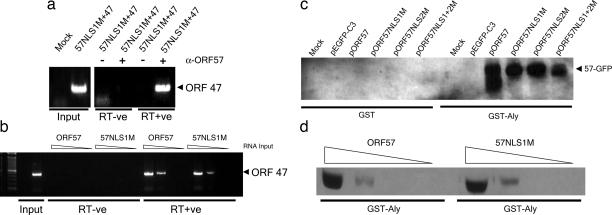
We next sought to establish whether the ORF57 nucleolar localization mutants were still able to bind viral mRNA and interact with cellular mRNA export proteins. If the mutant ORF57 proteins lacked these properties, this finding would offer an alternative explanation for the observed failure to export viral mRNA. First, to test whether ORF57 NLS mutants were still able to associate with HVS mRNA transcripts, RNA immunoprecipitation (RNA-IP) assays were performed. Initially, we confirmed that WT ORF57 interacted with ORF47 (a late structural gene) mRNA by RNA-IP in HVS A11-S4-infected OMK cells (see Figs. 7 and 8). Next, a vector expressing HVS ORF47 was transfected into 293T cells either alone or in addition to a vector expressing the ORF57 NLS1 mutant, and total cell extracts were harvested in RNase-free lysis buffer. After immunoprecipitation with an ORF57-specific polyclonal antibody, samples were treated with proteinase K, and total RNA was phenol/chloroform-extracted for DNase treatment, reverse transcription, and nested PCR. RNA extracted from transfected 293T total cell lysates served as negative and positive controls. ORF57 immunoprecipitation of extracts from 293T cells that had been transfected with ORF47 and ORF57 NLS1 mutant displayed a clear interaction between ORF57 NLS1 mutant and the viral ORF47 mRNA, when compared with no-antibody controls (Fig. 4a), this PCR product was also absent in the reverse transcription negative control, indicating no DNA contamination. Moreover, to determine whether WT ORF57 and ORF57NLS1M had similar RNA-binding affinities, RNA-IPs were repeated by using 10-fold dilutions of extracted RNA. The results showed that both WT ORF57 and ORF57NLS1M bind RNA at similar dilutions (Fig. 4b). These data clearly show that the ORF57 NLS1 mutant, although unable to traffic to the nucleolus, retains the ability to bind intronless viral mRNA. Second, GST pull-downs were performed to determine whether the ORF57 NLS mutants maintained the ability to interact with nuclear export factors. Recombinant GST-Aly was expressed in Escherichia coli, purified on GST-agarose beads, and used in pull-downs with total cell lysate from 293T cells transfected with the WT or mutant ORF57 constructs. GST alone served as a negative control. Western blot analysis revealed that GST-Aly binds to full-length ORF57 as described in ref. 17. Moreover, all three of the ORF57 NLS point mutants were able to interact with Aly in vitro (Fig. 4c). Furthermore, to determine whether WT ORF57 and ORF57NLS1M have similar binding affinities with Aly, GST pull-downs were again performed. GST-Aly bound to glutathione-agarose beads was incubated with 10-fold dilutions of total cell lysate from 293T cell transfected with WT ORF57 or ORF57NLS1M constructs. The results showed that both WT ORF57 and ORF57NLS1M bind Aly at similar dilutions (Fig. 4d). This result confirmed that although pORF57NLS1M was unable to localize to the nucleolus, it had not lost the ability to interact with Aly.
Fig. 4.
The ORF57 NLS1 mutant retains the ability to bind viral RNA and interact with mRNA export proteins with similar affinity to WT ORF57. (a) 293T cells were transfected with the labeled expression vectors and cultured for 24 h. After UV cross-linking, RNA-IPs were performed by using an antibody to ORF57, and nested RT-PCR was carried out on the extracted RNA for ORF47 mRNA. Total RNA extracted from mock-transfected and ORF47/ORF57-transfected 293T cells was used as a negative and positive control for the RT-PCR (Input). (b) RNA-IPs were repeated to compare the binding affinities of WT ORF57 and ORF57NLS1M. Nested RT-PCRs were performed by using 10-fold dilutions of extracted RNA (10, 1, 0.1, and 0.01 μl). (c) BL-21 cells were transformed with either pGEX-4T or pGEX-Aly, and expression of the fusion protein was induced. GST-Aly and control GST protein was bound to glutathione-agarose beads and purified by washing before incubating bound beads with total cell lysate from 293T cells transfected with the labeled constructs. Beads were washed, and bound proteins were analyzed by Western blot using a GFP-specific antibody. (d) GST pull-down assays were repeated to compare the binding affinity of GST-Aly with WT ORF57 and ORF57NLS1M. GST-Aly bound to glutathione-agarose beads was incubated with 10-fold dilutions (1,000, 100, 10, and 1 μl) of total cell lysate from 293T cells transfected with WT ORF57 or ORF57NLS1M constructs.
Retargeting of Nucleolar Localization with a Heterologous NoLS Reinstates ORF57-Mediated Viral mRNA Nuclear Export.
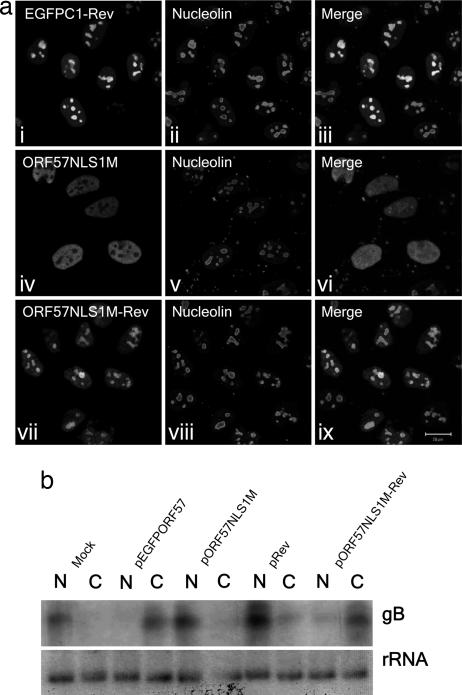
The observed loss of ORF57-mediated viral mRNA nuclear export in ORF57 NLS mutants that are unable to traffic to the nucleolus is not caused by a loss of binding to the viral mRNA or a failure to interact with cellular export factors. Therefore, we set out to demonstrate a direct role for the nucleolus in the ORF57-mediated export of intronless viral mRNA. Because the only phenotypic change we have observed in the 57NLS1M mutant from WT ORF57 is the inability to traffic to the nucleolus, it was of interest to determine whether reconstitution of nucleolar localization would be sufficient to restore ORF57NLS1M-mediated viral mRNA export. The HIV-1 Rev protein contains an extensively characterized minimal NoLS (21, 22); this 16-aa region was inserted at the C terminus of the ORF57 NLS1 mutant vector to create pORF57NLS1M-Rev. In addition, pEGFPC1-Rev expressing the Rev NoLS fused to GFP alone was engineered. Immunofluorescence microscopy performed on transiently transfected HeLa cells confirmed that pEGFPC1-Rev was targeted to the nucleolus (Fig. 5a i–iii). As reported earlier, the ORF57 NLS1 mutant displays a nuclear localization pattern but is absent from the nucleolus (Fig. 5a iv–vi); however, the ORF57NLS1M-Rev protein readily localized to both the nucleus and the nucleolus, reminiscent of the WT ORF57 protein (Fig. 5a vii–ix), confirming that the C-terminal insertion of the Rev NoLS had successfully restored the ORF57 NLS1 mutant's ability to traffic to the nucleolus.
Fig. 5.
Insertion of the HIV-1 Rev NoLS at the C terminus of pORF57NLS1M reconstitutes nucleolar localization and restores viral mRNA export. (a) HeLa cells were cultured on glass coverslips to 80% confluency and transfected with the labeled constructs. Twenty-four hours after transfection, cells were fixed, and immunofluorescence was carried out by using a nucleolin-specific polyclonal antibody. (b) 293T cells were cultured in 35-mm plates to 80% confluency and transfected with the respective ORF57–GFP constructs plus pBK-gB. Cells were incubated at 37°C for 24 h, and total RNA was extracted from nuclear and cytoplasmic fractions by using a PARIS kit. RNA was separated on a denaturing agarose gel, and Northern blotting was carried out by using a gB-specific radiolabeled probe. A probe to the 18S subunit of ribosomal RNA was used as a loading control.
Having established that ORF57NLS1M-Rev was able to traffic to the nucleolus, we then asked: Does restoration of nucleolar trafficking also restore the defect in ORF57-mediated viral mRNA nuclear export? 293T cells were transfected with a gB expression vector in addition to mock control, pORF57–GFP, pORF57NLS1M, and pORF57NLS1M-Rev. Twenty-four hours after transfection, total RNA was extracted from the nuclear and cytoplasmic cell fractions. The RNA from each fraction was then analyzed by Northern blot using a gB-specific radiolabeled probe. As reported earlier, gB mRNA is retained in the nuclear fraction in the mock control but exported into the cytoplasm when cotransfected with ORF57–GFP. The ORF57 NLS1 mutant lacks this export function; however, data shown in Fig. 5b clearly show that ORF57-mediated mRNA nuclear export function is restored in the presence of the ORF57NLS1M-Rev protein. These data show that nucleolar trafficking of a key viral mRNA export protein is an essential event in viral mRNA nuclear export.
Discussion
The mechanisms that are responsible for trafficking of a protein to subnuclear structures, such as the nucleolus, are not clearly defined. Several cellular and viral NoLSs have now been identified and characterized, although no obvious consensus sequence has been described. At present, most of the reported NoLSs are rich in arginine and lysine residues and invariably encompass a pat4 or pat7 NLS (3, 23–25). We have identified two distinct NLSs within ORF57, either of which is necessary and sufficient for ORF57 nuclear localization. The first NLS is a classical pat4 motif (KRPR), and the second consists of 10 amino acids, five of which are basic residues (RRPRSPFRKP). Uniquely, either NLS is sufficient for nuclear localization, but both NLSs are essential to traffic ORF57 to the nucleolus. How exactly the two NLSs facilitate ORF57 nucleolar localization is unclear. Current thinking states that proteins that localize to the nucleolus do so by means of an interaction with other nucleolar components (3, 26–28). Therefore, one possible explanation is that either NLS is capable of nuclear import by means of an interaction with importins. However, the remaining NLS is then required for nucleolar targeting by means of an as-yet- unidentified nucleolar component. This protein has yet to be identified, but it is possible that its identification may be accomplished by comparing the nucleolar interactions between WT ORF57 and 57NLS1M. Alternatively, the close proximity of the two NLSs may function in an additive manner to drive ORF57 into the nucleolus. To our knowledge, there are no examples of two distinct NLSs functioning in tandem to facilitate nucleolar localization; therefore, this nucleolar targeting mechanism is unique. The importance of nucleolar targeting to ORF57 function is highlighted when a heterologous NoLS is placed distal to the mutated ORF57 NoLS. This modification fully rescues the mRNA nuclear export function of the ORF57 NLS1 mutant and suggests that it is the actual localization of ORF57 in the nucleolus that is critical for its function in viral mRNA nuclear export, rather than a specific feature of the ORF57 NoLS per se.
An increasing number of virus proteins, including examples from retroviruses, DNA/RNA viruses, and plant viruses, have been reported to exhibit nucleolar localization (9, 22, 29–35). However, no specific function has been assigned to the nucleolar localization of any of these proteins. It is likely that these viruses are utilizing the plurifunctional nature of the nucleolus to enhance virus gene function and replication or subvert host cell function. Of all virus proteins known to localize to the nucleolus, HIV-1 Rev is the best characterized. The function of Rev is to promote the export of the unspliced 9-kb and singly spliced 4-kb HIV-1 RNAs out of the nucleus (36–39), and, as such, Rev is functionally similar to the ORF57 protein from herpesviruses. Although the actual localization of Rev itself to the nucleolus has yet to be shown to be essential for its role in HIV-1 replication, it appears that the nucleolus has a fundamental role to play in HIV-1 replication. Some elegant experiments using a hammerhead ribozyme targeted to the nucleolus and specific for HIV-1 RNA showed that HIV-1 RNA trafficking through the nucleolus is essential for HIV-1 replication (40). Similarly, nucleolar targeting of a RNA-based inhibitor of HIV-1 Tat (a second HIV-1 nucleolar protein) was also able to significantly decrease HIV-1 replication, confirming that nucleolar targeting of Tat is required for HIV-1 replication (41).
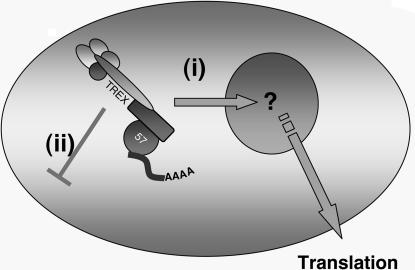
The data reported here describe a direct functional role for nucleolar trafficking of a viral protein, specifically in viral mRNA nuclear export. We have generated specific ORF57 point mutants that retain the ability to localize to the nucleus, bind viral mRNA, and interact with nuclear export factors; however, they do not localize to the nucleolus. These mutants are unique in the fact that they retain every feature of the WT protein apart from localizing to the nucleolus. Intriguingly, these data highlight a previously unrecognized and essential role for the nucleolus in ORF57-mediated viral mRNA nuclear export (Fig. 6), a role that was confirmed by rescuing the ORF57 point mutant's nucleolar trafficking defect by means of insertion of the HIV-Rev NoLS at its C terminus.
Fig. 6.
Proposed model for viral mRNA export in HVS. These data led to the findings that ORF57 trafficking through the nucleolus is essential for viral mRNA nuclear export. WT ORF57 is able to traffic through the nucleolus, bind intronless viral mRNAs, and associate with cellular mRNA export proteins to achieve viral mRNA nuclear export (i). However, ORF57 mutants incapable of nucleolar trafficking are unable to export bound viral mRNA, even though assembly of the export complex appears to be unperturbed (ii).
There is a growing precedence for the nucleolus to have regulatory function in mRNA export in eukaryotic cells. For example, in fission yeast, the nucleolus is essential for the nuclear export of intron-containing mRNA transcripts (42). Furthermore, the human nucleolar proteome currently contains in excess of 30 proteins that are known to be involved mRNA modification and export, strongly suggesting that the nucleolus has an as-yet-unidentified role in cellular mRNA modification or export. The redistribution of nuclear export proteins to the nucleolus in HVS-infected cells, and, specifically, the observation that hTREX colocalizes with ORF57 in the nucleolus, poses the question: Is the nucleolus acting as an assembly point for viral messenger ribonucleoproteins? Although this scenario remains a possibility, our data show that the ORF57 NLS1 mutant retains the ability to interact with components of hTREX (namely, Aly), indicating that the viral mRNA export complex and ORF57 are more likely to assemble in the nucleoplasm and then shuttle through the nucleolus before nuclear export.
Presumably, there is some advantage for the herpesvirus to colocalize its mRNA export protein and cofactors inside the nucleolus, although exactly what this advantage may be remains largely unknown. A number of different RNAs are known to be processed during nucleolar trafficking. Examples include the 2′-O-methylation and/or pseudouridylation of small nucleolar RNAs (43); modification/processing of N-myc, c-myc, and myoD1 mRNA (44); U6 small nuclear RNA (45); the signal recognition particle (46); telomerase RNA (47); and the RNase P RNA (48). It is possible that the herpesvirus mRNA is also being modified in a similar fashion. Alternatively, nucleolar trafficking of viral messenger ribonucleoproteins could allow the virus to avoid surveillance mechanisms that may otherwise degrade foreign viral intronless mRNAs before translation.
To summarize, our results establish that the nucleolus has an essential role in ORF57-mediated viral mRNA nuclear export. We describe nucleolar localization of ORF57 and show that ORF57 also promotes the redistribution of key mRNA export proteins to the nucleolus. ORF57 nucleolar localization occurs by means of a previously unrecognized NoLS that is composed of a pat4 NLS and a second NLS, either of which is capable of ORF57 nuclear import, but both of which are required for trafficking of ORF57 to the nucleolus. ORF57 point mutants that abrogate nucleolar trafficking also render ORF57 nonfunctional for viral mRNA nuclear export, a phenotype that was rescued by the insertion of a nonrelated NoLS at the C terminus of the ORF57 mutant. Together, these data describe a direct functional role for nucleolar targeting of a viral protein and warrant further investigation into the essential events occurring during ORF57 nucleolar trafficking and how they serve to regulate herpesvirus mRNA nuclear export. Moreover, these findings serve to highlight the plurifunctional role of the nucleolus in RNA processing.
Methods
Plasmids.
Details of plasmid construction and oligonucleotides are available on request.
Cell Culture and Transfection.
HeLa, HEK-293 (HEK 293T), and OMK cells were cultured in DMEM (Invitrogen, Paisley, U.K.) supplemented with 10% FCS (Invitrogen), glutamine, and penicillin-streptomycin. Plasmid transfections were performed by using Lipofectamine 2000 (Invitrogen) per the manufacturer's instructions.
Fluorescence Microscopy.
Immunofluorescence on adherent cells was carried out as described in ref. 17. Briefly, cells were grown on glass coverslips, washed in PBS, and fixed by successive 10-min incubations with formaldehyde [4% (vol/vol) in PBS] and Triton X-100 [1% (vol/vol) in PBS]. Cells were washed three times in PBS and blocked in PBS containing BSA [1% (wt/vol), Sigma-Aldrich, Poole, U.K.] for 1 h at 37°C. Primary and secondary antibodies were diluted 1:100 in PBS/1% BSA solution and incubated sequentially for 1 h at 37°C in the dark with 5× PBS washes carried out after incubation with the primary and secondary antibody. Coverslips were mounted in Vectorshield mounting medium (Vector Laboratories, Peterborough, U.K.), and staining was visualized on an upright LSM 510 META Axioplan 2 microscope (Carl Zeiss, Berlin, Germany) using LSM Imaging software (Carl Zeiss).
Immunoprecipitations.
GST pull-downs and coimmunoprecipitations, in addition to subsequent protein analysis by SDS/PAGE and Western blot, were carried out as described in ref. 17. RNA-IPs were carried out as follows: ≈2 × 107 293T cells were washed with ice-cold PBS and UV-irradiated (at 900 mJ/cm2 for 4 min) in a Stratalinker 2400 (Stratagene, Amsterdam, The Netherlands) to cross-link protein and RNA. Cells were scraped, transferred to an RNase-free tube, and pelleted at 300 × g for 3 min. Cell pellets were resuspended in 2 ml of TAP lysis buffer [10 mM Hepes/10 mM KCl/2 mM MgCl2/0.5% (vol/vol) Nonidet P-40/5% (vol/vol) glycerol/protease inhibitor mixture (Roche, Lewes, U.K.)/RNase Out (1 μl/ml−1, Invitrogen)]. Cells were incubated on ice for 30 min with occasional vortexing to lyse cells. Protein A-Sepharose beads were prepared, coupled to the desired antibody (10 μl of antibody per immunoprecipitation), and added to the cleared cell lysate described above; samples were mixed end-over-end for 2 h at 4°C. Protein A beads were then washed five times with NT2 wash buffer [50 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mM MgCl2/0.05% (vol/vol) Nonidet P-40], and RNA was eluted by incubating beads at 55°C for 15 min in 100 μl of NT2 buffer containing SDS [0.1% (vol/vol)] and 0.5 mg/ml proteinase K. Beads were pelleted, and the supernatant was transferred to a fresh RNase-free tube. RNA was extracted by using TRIzol (Invitrogen), and precipitation was aided by the addition of 20 μg of glycogen (Sigma-Aldrich). Any contaminating DNA was removed from RNA samples by using the DNA-free kit (Ambion, Huntingdon, U.K.) per the manufacturer's instructions. Nested RT-PCR (SuperScript II, Invitrogen) was carried out on each RNA sample to identify any positive interactions; total RNA extractions from the relevant samples served as negative and positive controls.
Northern Blots.
Nuclear and cytoplasmic RNA for Northern blots was extracted from transiently transfected cells by using the PARIS kit (Ambion) per the manufacturer's instructions. Northern blots were carried out as described in ref. 14. Briefly, nuclear and cytoplasmic RNAs were separated by electrophoresis on 1% denaturing formaldehyde-agarose gel. The RNA was transferred to Hybond-N membrane (Amersham Pharmacia, Chalfont St. Giles, U.K.) and hybridized with 32P-radiolabeled random-primed probes specific for gB and 18S rRNA.
Supplementary Material
Acknowledgments
We thank Robin Reed (Harvard University, Boston, MA) for the generous gift of antibody reagents, Michael Malim (Kings College, London, U.K.) for provision of the HIV-Rev expression construct, and David Matthews and Julian Hiscox for useful discussions. This work was supported by the Biotechnology and Biological Sciences Research Council.
Abbreviations
- NoLS
nucleolar localization signal
- NLS
nuclear localization signal
- HVS
herpesvirus saimiri
- hTREX
human TREX
- OMK
owl monkey kidney
- gB
glycoprotein B
- RNA-IP
RNA immunoprecipitation.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office. J.U.J. is a guest editor invited by the Editorial Board.
References
- 1.Misteli T, Spector DL. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 2.Shaw PJ, Jordan EG. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 3.Carmo-Fonseca M, Mendes-Soares L, Campos I. Nat Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 4.Olson MO, Dundr M, Szebeni A. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 5.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 7.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiscox JA. Arch Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrecht JC, Nicholas J, Biller D, Cameron KR, Biesinger B, Newman C, Wittmann S, Craxton MA, Coleman H, Fleckenstein B, et al. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boshoff C, Chang Y. Annu Rev Med. 2001;52:453–470. doi: 10.1146/annurev.med.52.1.453. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 13.Boyne JR, Whitehouse A. Clin Microbiol Infect. 2006;12:110–117. doi: 10.1111/j.1469-0691.2005.01317.x. [DOI] [PubMed] [Google Scholar]
- 14.Whitehouse A, Cooper M, Meredith DM. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin DJ, Hall KT, Stevenson AJ, Markham AF, Whitehouse A. J Virol. 1999;73:10519–10524. doi: 10.1128/jvi.73.12.10519-10524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin DJ, Whitehouse A. J Biol Chem. 2001;276:19905–19912. doi: 10.1074/jbc.M009513200. [DOI] [PubMed] [Google Scholar]
- 17.Williams BJ, Boyne JR, Goodwin DJ, Roaden L, Hautbergue GM, Wilson SA, Whitehouse A. Biochem J. 2005;387:295–308. doi: 10.1042/BJ20041223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper M, Goodwin DJ, Hall KT, Stevenson AJ, Meredith DM, Markham AF, Whitehouse A. J Gen Virol. 1999;80 (Part 5):1311–1316. doi: 10.1099/0022-1317-80-5-1311. [DOI] [PubMed] [Google Scholar]
- 20.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 21.Cochrane AW, Perkins A, Rosen CA. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota S, Siomi H, Satoh T, Endo S, Maki M, Hatanaka M. Biochem Biophys Res Commun. 1989;162:963–970. doi: 10.1016/0006-291x(89)90767-5. [DOI] [PubMed] [Google Scholar]
- 23.Cokol M, Nair R, Rost B. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Bustos J, Heitman J, Hall MN. Biochim Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- 25.Macara IG. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Mol Cell Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagahama M, Hara Y, Seki A, Yamazoe T, Kawate Y, Shinohara T, Hatsuzawa K, Tani K, Tagaya M. Mol Biol Cell. 2004;15:5712–5723. doi: 10.1091/mbc.E04-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheer U, Hock R. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 29.Cullen BR, Hauber J, Campbell K, Sodroski JG, Haseltine WA, Rosen CA. J Virol. 1988;62:2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews DA, Russell WC. J Gen Virol. 1998;79(Part 7):1671–1675. doi: 10.1099/0022-1317-79-7-1671. [DOI] [PubMed] [Google Scholar]
- 31.Pyper JM, Clements JE, Zink MC. J Virol. 1998;72:7697–7702. doi: 10.1128/jvi.72.9.7697-7702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R, Haseltine WA, Rosen CA. J Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siomi H, Shida H, Maki M, Hatanaka M. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. J Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada H, Jiang YM, Zhu HY, Inagaki-Ohara K, Nishiyama Y. J Gen Virol. 1999;80:2157–2164. doi: 10.1099/0022-1317-80-8-2157. [DOI] [PubMed] [Google Scholar]
- 36.Cochrane AW, Chen CH, Rosen CA. Proc Natl Acad Sci USA. 1990;87:1198–1202. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Agostino DM, Ciminale V, Pavlakis GN, Chieco-Bianchi L. AIDS Res Hum Retroviruses. 1995;11:1063–1071. doi: 10.1089/aid.1995.11.1063. [DOI] [PubMed] [Google Scholar]
- 38.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 39.Zapp ML, Green MR. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 40.Michienzi A, Cagnon L, Bahner I, Rossi JJ. Proc Natl Acad Sci USA. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michienzi A, Li S, Zaia JA, Rossi JJ. Proc Natl Acad Sci USA. 2002;99:14047–14052. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ideue T, Azad AK, Yoshida J, Matsusaka T, Yanagida M, Ohshima Y, Tani T. J Cell Sci. 2004;117:2887–2895. doi: 10.1242/jcs.01155. [DOI] [PubMed] [Google Scholar]
- 43.Weinstein LB, Steitz JA. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 44.Bond VC, Wold B. Mol Cell Biol. 1993;13:3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tycowski KT, You ZH, Graham PJ, Steitz JA. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 46.Jacobson MR, Pederson T. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell JR, Wood E, Collins K. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 48.Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.