Abstract
Myxobacteria are single-celled, but social, eubacterial predators. Upon starvation they build multicellular fruiting bodies using a developmental program that progressively changes the pattern of cell movement and the repertoire of genes expressed. Development terminates with spore differentiation and is coordinated by both diffusible and cell-bound signals. The growth and development of Myxococcus xanthus is regulated by the integration of multiple signals from outside the cells with physiological signals from within. A collection of M. xanthus cells behaves, in many respects, like a multicellular organism. For these reasons M. xanthus offers unparalleled access to a regulatory network that controls development and that organizes cell movement on surfaces. The genome of M. xanthus is large (9.14 Mb), considerably larger than the other sequenced δ-proteobacteria. We suggest that gene duplication and divergence were major contributors to genomic expansion from its progenitor. More than 1,500 duplications specific to the myxobacterial lineage were identified, representing >15% of the total genes. Genes were not duplicated at random; rather, genes for cell–cell signaling, small molecule sensing, and integrative transcription control were amplified selectively. Families of genes encoding the production of secondary metabolites are overrepresented in the genome but may have been received by horizontal gene transfer and are likely to be important for predation.
Keywords: evolution of signaling, genome expansion, multicellular development
Myxobacteria are one of nature's explorations of communal living. These soil-dwelling, single-celled prokaryotes move and feed in predatory groups. Myxococcus xanthus, whose lifecycle is shown in Fig. 1, constructs species-specific multicellular structures called fruiting bodies and differentiates spores within them. Growth and sporulation alternate according to the availability of nutrient or prey. Nutrient limitation initiates fruiting body development and sporulation, whereas nutrient availability leads spores to germinate and energizes growth and cell movement by gliding. At high cell density, gliding of the long rod-shaped growing cells is constrained by interactions between the cells. Cooperative interactions are orchestrated by the cell-to-cell exchange of the soluble A signal (1) and the contact-mediated C signal (reviewed in ref. 2). The C signal network controls movement of the rod-shaped cells and regulates gene expression until the cells differentiate into spores that are unable to move on their own (3). The regulatory network, although relatively simple in design, produces complex multicellular development with true cellular differentiation. The ecological success of the myxobacterial lifestyle is measured by the millions of myxobacterial cells per gram of cultivated soil and by the fact that their 50 species are found in topsoils around the earth (4).
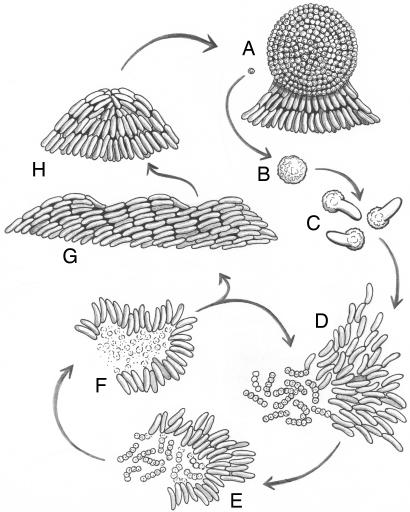
Fig. 1.
The lifecycle of M. xanthus. A swarm (a group of moving and interacting cells) can have either of two fates depending on their environment. The fruiting body (A) is a spherical structure of ≈1 × 105 cells that have become stress-resistant spores (B). The fruiting body is small (0.10 mm high) and sticky, and its spores are tightly packed. When a fruiting body receives nutrients, the individual spores germinate (C) and thousands of M. xanthus cells emerge together as an “instant” swarm (D). When prey is available (micrococci in the figure), the swarm becomes a predatory collective that surrounds the prey. Swarm cells feed by contacting, lysing, and consuming the prey bacteria (E and F). Fruiting body development is advantageous given the collective hunting behavior. Nutrient-poor conditions elicit a unified starvation stress response. That response initiates a self-organized program that changes cell movement behavior, leading to aggregation. The movement behaviors include wave formation (G) and streaming into mounded aggregates (H), which become spherical (A). Spores differentiate within mounded and spherical aggregates. We use the term “swarming” in its general sense to denote a process “in which motile organisms actively spread on the surface of a suitably moist solid medium” (81).
The sequence of the recently finished M. xanthus genome revealed a single circular chromosome of 9,139,763 bp (GenBank accession no. CP000113). That large size compared with other bacteria raises the questions of how and why genomes enlarge. It has been suggested that large bacterial genomes correlate with a variable lifestyle and a small effective population size (5–7). For example, the loss of genes from Buchnera aphidicola is attributed to a symbiotic adaptation with aphids (8). Agrobacterium tumefaciens and Sinorhizobium melliloti acquired large plasmids as they became plant pathogens and symbionts. How might the large size of the M. xanthus genome be related to its multicellular lifestyle?
Results and Discussion
Genome Expansion.
The evolutionary origin of M. xanthus lies within the δ subgroup of proteobacteria, according to the sequence of its 16S ribosomal RNA (9). All other sequenced δ-proteobacteria (eight at this time: Anaeromyxobacter dehalogenans, Bdellovibrio bacteriovorus, Desulfotalea psychrophila, Desulfovibrio desulfuricans, Desulfovibrio vulgaris, Geobacter metallireducens, Geobacter sulfurreducens, and Pelobacter carbinolicus) have genome sizes that range from 3.66 to 5.01 Mb. Because M. xanthus is 9.14 Mb there seems to have been an enlargement by 4–5 Mb. Genome expansion specific to the lineage of myxobacteria is strongly suggested by the almost identical genome sizes of the M. xanthus-related Stigmatella aurantica and Stigmatella erecta, estimated as 9.5 and 9.8 Mb, respectively (10). Among possible contributors to expansion, the acquisition of significant amounts of noncoding DNA is ruled out by the high density of coding sequences in M. xanthus, evident in Fig. 2, layers 1 and 2. More than 90% of the genome consists of protein coding sequences (CDS) with predicted products averaging 376 aa. Plasmid acquisition is ruled out because the DNA is found as a single chromosome of 9.14 Mb with a single origin of replication (base pair 1 in Fig. 2). Some of the expansion that is evident results from extensive gene duplication. For M. xanthus, comparisons of the 7,388 predicted CDS to each other using BLASTP and hidden Markov models (HMM, PFAM, and TIGRFAM) (11) indicate that 3,542 CDS, or 48% of the proteome, constitute 872 families (having at least two members) of paralogous genes. Duplications provide the raw material for the evolution of new gene functions (12, 13), and global studies have borne out the importance of duplications in bacteria (14, 15).
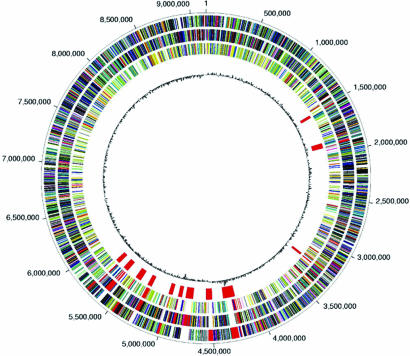
Fig. 2.
Genome map: a single circle. The entire genome is summarized in five layers. The two outermost layers represent the genes expressed in the clockwise direction (layer 1) and the counterclockwise direction (layer 2). The role of genes such as fatty acid and phospholipids, metabolism, cell envelope, amino acid biosynthesis, DNA metabolism, protein synthesis, central intermediary metabolism, energy metabolism, and regulatory functions are color-coded. Layer 3 shows the lineage-specific duplications that are discussed in Results. The enzymes that catalyze production of secondary metabolites are in layer 4. Base pair 1 was assigned to the predicted origin of replication, located by GC nucleotide skew (82) among genes in the dnaA, dnaN, recF, and gyrA region. Layer 5 plots the GC skew.
To identify the duplications that appeared during the divergence of myxobacteria from other δ-proteobacteria, the entire M. xanthus genome was compared with a reference set consisting of all of the genes in all sequenced genomes available in 2005 (J. Badger and J.E., unpublished data). The reference set for this study included four δ-proteobacteria that had been sequenced by 2005: specifically B. bacteriovorus, De. vulgaris, G. sulfurreducens, and D. psychrophila. The comparison revealed that 1,153 CDS at least, or 15.6% of the M. xanthus proteome, belong to paralogous groups of proteins that are more closely related to one another than to any protein from any other sequenced organism. We consider such duplications to be lineage-specific, assuming that they duplicated and differentiated in the immediate ancestors of M. xanthus. The lineage-specific duplications are indicated in layer 3 of Fig. 2; they are distributed at roughly equal density around the whole chromosome. Table 1 identifies the largest families of lineage-specific duplications according to their cellular function. The genomic data summarized in Table 1 were derived from the complete list of CDS with their annotations. The data are presented in Table 2, which is published as supporting information on the PNAS web site.
Table 1.
Paralogs and lineage-specific duplications
| Functional role categories | No. of genes in role category | No. of genes in paralogous families | No. of genes in lineage-specific duplication clusters | Expected no. of duplications, if by chance |
|---|---|---|---|---|
| Unknown function/general | 608 | 391 | 103 | 95 |
| Unknown function/enzymes of unknown specificity | 484 | 371 | 52 | 75 |
| Regulatory functions/protein interactions | 300 | 248 | 157 | 47 |
| Cell envelope/other | 550 | 204 | 78 | 85 |
| Signal transduction/two-component systems | 258 | 202 | 137 | 40 |
| Regulatory functions/DNA interactions | 209 | 180 | 70 | 33 |
| Transport and binding proteins/unkown substrate | 150 | 129 | 40 | 23 |
| Protein fate/degradation of proteins, peptides, and glycoproteins | 146 | 106 | 40 | 23 |
| Cell envelope/biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | 109 | 79 | 7 | 17 |
| Transport and binding proteins/cations and iron-carrying compounds | 101 | 73 | 26 | 16 |
| Energy metabolism/electron transport | 108 | 70 | 27 | 17 |
| EBP | 53 | 51 | 26 | 8 |
| STPK | 97 | 97 | 83 | 15 |
| Cellular processes/chemotaxis and motility | 99 | 66 | 46 | 15 |
| Protein fate/protein folding and stabilization | 79 | 60 | 21 | 12 |
| Transport and binding proteins/other | 69 | 59 | 17 | 11 |
| DNA metabolism/DNA replication, recombination, and repair | 106 | 58 | 4 | 17 |
The number of genes in the largest paralogous families in the M. xanthus genome are tabulated by role category (column 1). For each category, the second column shows the total number of genes in M. xanthus. The third column shows the number of genes that belong to paralogous families. The fourth column shows the number of gene clusters in the category that are lineage-specific duplications. The fifth column shows the expected (whole) number of duplicated genes, assuming that every gene in the category has the same probability of duplication.
The 15.6% of the proteome representing lineage-specific duplications would account for 1.4 Mb of the discrepancy between myxobacteria and the other δ-proteobacteria. Some of the remaining 2.6–3.6 Mb of expansion probably represents lineage-specific duplications that were not detected because of the stringency of the criteria used for determining membership in a family of paralogs. Also, at least 1.4 Mb of expansion may arise from horizontal gene transfer (HGT). HGT has played a significant role increasing the size of the γ-proteobacterial genomes (7, 16). Moreover, substantial HGT was recently proposed for B. bacteriovorus, which is a prey-consuming δ-proteobacterium like M. xanthus (17). Several methods for detecting horizontally transferred genes have been suggested. One method, used to identify the pathogenicity islands of Escherichia coli, looks for piece-wise variations of the nucleotide sequence signature because signatures tend to homogeneity around the chromosome (18, 19). Horizontally transferred segments, which initially have the donor signature, adopt with time the signature of their new lineage (20–22). Before amelioration, horizontally transferred genes can be detected by an unusual signature compared with the rest of the chromosome. However, scans around the entire M. xanthus genome revealed neither GC nucleotide skew nor signature transitions that would delineate the two edges of a segment transferred by HGT, other than those at the edges of the stable RNA genes, which vary because of selection, not to HGT (22). This failure to detect edge pairs parallels findings in B. bacteriovorus (23).
The many genes encoding enzymes of secondary metabolism in M. xanthus seem likely to have been acquired by HGT for reasons other than pairs of sequence discontinuities. Most of these genes are clustered between 4.4 and 5.8 Mb clockwise from the replication origin, with another set of clusters between 1.5 and 3.5 Mb (Fig. 2, layer 4). Although these enzymes are not sequence paralogs and are not in Table 1, these modular enzymes are duplicated in terms of their individual catalytic functions (24). Because these gene clusters constitute 8.6% of the M. xanthus genome, it has about twice the capacity for producing polyketides and mixed polyketide–polypeptides of either Streptomyces coelicolor or Streptomyces avermitilis, whose genomes are similar in size to M. xanthus (24, 25). Because the genes are clustered and (functionally) duplicated, but lack sequence discontinuities, we conclude that searching for pairs of signature discontinuities limits recognition of HGT.
One-third of the M. xanthus CDS have their four strongest BLAST hits (with cutoff e values <1e-10) outside the δ-proteobacteria. This finding negates the expectation of vertical inheritance. A similar observation made in B. bacteriovorus (23) was interpreted as a sign of ancient HGT by incorporation of undegraded prey DNA into the Bdellovibrio genome (17). But this hypothesis seems not to apply to M. xanthus for several reasons. First, HGT should be rare by virtue of its mechanism; it seems implausible that one-third of total genes should be so acquired. Second, M. xanthus is thought to feed on a wide range of bacteria in soil (discussed below in Predation), and many of its prey would be expected to have a different nucleotide signature. Their edges should have been detected, yet none were. Third, because M. xanthus was first isolated from soil in 1941 (26), some predatory HGT should, by the Gophna hypothesis (17), have been quite recent and thus detected. Fourth, considering the several periplasmic restriction endonucleases found in myxobacteria (27), we think it unlikely that gene-size fragments could survive and give HGT. Rather than ancient HGT, we find the hypothesis of rapid amelioration (19) a better explanation for the paucity of pairs of signature edges. Because the process of gene duplication would be expected to ameliorate the new copy, signature edges might thus be obscured.
Lineage-Specific Gene Duplications.
As mentioned, the many lineage-specific duplications observed are distributed all around the genome. Moreover, they play many different functional roles in M. xanthus, according to the list of functional categories that have significant numbers of lineage-specific duplications in Table 1 (28, 29). The genes seem not to have been duplicated at random, and the duplications are out of proportion to the number of genes in the various role categories, as shown by comparing the number observed with the number expected (if random) in Table 1. Some types of CDS seem not to have expanded relative to the other δ-proteobacteria: genes encoding the enzymes of DNA metabolism and the enzymes of cell envelope synthesis and degradation were duplicated less than the chance expectation (Table 1). Unknown functions/general and enzymes of unknown specificity were duplicated at the chance rate, as might have been expected for an all-encompassing category. By contrast, regulatory functions, serine–threonine protein kinases, σ54 enhancer binding proteins (EBPs), chemosensory, and motility have been duplicated more frequently than their genomic abundances would have predicted. The higher frequencies suggest that the acquisition of a new function gave them a selective advantage and thus expanded the genome. To evaluate the likelihood of this course of events, the biochemistry of several frequently duplicated proteins was examined.
STPKs.
Many of the 97 M. xanthus STPKs, which are products of the STPK genes, are found among the lineage-specific duplications in Table 1. Multiple STPK genes are not likely to have been inherited from their δ-proteobacterial precursor because they are rare: G. sulfurreducens has none, B. bacteriovorus has one, De. vulgaris has three, and D. psychrophila has two potential STPK paralogs (Table 2). Most likely the many duplications occurred as the myxobacteria were branching from their precursor. Twenty of the STPKs were determined to be essential for fruiting body development and sporulation by deletion analysis of 94 of the 97 STPK genes (30, 31). However, because this screen was carried out under a single nutritional regime, it is likely that other STPK genes are essential under other conditions, as was observed in one study of essential developmental genes (32).
Twenty-two STPK genes are organized in pairs that are adjacent or separated by fewer than four genes. Five of these gene pairs are clearly duplications because the genes are immediately adjacent and are oriented in the same direction (Fig. 4, which is published as supporting information on the PNAS web site). Pkn7 (MXAN2910) and Pkn11 (MXAN2911), for example, are adjacent and oriented in the same direction (M. Inouye and S. Inouye, unpublished observations). Another pair of adjacent STPKs, Pkn6 (MXAN2550) and Pkn5 (MXAN2549), belong to the same sequence subclass of STPKs, but the genes are oriented in opposite directions. Duplication and subsequent divergence of STPK specificity could have generated new regulatory elements.
σ54 Activator Proteins.
Many developmentally regulated genes in M. xanthus are expressed from σ54 promoters. Such promoters always require an activator protein, of which NtrC is the prototype, to form an open polymerase–promoter complex in which transcription is initiated (33–35). These activator proteins bind to enhancers, which are regulatory DNA sequences either upstream or downstream from the promoters; consequently, they are often known as EBPs. These proteins constitute another large family of lineage-specific paralogs (Table 1). M. xanthus has 53 EBP genes, and Table 1 indicates that at least half arose as lineage-specific duplications. A few may have been inherited from their δ-proteobacterial ancestor because G. sulfurreducens has 18, D. psychrophila has 8, and B. bacteriovorus has 5 potential paralogs, whereas no potential paralogs were detected in De. vulgaris (Table 2). Most EBPs are components of signal transduction circuits that respond to environmental cues. They have a common organization with a central ATPase domain responsible for ATP hydrolysis and interaction with the σ54 factor, a C-terminal DNA-binding domain, and an N-terminal sensory domain that regulates the ATPase activity of the central domain in response to sensory stimuli (36, 37).
Twelve of the EBPs in M. xanthus have a forkhead-associated (FHA) domain as their N-terminal sensory unit. The FHA domain is essential for the EBP that is encoded by MXAN4899 (38). Knockout mutations of MXAN4899 disrupt the pattern of developmental gene expression, alter fruiting body development, and block sporulation (38). The mutant phenotypes pointed to a specific role for MXAN4899 in the C signal transduction pathway. FHA domains are phosphothreonine-specific recognition domains involved in specific phosphorylation-dependent protein–protein interactions. An FHA domain would thus couple the sensory activity of a cognate STPK to the expression of σ54-dependent developmental genes (39). Other EBP genes are found to be next to an STPK gene; often, adjacent EBP/STPK gene pairs turn out to be cognate proteins. MXAN4899 and the EBP/STPK pairs provide evidence that STPKs can activate transcription, a concept recently proposed on theoretical grounds for a metabolic pathway in St. coelicolor (40).
Two-Component Systems.
The most frequent N-terminal sensory sequences of the EBPs in M. xanthus are CheY-like receiver domains. Receiver domains in bacteria are normally found in cognate pairs with a sensor histidine protein kinase (HPK) for two-component signal transduction (41) systems that respond to a broad range of extracellular or intracellular signals. The presence of these pairs in M. xanthus and in the other δ-proteobacteria suggests that most of the σ54 activators belong to two-component systems. The M. xanthus genome encodes 137 sensor and hybrid histidine kinases, which is far more than any of the other δ-proteobacteria: G. sulfurreducens has 21 sensor and hybrid histidine kinase paralogs, B. bacteriovorus has 6, De. vulgaris has 4, and D. psychrophila has 7 potential paralogs (Table 2). Some of the M. xanthus HPKs have additional sensory or output domains; there are PAS domains, which are capable of sensing the redox state (42) or of responding to light (43). There are GAF domains, which may bind cAMP/cGMP (41), and HAMP domains, which convey signals from input domains to output modules in chemotaxis receptors (44). GAF domains may be involved in sensing, producing, or degrading cyclic nucleotides, which could be global regulators in M. xanthus, although they have not yet been experimentally explored.
Several two-component gene pairs encoded by adjacent HPK and response regulator genes have previously been described in M. xanthus, including sasS/sasR (45, 46), pilS/pilR (47), the mrp genes (48), and the esp genes (49). To map more of the two-component systems in M. xanthus, the genes that neighbor each EBP were examined. Indeed, 21 among the >50 σ54 EBPs were found to neighbor a HPK (Table 3, which is published as supporting information on the PNAS web site). Twelve of the 21 EBP/HPK pairs are immediately next to each other in the genome. Another 24 of the EBP genes are neighbors of one or more genes that encode STPKs (their clustering is shown in Table 3). Expectations for a uniform nonclustered (Poisson) distribution of HPK or a STPK gene were compared with the observed distribution in Table 4, which is published as supporting information on the PNAS web site, revealing a strong tendency for the EBP genes to cluster with either an STPK or an HPK gene. The cluster intervals often included other types of regulatory components: extracytoplasmic function (ECF) σ factors (50, 51) or response regulators, as shown in Table 3. CarQ is one of those ECF σ factors; it regulates the production of protective carotenoids in response to exposure to blue light (52, 53). The observed linkages suggest that some EBP, STPK, HPK, and ECF σ factors can work together in complex regulatory units.
Regulatory Network Design.
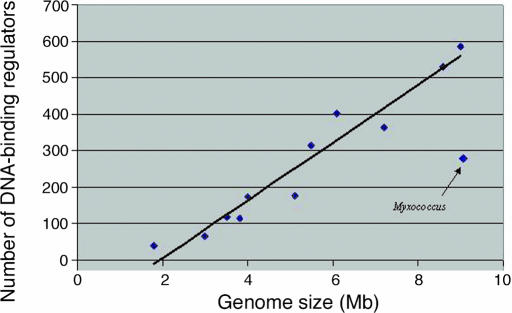
In bacteria, DNA-binding proteins that also bind a small ligand molecule are the most common transcriptional regulators (54, 55). The amount of ligand bound controls the level of transcriptional activity. The LacI, TetR, AraC, GntR, AsnC, and LuxR proteins exemplify such “one-component regulators” (55). In light of the capacity of M. xanthus to adapt to a fluctuating environment, one might have anticipated finding many one-component regulators in its genome. However, regulators of the IclR, LacI, ROK, and DeoR families are missing entirely in M. xanthus, whereas regulators of the AraC, GntR, AsnC, and LuxR families are conspicuously underrepresented. As shown in Fig. 3, one-component regulators are considerably less abundant in M. xanthus than in other soil bacteria, considering their genome size. The number for M. xanthus is less than half the expected number.
Fig. 3.
The number of one-component, DNA-binding response regulators depends on genome size. The number of one-component, DNA-binding response regulators is plotted vs. genome size for 13 completely sequenced soil bacteria. The linear correlation between genome size and number of DNA-binding transcriptional regulators (excluding M. xanthus) is 97% over the range from 1.8 Mb for Thermatoga maritima to 9.0 Mb for St. avermitilis. St. coelicolor A3 (2) (8.6 Mb), Burkholderia pseudomallei K96243 (7.2 Mb), Pseudomonas putida KT 2440 (6.1 Mb), Bacillus cereus ATCC 14579 (5.5 Mb), Shewanella oneidensis MR-1 (5.1 Mb), Caulobacter crescentus CB 15 (4.0 Mb), G. sulfurreducens PCA (3.8 Mb), De. vulgaris Hildenborough (3.5 Mb), and Deinococcus radiodurans R1 (3.0 Mb) are also included.
Underrepresentation of one-component regulators contrasts with an abundance of multicomponent regulatory pathways, which provide sensory inputs to several steps of the pathway. As described above, M. xanthus has complex regulatory pathways that involve STPKs and sensor histidine kinases linked to σ54 activators. Some pathways that include ECF σ factors, which have their own sensory inputs, are linked to those pathways. Moreover, these multicomponent systems are in abundance (Table 1). Each pathway that includes two or more proteins having sensory sites evidently has the ability to integrate several signals, some from external stimuli and some from metabolism. The “quorum-sensing pathway” found in Vibrio harveyi is an example (56). Multicomponent signaling pathways parallel, in terms of signal integration, the pathways that regulate multicellular development in eukaryotes (57).
Predation.
How can M. xanthus feed efficiently on the proteins of such a wide variety of bacteria and yeasts (4)? The first clue was the astonishing number of polyketide and nonribosomal polypeptide synthases in the genome (indicated in Fig. 2, layer 4). Also, M. xanthus may be resistant to cephalosporin-like antibiotics because it encodes isopenicillin-N-epimerase and cephalosporin hydroxylase. Secreting inhibitors to which M. xanthus is resistant would tend to inhibit the growth of competitors, as observed (58), and to weaken potential prey (59). A second genomic clue is that M. xanthus has genes for the synthesis of all of the amino acids, but it lacks the ilvC and ilvD genes, which are necessary for the biosynthesis of leucine, isoleucine, and valine. Nutritional studies showed requirements for the branched-chain amino acids (60). Inasmuch as those required amino acids account for one-fifth of the amino acids found in average proteins, predation seems to have become a reliable alternative to biosynthesis.
M. xanthus culture fluids are known to lyse cell walls (61), and the extracellular lytic and proteolytic activities probably account for the increase in the rate of M. xanthus growth observed on casein at high cell density (62). In addition to evidence for extracellular lysis, lysis is observed to follow direct physical contact with prey cells (63, 64), as illustrated in Fig. 1 D–F. Altogether, 14 M. xanthus proteins should be able to hydrolyze peptidoglycan. After the prey cell wall has been breached, proteolysis could occur, and M. xanthus has 146 putative proteases and metalloproteases (Table 1, protein fate/degradation category). Consistent with feeding by contact, half of those proteases should be either periplasmic or secreted to the cell surface, according to their signal peptides and other domains normally associated with secreted proteins. At least 25 proteases are cytoplasmic, but regulated, like lonV and lonD (65–67). As observed for mitochondria and for E. coli, M. xanthus could transfer polypeptides generated by an initial protease digestion to its regulated FtsH- and ClpP-like proteases.
We suggest that M. xanthus has protein-digesting machines dedicated to feeding. One piece of evidence for the suggestion is that many of its chaperone/protease genes are repeated: lon (two copies), groEL (two copies), ftsH (two copies), hsp90 (two copies), dnaJ (three copies), clpX (four copies), clpA/B (six copies), and dnaK (15 copies). In E. coli and mitochondria, the products of these genes are thought to degrade misfolded and damaged proteins, allowing their amino acids to be recycled (68, 69), but in M. xanthus they could be used for feeding. Second, an examination of the whole genome for genes with codon-usage frequencies that are similar to the genes encoding ribosomal proteins (the mark of “highly expressed genes”) indicates that many M. xanthus ATP-dependent proteases and many chaperones were highly expressed (70). They are as highly expressed by M. xanthus as its tricarboxylic acid cycle enzymes and electron transport proteins. This finding suggests that the chaperones and the ATP-dependent proteases are parts of multiprotein assemblies that take in folded proteins from prey cytoplasm into a periplasmic chaperone, which denatures and then digests them. The amino acid end products would be released to the cytoplasm to enter the tricarboxylic acid cycle for energy generation or to be activated for polypeptide synthesis. The E. coli FtsH protein is an integral inner membrane protein that projects its ATPase domain into the periplasm. If the M. xanthus FtsH protein, one of its most highly expressed proteins (70), is similar, it would be in a position to draw denatured prey proteins into its protease cavity (71). δ-Proteobacteria generally possess large, multiprotein networks in their periplasm involved in generating energy, like the hydrogen oxidase complex of De. vulgaris that couples to cytoplasmic sulfate reduction (72). According to this view, by sequestering proteolysis to the interior of protease cavities (68, 69) in their periplasms, the cells avoid destroying their own proteins. There is evidence that the E. coli DnaK protein presents partially unfolded proteins to FtsH protein (73). M. xanthus may need several DnaK proteins to present the wide variety of proteins found in prey. Thus, the multiprotein complexes proposed would be the molecular mouths and digestive tract of the cells.
Gene duplication made a major contribution to the myxobacterial lineage-specific expansion from a smaller ancestral δ-proteobacterium. Duplications were followed by divergence of the new gene copies, endowing them with new specificities. Genes were not duplicated at random: some gene functions were not duplicated at all, whereas genes for cell–cell signaling, small-molecule sensing, and multicomponent transcriptional control were amplified preferentially. M. xanthus has less than half the expected number of one-component transcriptional regulators for a genome of its size, and they seem to have been replaced by multicomponent regulators. A multicomponent pathway that has two or more proteins with sensory sites has the ability to integrate signals. Some signals may come from outside, and others may come from within the cell to register its metabolic state. These findings strongly suggest that the duplicated and diverged genes enabled evolution of the complex signaling required for the multicellular lifestyle of myxobacteria.
Materials and Methods
Sequencing.
M. xanthus, strain DK1622, was initially sequenced 4.5-fold by Monsanto and released to the academic community in April 2001. The Institute for Genomic Research completed the sequence by additional random sequencing, assembled it, and filled the residual gaps by directed sequencing (74).
Gene Identification.
The ORFs most likely to encode proteins were identified by GLIMMER (75), and each translated gene was searched against The Institute for Genomic Research nonidentical amino acid sequence database by using BLAST-Extend-Repraze (http://ber.sourceforge.net). The PFAM (76) and TIGRFAM (11) libraries of hidden Markov models were also searched. Sequence signatures, domains, or functional sites were predicted by using PROSITE (77), SignalP (78), TMHMM (79), and COG (80). Search results were examined for initiator codons and to identify any errors in sequence by comparison with the traces. Overlapping genes were manually resolved by using initiation codons or by retaining the one with sequence similarity to another protein. The final genome is predicted to encode 7,388 proteins.
Identification of Lineage-Specific Duplications.
To identify the paralogous gene families that have expanded subsequent to the presumed divergence of myxobacteria from other δ-proteobacteria, the entire genome was compared with a reference set consisting of all of the genes in all sequenced genomes by using the program Automated Phylogenetic Inference System (APIS; J. Badger and J.E. unpublished data), which generates phylogenetic trees for each gene.
Supplementary Material
Acknowledgments
We thank Carol Berthiaume for artistic rendering of the Myxococcus lifecycle; D. E. Whitworth for verifying the list of two-component systems before his publication; and the following members of the Myxococcus community who helped annotate the genome: J. S. Jakobsen, L. J. Shimkets, R. Welch, M. S. Avadhani, W. P. Black, H. B. Bode, P. J. Bonner, J. M. Buchner, V. Bustamante, A. Castañeda-García, M. Chavira, P. J. A. Cock, P. Curtis, M. E. Diodati, X. Y. Duan, A. Garza, R. E. Gill, J. C. Haller, P. Hartzell, P. I. Higgs, D. A. Hodgson, S. Inouye, L. Jelsbak, B. Julien, A. C. Karls, J. Kirby, L. Kroos, Y. Li, A. Lu, R. Lux, X. Ma, R. Müller, J. Muñoz-Dorado, H. Nariya, J. Pérez, V. D. Pham, R. Reid, J. J. Rivera, W. Shi, M. Singer, L. Søgaard-Andersen, D. Srinivasan, A. Treuner-Lange, L. Tzeng, P. Viswanathan, Z. Yang, D. R. Yoder, R. Yu, and D. R. Zusman. Initial 5-fold sequencing was carried out by Monsanto. The sequence was finished by The Institute for Genomic Research, supported by National Science Foundation Grant MCB-0236595. D.K. was supported by National Institute of General Medical Science Grant GM23441.
Abbreviations
- CDS
protein coding sequence
- HGT
horizontal gene transfer
- STPK
serine–threonine protein kinase
- EBP
enhancer binding protein
- HPK
histidine protein kinase
- ECF
extracytoplasmic function
- FHA
forkhead-associated.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. CP000113).
References
- 1.Plamann L, Li Y, Cantwell B, Mayor J. J Bacteriol. 1995;177:2014–2020. doi: 10.1128/jb.177.8.2014-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser D. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 3.Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, DC: Am Soc Microbiol; 1993. [Google Scholar]
- 4.Reichenbach H. In: Myxobacteria II. Dworkin M, Kaiser D, editors. Washington, DC: Am Soc Microbiol; 1993. pp. 13–62. [Google Scholar]
- 5.Lynch M, Conery JS. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 6.Konstantinidis KT, Tiedje JM. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerat E, Daubin V, Ochman H, Moran N. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamas I, Klasson L, Canback B, Naslund A, Ericksson A-S, Wernegreen J, Sandstrom J, Moran N, Andersson S. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 9.Shimkets L, Woese CR. Proc Natl Acad Sci USA. 1992;89:9459–9463. doi: 10.1073/pnas.89.20.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimkets LJ. In: Myxobacteria II. Dworkin M, Kaiser D, editors. Washington, DC: Am Soc Microbiol; 1993. pp. 85–107. [Google Scholar]
- 11.Haft D, Loftus B, Richardson D, Yang F, Eisen J, Paulsen I, White O. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 13.Kimura M, Ohta T. Proc Natl Acad Sci USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pushker R, Mira A, Rodriguez-Valera F. Genome Biol. 2004;5:R27. doi: 10.1186/gb-2004-5-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gevers D, Vandepoele K, Simillon C, Van de Peer Y. Trends Microbiol. 2004;12:148–154. doi: 10.1016/j.tim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Boucher Y, Douady CJ, Papke R, Walsh D, Boudreau M, Nesbo C, Case R, Doolittle WF. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 17.Gophna U, Charlebois R, Doolittle WF. Trends Microbiol. 2006;14:64–69. doi: 10.1016/j.tim.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Karlin S, Mrazek J, Campbell AM. J Bacteriol. 1997;179:3899–3913. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlin S. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 20.Lam H, Winkler M. J Bacteriol. 1992;174:6033–6045. doi: 10.1128/jb.174.19.6033-6045.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence J, Ochman H. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 22.Eisen JA. Curr Opin Microbiol. 2000;3:475–480. doi: 10.1016/s1369-5274(00)00125-9. [DOI] [PubMed] [Google Scholar]
- 23.Rendulic S, Jagtap P, Rosinus A, Eppinger E, Barr C, Lanz C, Keller H, Lambert C, Evans K, Goesman A, et al. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 24.Bode H, Muller R. J Ind Microbiol Biotechnol. 2006;33:577–588. doi: 10.1007/s10295-006-0082-7. [DOI] [PubMed] [Google Scholar]
- 25.Reichenbach H, Höfle G. Biotech Adv. 1993;11:219–277. doi: 10.1016/0734-9750(93)90042-l. [DOI] [PubMed] [Google Scholar]
- 26.Beebe JM. J Bacteriol. 1941;42:193–223. doi: 10.1128/jb.42.2.193-223.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer H, Reichenbach H. J Bacteriol. 1978;136:708–713. doi: 10.1128/jb.136.2.708-713.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen JA, Fraser CM. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- 29.Jordan I, Makarova K, Spouge J, Wolf Y, Koonin E. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muñoz-Dorado J, Inouye S, Inouye M. Cell. 1991;67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 31.Inouye S, Jain R, Ueki T, Nariya H, Xu C, Hsu M, Fernandez-Luque BA, Munoz-Dorado J, Farez-Vidal E, Inouye M. Microb Comp Genomics. 2000;5:103–120. doi: 10.1089/10906590050179783. [DOI] [PubMed] [Google Scholar]
- 32.Jakobsen JS, Jelsbak L, Jelsbak L, Welch R, Cummings C, Goldman B, Stark E, Slater SC, Kaiser D. J Bacteriol. 2004;186:4361–4368. doi: 10.1128/JB.186.13.4361-4368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham DL, Szeto D, Keener J, Kustu S. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 34.Sasse-Dwight S, Gralla JD. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 35.Wedel A, Kustu S. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 36.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studholme D, Dixon R. J Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jelsbak L, Givskov M, Kaiser D. Proc Natl Acad Sci USA. 2005;102:3010–3015. doi: 10.1073/pnas.0409371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroos L. Proc Natl Acad Sci USA. 2005;102:2681–2682. doi: 10.1073/pnas.0500157102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bibb MJ. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 41.Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. Washington, DC: Am Soc Microbiol; 1995. [Google Scholar]
- 42.Taylor BL, Zhulin IB. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponting CP, Aravind L. Curr Biol. 1997;7:R674–R678. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 44.Szurmant H, Ordal GW. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C, Kaplan HB. J Bacteriol. 1997;179:7759–7767. doi: 10.1128/jb.179.24.7759-7767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo D, Wu Y, Kaplan HB. J Bacteriol. 2000;182:4564–4571. doi: 10.1128/jb.182.16.4564-4571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu SS, Wu J, Kaiser D. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun H, Shi W. J Bacteriol. 2001;183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho K, Zusman DR. Mol Microbiol. 1999;34:714–725. doi: 10.1046/j.1365-2958.1999.01633.x. [DOI] [PubMed] [Google Scholar]
- 50.Bentley S, Chater K, Cerdeno-Tarraga A, Challis G, Thomson N, James K, Harris D, Quail M, Keiser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 51.Helmann JD. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 52.Moreno A, Fontes M, Murillo FJ. J Bacteriol. 2001;183:557–569. doi: 10.1128/JB.183.2.557-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitworth D, Bryan S, Berry A, McGowan S, Hodgson DA. J Bacteriol. 2004;186:7836–7846. doi: 10.1128/JB.186.23.7836-7846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babu M, Teichmann SA. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulrich LE, Koonin EV, Zhulin IB. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 57.Levine M, Davidson EH. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiegna F, Velicer GJ. PLoS Biol. 2005;3:e370. doi: 10.1371/journal.pbio.0030370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chater KF, Hopwood DA. In: Genetics of Bacterial Diversity. Hopwood DA, Chater KF, editors. London: Academic; 1989. pp. 129–150. [Google Scholar]
- 60.Bretscher AP, Kaiser D. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sudo S, Dworkin M. J Bacteriol. 1972;110:236–245. doi: 10.1128/jb.110.1.236-245.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenberg E, Keller KH, Dworkin M. J Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McBride MJ, Zusman DR. FEMS Microbiol Lett. 1996;137:227–231. doi: 10.1111/j.1574-6968.1996.tb08110.x. [DOI] [PubMed] [Google Scholar]
- 64.Zhang H, Rao N, Shiba T, Kornberg A. Proc Natl Acad Sci USA. 2005;102:13416–13420. doi: 10.1073/pnas.0506520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tojo N, Inouye S, Komano T. J Bacteriol. 1993;175:2271–2277. doi: 10.1128/jb.175.8.2271-2277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tojo N, Inouye S, Komano T. J Bacteriol. 1993;175:4545–4549. doi: 10.1128/jb.175.14.4545-4549.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hager E, Tse H, Gill RE. Mol Microbiol. 2001;39:765–780. doi: 10.1046/j.1365-2958.2001.02266.x. [DOI] [PubMed] [Google Scholar]
- 68.Gottesman S. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 69.Sauer R. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karlin S, Brocchieri L, Mrazek J, Kaiser D. Proc Natl Acad Sci USA. 2006;103:11352–11357. doi: 10.1073/pnas.0604311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ito K, Akiyama Y. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 72.Heidelberg J, Seshadri R, Haveman S, Hemme C, Paulsen I, Kolonay J, Eisen J, Ward N, Methe B, Brinkac L, et al. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- 73.Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B. FEBS Lett. 2002;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- 74.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist C, Hofmann K, Bairoch A. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bendtsen J, Nielsen H, von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 79.Krogh A, Larson B, von Heijne G, Sonnhammer E. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 80.Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, et al. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singleton P, Sainsbury D. Dictionary of Microbiology and Molecular Biology. Chichester, UK: Wiley; 2001. [Google Scholar]
- 82.Lobry JR. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.