Abstract
Much research has implicated the striatum in motor learning, but the underlying mechanisms have not been identified. Although NMDA receptor (NMDAR)-dependent long-term potentiation has been observed in the striatum, its involvement in motor learning remains unclear. To examine the role of striatal NMDAR in motor learning, we created striatum-specific NMDAR1 subunit knockout mice, analyzed the striatal anatomy and neuronal morphology of these mice, evaluated their performance on well established motor tasks, and performed electrophysiological recordings to assay striatal NMDAR function and long-term synaptic plasticity. Our results show that deleting the NMDAR1 subunit of the NMDAR specifically in the striatum, which virtually abolished NMDAR-mediated currents, resulted in only small changes in striatal neuronal morphology but severely impaired motor learning and disrupted dorsal striatal long-term potentiation and ventral striatal long-term depression.
Keywords: long-term potentiation, NMDA receptor, knockout, RGS9-2
The basal ganglia are known to control voluntary behavior, but extensive evidence shows that they also play a role in learning and memory. In particular, the striatum, a major nucleus of the basal ganglia, has been implicated in the acquisition of instrumental responses and habit formation (1, 2). Various motor learning tasks, such as chaining of motor sequences, visuomotor skill acquisition, instrumental lever-pushing, and serial reaction-time tests all involve the striatum (3,4,5–6).
Striatal learning is thought to depend on neuronal modification through alterations in neuronal ensemble activity and synaptic plasticity (1, 7). Several studies have identified changes in ensemble activity patterns during procedural learning tasks (1, 8). In a rotarod motor skill learning paradigm, mice exhibited changes in neuronal ensemble activity that varied in pattern through the different phases of learning (9). In addition to ensemble changes, synaptic plasticity in the striatum has also been detected. Both long-term depression (LTD) and long-term potentiation (LTP) in the dorsal striatum (10,11–12) and LTD in the ventral striatum or nucleus accumbens (NAc) have been reported by previous studies (13). Synaptic plasticity in the ventral striatum has been linked mainly to behavioral sensitization and appetitive Pavlovian learning (14,15–16). Corticostriatal LTP, on the other hand, has been implicated in instrumental learning, with the degree of potentiation correlating with the time required for acquisition of a lever-pressing skill (4). Striatal synaptic plasticity involves a variety of neurotransmitters, including glutamate (10, 17–19), and pharmacological inhibition of striatal NMDAR activity interferes with instrumental learning (12, 20).
Despite the mounting evidence for a critical role for the striatum in motor learning and significant progress in our understanding of striatal synaptic plasticity, there is little evidence directly linking any particular mechanism of striatal synaptic plasticity with a particular type of motor learning. In this study, we employ a genetic mouse model that lacks NMDAR1, a subunit of NMDAR, specifically in the striatum and show that the ablation of striatal NMDAR activity impairs motor learning, along with a corresponding deficit in striatal synaptic plasticity.
Results
Generation of Knockout (KO) Mice.
The striatum-specific Cre mouse was made by employing the restricted expression pattern of RGS9-2 protein, the product of a splice variant of the RGS9 gene that is expressed predominantly in the striatum (21). A cre gene was inserted at the 3′ end of the RGS9 gene (Fig. 1A). To confirm that Cre was expressed mainly in the striatum, the RGS9-cre mouse was crossed with the ROSA26 reporter mouse, which has a functional lacZ gene only in cells where a sequence-specific recombination by Cre has occurred (22). Progenies containing both RGS9-cre and ROSA26 genes from postnatal day 8 (P8) to P90 were histochemically processed for β-galactosidase activity. Recombination was detected in the striatum as early as P8. The olfactory tubercle also exhibited recombination, but no signal was detected in the cerebellum and substantia nigra (Fig. 1B).
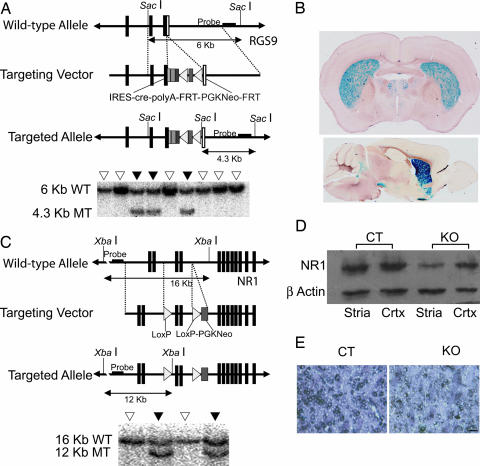
Fig. 1.
Generation of RGS9-cre and NMDAR1-loxP mice. (A) Targeting construct for the RGS9-cre mouse. Filled boxes represent exons 17 and 18 and the translated region of exon 19. Open boxes represent the untranslated region of exon 19. A cassette containing an IRES, cre coding sequence, polyA tail, neomycin resistance gene driven by the PGK promoter flanked by FRT sequences was placed downstream of the translated region of exon 19. Also shown is a representative Southern blot analysis of the transfected ES cell colonies. WT, wild-type locus; MT, mutant locus; filled inverted triangles, targeted clone; open inverted triangles, untargeted clone. (B) β-galactosidase histochemistry of adult mice produced by crossing RGS9-cre with ROSA26 reporter mice. Activity of β-galactosidase was present predominantly in the striatum (Upper), with expression also in the olfactory tubercles (Lower), indicating that Cre expression was restricted to those areas. Note the absence of recombinase activity in the cerebellum. (C) Targeting construct for the NMDAR1-loxP mouse. LoxP sequences flank exons 9 and 10 of the NMDAR1. A PGK-neo cassette was placed after the second loxP. Also shown is representative Southern blot analysis of the transfected ES cell colonies with and without homologous incorporation of the targeting construct. (D) Western blot analysis with NMDAR1 antibody showing an approximate 65% reduction (n = 2 per genotype) of NMDAR1 expression in striata of KO mice relative to CT mice. Cortical NMDAR1 levels were unaffected. CT, control mouse; KO, knockout mouse; Stria, striatum; Crtx, cortex. (E) In situ hybridization with an NMDAR1 probe showed much less NMDAR1 mRNA in the striatum of KO than CT mice. (Scale bar: 20 μm.)
To make the NMDAR1-loxP mouse, we used a targeting construct that allowed for the Cre-directed removal of exons 9 and 10 of the NMDAR1 gene (23) (Fig. 1C). RGS9-cre/NMDAR1-loxP double-heterozygous and NMDAR1-loxP heterozygous or homozygous mice were crossed to produce knockouts of striatal NMDAR1. Western blot analysis showed a large reduction of NMDAR1 protein in the striatum, with residual amounts likely from glial cells that presumably do not express Cre or from incoming projections from the cortex and other brain regions (21) (Fig. 1D). In situ hybridization also showed a reduction of NMDAR1 in the striatum (Fig. 1E).
Anatomy.
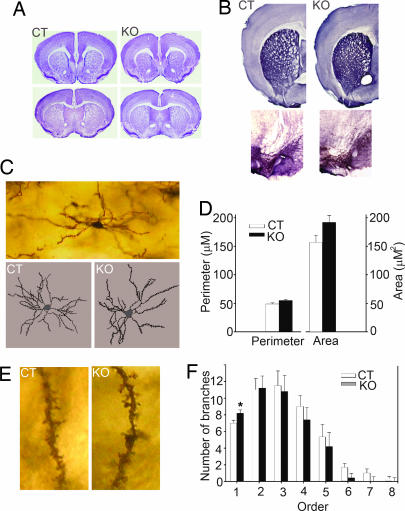
KO mice did not exhibit any gross anatomical alterations in the cortex and striatum, as determined by Nissl staining (n = 3 per genotype; Fig. 2A). Immunohistochemistry performed with a tyrosine hydroxylase antibody showed that dopaminergic neurons and dopaminergic innervations to the caudate putamen were intact in KO mice (n = 3 per genotype; Fig. 2B). Analysis of tracings of medium spiny neurons processed with the Golgi stain (n = 3 per genotype; Fig. 2C) showed that KO mice had neuronal cell bodies of mainly normal size (perimeter, P = 0.06; area, P = 0.08; Fig. 2D). Spine count and spine density were similar in KO and control (CT) mice (spine, P = 0.33; spine density, P = 0.67; Fig. 2E). KO mice had a significantly higher number of primary dendrites (KO = 8.2, CT = 7, P = 0.048; Fig. 2F).
Fig. 2.
Anatomical analysis. (A) Nissl staining showed that KO mice had no gross anatomical abnormalities in the cortex and dorsal or ventral striatum. (B) Dopaminergic innervations to the striatum (Upper) and dopaminergic neurons in the substantia nigra (Lower) also were normal in KO compared with CT mice. (C) A representative medium spiny neuron at ×10 magnification and representative traces of CT and KO medium spiny neurons produced at ×40 magnification. (D) Cell body size of medium spiny neurons of CT and KO mice were similar. (E) No differences in spine count and density were detected between CT and KO mice. Representative second-order dendrites with spines are shown at ×60 magnification. (F) KO mice had significantly more primary dendrites.
Behavioral Tests.
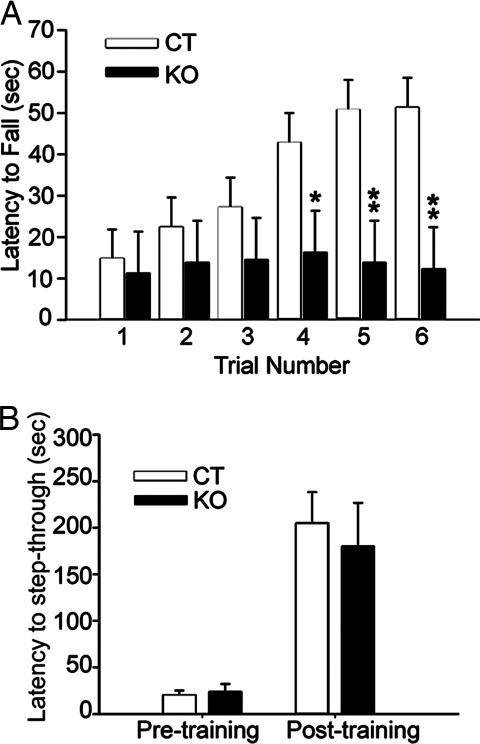
Adult mice (7 KO and 14 CT) were indistinguishable from one another on the basis of visual inspection for overt abnormalities. Mice were also tested on the accelerating rotarod (24) for three trials on one day and then again 1 week later. ANOVA results indicated a significant interaction between genotype and trial [F (5, 100) = 3.23, P ≤ 0.01; Fig. 3A]. Post hoc pairwise comparisons showed that KO mice were able to run on the rotarod equally as well as CT mice during trials 1–3. However, CT animals exhibited increased duration on the rotating rod with successive trials, whereas KO animals did not, providing evidence of impaired motor learning in KO mice. The performance of CT mice was significantly better than that of KO animals during the last three trials (P = 0.002–0.032).
Fig. 3.
Behavioral tests. (A) Rotarod test for motor learning. CT and KO mice performed equally well during the first three trials of the accelerated rotarod motor test. Time spent on the rotarod increased significantly in CT mice during trials 4–6, but no such increase was observed in KO mice, indicating a motor learning deficit in KO mice. Overall performance during the last three trials was significantly different between the CT and KO groups. ∗, P < 0.05; ∗∗, P < 0.005. (B) Inhibitory avoidance. CT and KO mice had similar latency to crossing into the dark chamber before training with the administration of a shock. Both learned from the one-trial footshock and had similar delayed latency of step-through into the dark chamber 24 h after training.
As a control to determine whether learning involving brain regions other than the striatum was affected by striatal-specific deletion of the NMDAR1 gene, an inhibitory avoidance test thought to involve the amygdala and hippocampus (25, 26) was performed on 8 KO and 16 CT mice. CT and KO mice took equal time to cross into the dark chamber before training (P = 0.70). Both groups learned equally well to avoid the context in which shock was administered and had similar retention times (P = 0.66; Fig. 3B).
KO mice also had normal locomotive activity. In the open-field test, no significant differences in horizontal activity and total distance traveled during the 15-min testing period were observed between CT and KO mice (P = 0.30 for horizontal activity; P = 0.55 for total distance).
Measurement of NMDAR-Mediated Currents.
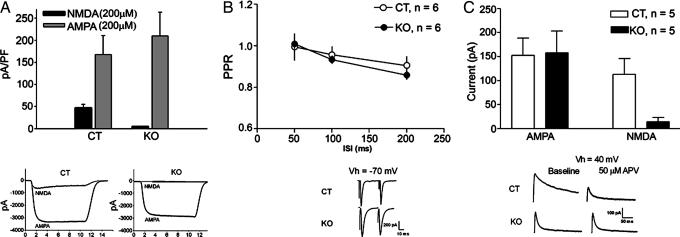
To confirm that NMDAR function is lost in KO mice, NMDAR-mediated currents were examined by using whole-cell recording. Neurons suitable for whole-cell recording were isolated from both mouse lines (n = 12 neurons from four CT mice and 19 neurons from four KO mice). The density of current activated by 200 μM NMDA + 10 μM glycine was greatly reduced in the KO vs. CT mice (P ≤ 0.05, unpaired t test; Fig. 4A). The majority of neurons from KO mice showed no detectable current in response to this agonist, whereas every neuron from the CT group exhibited NMDAR-mediated current. The NMDAR antagonist dl-2-amino-5-posphonovaleric acid (DL-APV) blocked the current activated by NMDA + glycine (data not shown). Application of 200 μM α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) in the presence of 100 μM cyclothiazide produced robust, nondesensitizing currents in all neurons tested. There was no significant difference between the two groups in the density of current activated by AMPA + cyclothiazide (P ≥ 0.05).
Fig. 4.
Current density and basal synaptic physiology. (A) In isolated striatal neurons, the density of AMPAR-mediated current is similar in CT and KO mouse neurons, whereas KO mice exhibited greatly reduced NMDAR-mediated current density in comparison with CT mice. (B) Paired pulse ratio of EPSCs was similar in CT and KO mice. (C) AMPAR-mediated EPSCs from CT and KO mice were comparable, but NMDAR-mediated EPSCs were nearly abolished in KO mice.
To examine basal synaptic function, paired pulse ratio [defined as excitatory postsynaptic current (EPSC) 2/EPSC1)] was measured by giving two pulses (separated by 50, 100, or 200 ms) every 20 s. Repeated-measures ANOVA with group as the between-subjects factor and interstimulus interval (ISI) as the within-subject factor indicated no differences between CT and KO mice at any of the ISIs (no interaction between group and ISI, F <1; Fig. 4B). EPSC amplitudes were measured at +40 mV in the absence and presence of APV to determine the contribution of AMPA receptors (AMPARs) and NMDARs to transmission. The AMPAR-mediated EPSC was measured after 5 min of APV application; the NMDAR-mediated current amplitude was obtained by subtracting the AMPAR-mediated response from the baseline response. There was no difference between groups in the amplitude of AMPAR-mediated EPSCs (P > 0.05); however, there was a significant difference in NMDAR-mediated EPSCs (P < 0.05; Fig. 4C).
Synaptic Plasticity.
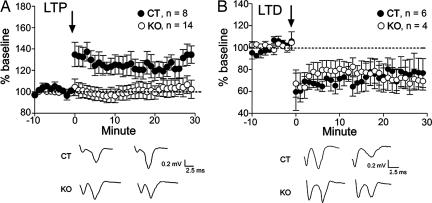
To determine whether a specific form of striatal synaptic plasticity is associated with motor learning, we examined NMDAR-dependent LTP in dorsomedial striatum in CT and KO mouse slices by using field potential recording. There was no difference between CT and KO mice in baseline population spike amplitude (0.69 ± 0.07mV, n = 8 slices from four KO mice; 0.58 ± 0.04mV, n = 14 slices from four CT mice; P ≥ 0.05). Slices from CT mice exhibited LTP (after high-frequency stimulation at 50 Hz; population spike amplitude = 135 ± 9% of baseline, P ≤ 0.05; Fig. 5A), whereas slices from KO mice showed no evidence of LTP (100 ± 7% of baseline, P ≥ 0.05). In slices from CT mice, LTP was blocked when 50 μM APV was added to the solution (four slices, 85 ± 13% of baseline, P ≥ 0.05). To determine whether NMDAR1 deletion has any effect on striatal plasticity that does not require NMDAR activation, we also used field potential recording to examine LTD in the dorsolateral striatum. Slices from both CT and KO mice showed LTD (CT, n = 6 slices from two mice, 72 ± 9% of baseline, P < 0.05; KO, n = 4 slices from one mouse, 73 ± 8% of baseline, P < 0.05; Fig. 5B).
Fig. 5.
Disruption of LTP, but not LTD, in dorsal striatum. (A) Field potential recordings showed that the LTP induced in CT mice after high-frequency stimulation was absent in KO mice. (B) LTD in the dorsolateral striatum, which does not depend on NMDAR activation, was normal in KO mice. Arrows indicate the time point of high-frequency stimulation.
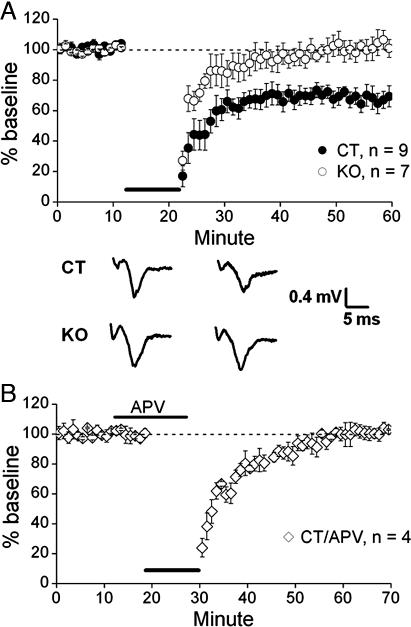
NMDARs are also known to be involved in LTD in the ventral striatum (27). To determine whether synaptic plasticity is altered in the ventral striatum of KO mice, we examined the induction of LTD in CT vs. KO mice in brain slices containing NAc. LTD could be reliably induced with moderate-frequency stimulation (10 Hz) in the NAc of CT mice (poststimulation population spike amplitude = 73.5 ± 4.0% of baseline, n = 9 slices from five CT mice), whereas LTD was absent in KO mice (100.1 ± 4.2% of baseline, n = 7 slices from four KO mice) (P < 0.0005, unpaired t test; Fig. 6A). The NMDAR antagonist APV (75 μM) prevented LTD induction in CT mice (100.5 ± 3.6% of baseline, four slices; Fig. 6B), further indicating that the LTD elicited in the NAc under our conditions is NMDAR-dependent.
Fig. 6.
Disruption of NMDAR-dependent LTD in ventral striatum. (A) Field potential recordings showed that the LTD induced in CT mice after moderate-frequency stimulation was absent in KO mice. (B) Reduction of LTD in CT mice in the presence of APV. The horizontal thick lines indicate the duration of 10-Hz frequency stimulation.
Discussion
We have shown that deleting NMDAR1 from the striatum, which effectively eliminated NMDA receptor function, abolished striatal LTP and impaired motor learning. By demonstrating parallel deficits in both striatal LTP and motor learning, we show that NMDAR-dependent striatal LTP is a likely synaptic substrate for this type of learning.
Our findings confirm the importance of the striatum in motor learning related to rotarod performance, which has often been attributed mainly to the cerebellum (28, 29). Recently published work has already questioned the critical role of the cerebellum in various types of motor learning. For example, motor learning in the rotarod test and the learning of motor timing in the eyeblink reflex test are unaffected by suppression of cerebellar LTD (30). Moreover, extensive recent work has linked the striatum to motor learning (9, 12, 20). Our results show that removal of the NMDAR1 subunit in the striatum alone is adequate to result in deficient motor learning. Although the striatum is capable of bidirectional changes in synaptic plasticity, the deficit in LTP that we observed implicates potentiation as the mode of synaptic change in the striatum responsible for at least the initial phase of motor learning.
The presence of LTD in the dorsal striatum in KO mice was not surprising because this form of plasticity does not require NMDAR activation (11, 31). Also expected was the elimination of LTD in the ventral striatum because, in our mutant mice, NMDAR1 is deleted in the entire striatum, including the ventral portion, and the ventral striatal LTD has been previously reported to be NMDAR-dependent (13, 27). There are, however, reasons to question the involvement of the ventral striatum in motor learning. Recent work (16) has shown that the ventral striatum is largely involved in Pavlovian approach behavior; in particular, NMDARs in the accumbens are required for the early consolidation of appetitive Pavlovian learning involving approaching stimuli predictive of rewards. Thus far, there is little if any evidence for a ventral striatal role in motor learning per se. More importantly, however, the absence of LTD in the ventral striatum suggests that in our KO mouse, appetitive learning involved in such addiction-related processes as incentive sensitization may also be disrupted (32).
The increased primary dendrites that we observed in the medium spiny neurons of our KO mice suggest a possible role of NMDAR activity in neuronal restructuring in the striatum. In the barrel field cortex, the number of primary dendrites on neurons has been shown to decrease gradually during development in normal mice (33). The role of NMDAR in cytoarchitecture also has been documented in another brain region. In the hippocampus, chronic blockade of NMDA receptors during the first 2 weeks of postnatal development results in more complex dendritic arborization of CA1 pyramidal cells (34). The increase in primary dendrites in our KO mice suggests a role for NMDARs in activity-dependent neuronal restructuring during development, most likely in the retraction and/or selective elimination of primary dendrites.
These findings have important implications for the underlying mechanisms of motor learning impairments seen in Parkinson's, dystonia, and Huntington's diseases that involve the corticostriatal circuitry. For example, in Huntington's disease, although glutamatergic excitotoxicity due to hypersensitivity of the NMDA receptor is thought to be involved in the loss of striatal neurons, some mouse models of the disease display instead an attenuated response to NMDAR agonists or a reduction in positive receptor regulation that indicates lower overall NMDAR activity (35,36,37,38,39–40). In light of our findings, a reduction in striatal NMDAR activity may well be involved in motor learning deficits reported to precede the onset of the hallmark symptoms of motor dysfunction in Huntington's disease (41,42–43).
The present findings contribute to an existing body of work on the roles of the NMDAR in various brain regions. Mouse models with cortex- and CA1 hippocampus-specific removal of the NMDAR1 subunit have already provided considerable information on somatosensory pattern development and synaptic plasticity in spatial memory (44, 45). In addition to elucidating the synaptic mechanism of striatal motor learning, our striatum-specific NMDAR1 knockout mice could potentially be a valuable tool for the study of other striatal functions.
Materials and Methods
Generation of RGS9-cre/NMDAR1-loxP Mice.
The two mouse lines, RGS9-cre and NMDAR1-loxP, were produced through gene-targeting methods and then crossed to produce the striatum-specific NMDAR1 KO. For detailed information on the targeting construct, breeding design, and genotyping, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
ROSA/RGS9-cre Mouse and LacZ Staining.
RGS9-cre mice produced were mated with ROSA26 reporter mice (22) purchased from The Jackson Laboratory (Bar Harbor, ME) (strain no. 3309). Mice carrying RGS9-cre and the ROSA26 reporter genes at P8–P90 were stained for β-galactosidase activity (23). Details of the methods used are provided in Supporting Materials and Methods.
Western Blot Analysis.
Proteins in striatal and cortical tissue lysate from KO and CT mice were separated on a 7.5% SDS/PAGE gel and analyzed by Western blot analysis using NMDAR1 and β-actin mouse antibodies (Chemicon, Temecula, CA) and rabbit anti-IgG antibody conjugated with HRP (Amersham Biosciences, Piscataway, NJ). Signals were developed with an ECL detection kit (Amersham Biosciences). The density of the bands was determined by ImageJ software (National Institutes of Health).
In Situ Hybridization.
The RNA probe template of NMDAR1 (46) was labeled using a digoxigenin RNA labeling kit with T3 polymerase (Roche Applied Science, Indianapolis, IN). The in situ hybridization protocol used was based on a method described in ref. 47, with some modifications as detailed in Supporting Materials and Methods. Detection of digoxigenin-labeled nucleic acids was performed with a digoxigenin nucleic acid detection kit (Roche Applied Science).
Nissl Staining.
Brain slices (30-μm thickness) were stained with a thionin-based Nissl stain as described previously (24).
Immunohistochemistry.
Brain coronal sections (50-μm thickness) from three mice per genotype were incubated with tyrosine hydroxylase antibody (Chemicon) and stained with Vectastain ABC peroxidase system and diaminobenzidine substrate kit (Vector Laboratories, Burlingame, CA).
Golgi Staining.
Brain tissues from three mice per genotype were processed with the Rapid Golgi staining kit (FD NeuroTechnologies, Ellicott City, MD), based on the manufacturer's protocol. Stained slices were sectioned at a thickness of 200 μm. Two medium spiny neurons of the striatum of each mouse were selected based on guidelines established previously (48). Neurons were traced using Neurolucida software (MicroBrightFields, Colchester, VT) at ×40 magnification. Analysis of neuronal tracings was performed with NeuroExplorer (MicroBrightFields).
Behavioral Tests.
The rotarod test was performed as described previously (24), except that mice were tested for three trials on each day for 2 days that were 1 week apart. Inhibitory avoidance was performed in an avoidance apparatus, with shocks administered at 0.2 mA for 1 s (49). Mice were tested for spontaneous activity levels in the open-field apparatus (AccuScan Instruments, Columbus, OH) (24). Data from the rotarod, inhibitory avoidance, and open-field tests were analyzed using two-way ANOVA with and without repeated measurements (24, 50).
Slice Preparation.
Coronal brain slices containing both striatum and cortex were prepared from mice that were between 3.5 and 4.5 weeks old (51) and were allowed to recover for 1 h at room temperature before recording was performed.
Whole-Cell Patch-Clamp Recording from Brain Slices.
Whole-cell recordings of synaptic currents and membrane potentials were performed as described previously (52). Details are provided in Supporting Material and Methods. EPSCs recorded during exposure of slices to NMDAR antagonist (APV, 50 μM) were used to measure AMPAR- and NMDAR-mediated synaptic current.
Field Potential Recording.
LTP and LTD in the dorsal striatum were measured after high-frequency stimulation of white matter near the striatum (53). LTD in the NAc with and without APV application was measured in parasagittal slices, using procedures published previously (54,55,56–57). Details are available in Supporting Materials and Methods.
Whole-Cell Recording in Isolated Neurons.
Corticostriatal slices were treated enzymatically, followed by trituration. AMPA and NMDA currents were recorded in isolated neurons (58). Details are given in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Enxiang Tao, Morgan Pence, Sungchil Yang, Emily Speer, Jun Su, and Shinichi Mitsui for their excellent technical assistance; Zia Rahman and Eric J. Nestler (both from University of Texas, Dallas, TX) for providing their genomic clones and partial sequences of RGS9; and Jang Ho Cha (Massachusetts General Hospital, Boston, MA) and William Greenough for helpful discussions. Work in the laboratory of Y.L. was supported by National Science Foundation Grant 9728742; National Institutes of Health (NIH) Grants AG17291, NS42356, and NS47692; the Dystonia Medical Research Foundation; the Bachmann–Strauss Dystonia and Parkinson Foundation; a start-up fund from the State of Illinois; the Beckman Institute for Advanced Science and Technology; and the Lucille P. Markey Charitable Trust. M.T.D. was partly supported by an NIH Systems and Integrative Biology training grant. F.Y. was partly supported by Saga Medical School, Saga, Japan. H.H.Y. and D.M.L. were supported by the Division of Intramural Clinical and Basic Research of the National Institute on Alcohol Abuse and Alcoholism/NIH. Y.W. was supported by private funds.
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
AMPA receptor
- APV
2-amino-5-phosphonovaleric acid
- CT
control
- EPSC
excitatory postsynaptic current
- KO
knockout
- LTD
long-term depression
- LTP
long-term potentiation
- NAc
nucleus accumbens
- NMDAR
NMDA receptor.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jog MS, Kubota Y, Connolly CI, Hillegaart V, Graybiel AM. Science. 1999;285:1745–1749. doi: 10.1126/science.286.5445.1745. [DOI] [PubMed] [Google Scholar]
- 2.Packard MG, Knowlton BJ. Annu Rev Neurosci. 2002;25:563–593. doi: 10.1146/annurev.neuro.25.112701.142937. [DOI] [PubMed] [Google Scholar]
- 3.Cromwell HC, Berridge KC. J Neurosci. 1996;16:3444–3458. doi: 10.1523/JNEUROSCI.16-10-03444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynolds JN, Hyland BI, Wickens JR. Nature. 2001;413:67–70. doi: 10.1038/35092560. [DOI] [PubMed] [Google Scholar]
- 5.Doyon J, Owen AM, Petrides M, Sziklas V, Evans AC. Eur J Neurosci. 1996;8:637–648. doi: 10.1111/j.1460-9568.1996.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y, Fukuyama H, Honda M, Okada T, Hanakawa T, Nakamura K, Nagahama Y, Nagamine T, Konishi J, Shibasaki H. Brain. 2000;123:790–799. doi: 10.1093/brain/123.4.790. [DOI] [PubMed] [Google Scholar]
- 7.Gerdeman GL, Partridge JG, Lupica CR, Lovinger DM. Trends Neurosci. 2003;26:184–192. doi: 10.1016/S0166-2236(03)00065-1. [DOI] [PubMed] [Google Scholar]
- 8.Barnes TD, Kubota Y, Hu D, Jin DZ, Graybiel AM. Nature. 2005;437:1158–1161. doi: 10.1038/nature04053. [DOI] [PubMed] [Google Scholar]
- 9.Costa RM, Cohen D, Nicolelis M. Curr Biol. 2004;14:1124–1134. doi: 10.1016/j.cub.2004.06.053. [DOI] [PubMed] [Google Scholar]
- 10.Calabresi P, Pisani A, Mercuri NB, Bernardi G. Eur J Neurosci. 1992;4:929–935. doi: 10.1111/j.1460-9568.1992.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 11.Lovinger DM, Tyler EC, Merritt A. J Neurophysiol. 1993;70:1937–1949. doi: 10.1152/jn.1993.70.5.1937. [DOI] [PubMed] [Google Scholar]
- 12.Palencia CA, Ragozzino ME. Neurobiol Learn Mem. 2004;82:81–89. doi: 10.1016/j.nlm.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Kombian SB, Malenka RC. Nature. 1994;368:242–246. doi: 10.1038/368242a0. [DOI] [PubMed] [Google Scholar]
- 14.Brebner K, Wong TP, Liu L, Liu Y, Campsall P, Gray S, Phelps L, Phillips AG, Wang YT. Science. 2005;310:1340–1343. doi: 10.1126/science.1116894. [DOI] [PubMed] [Google Scholar]
- 15.Martin M, Chen BT, Hopf FW, Bowers MS, Bonci A. Nat Neurosci. 2006;9:868–869. doi: 10.1038/nn1713. [DOI] [PubMed] [Google Scholar]
- 16.Dalley JW, Laane K, Theobald DE, Armstrong HC, Corlett PR, Chudasama Y, Robbins TW. Proc Natl Acad Sci USA. 2005;102:6189–6194. doi: 10.1073/pnas.0502080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernagut PO, Diguet E, Stefanova N, Biran M, Wenning GK, Canioni P, Bioulac B, Tison F. Neuroscience. 2002;114:1005–1017. doi: 10.1016/s0306-4522(02)00205-1. [DOI] [PubMed] [Google Scholar]
- 18.Centonze D, Gubellini P, Bernardi G, Calabresi P. Brain Res Brain Res Rev. 1999;31:1–5. doi: 10.1016/s0165-0173(99)00018-1. [DOI] [PubMed] [Google Scholar]
- 19.Calabresi P, Saiardi A, Pisani A, Baik JH, Centonze D, Mercuri NB, Bernardi G, Borrelli E. J Neurosci. 1997;17:4536–4544. doi: 10.1523/JNEUROSCI.17-12-04536.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin HH, Knowlton BJ, Balleine BW. Eur J Neurosci. 2005;22:505–512. doi: 10.1111/j.1460-9568.2005.04219.x. [DOI] [PubMed] [Google Scholar]
- 21.Rahman Z, Gold SJ, Potenz MN, Cowan CW, Ni YG, He W, Wensel TG, Nestler EJ. J Neurosci. 1999;19:2016–2026. doi: 10.1523/JNEUROSCI.19-06-02016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 23.Jin XL, Guo H, Mao C, Atkins N, Wang H, Avasthi PP, Tu YT, Li Y. Biochem Biophys Res Commun. 2000;270:978–982. doi: 10.1006/bbrc.2000.2532. [DOI] [PubMed] [Google Scholar]
- 24.Dang M, Yokoi F, McNaught KS, Jengelley TA, Jackson T, Li J, Li Y. Exp Neurol. 2005;196:452–463. doi: 10.1016/j.expneurol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 25.Malin EL, McGaugh JL. Proc Natl Acad Sci USA. 2006;103:1959–1963. doi: 10.1073/pnas.0510890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fin C, Schmitz PK, Da Silva RC, Bernabeu R, Medina JH, Izquierdo I. Eur J Pharmacol. 1994;271:227–229. doi: 10.1016/0014-2999(94)90284-4. [DOI] [PubMed] [Google Scholar]
- 27.Thomas MJ, Malenka RC, Bonci A. J Neurosci. 2000;20:5581–5586. doi: 10.1523/JNEUROSCI.20-15-05581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito M. Ann NY Acad Sci. 2002;978:273–288. doi: 10.1111/j.1749-6632.2002.tb07574.x. [DOI] [PubMed] [Google Scholar]
- 29.McCormick DA, Thompson RF. Science. 1984;223:296–299. doi: 10.1126/science.6701513. [DOI] [PubMed] [Google Scholar]
- 30.Welsh JP, Yamaguichi H, Zeng XH, Kojo M, Nakada Y, Takagi A, Sugimori M, Llinas RR. Proc Natl Acad Sci USA. 2005;102:17166–17171. doi: 10.1073/pnas.0508191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calabresi P, Maj R, Pisani A, Mercuri NB, Bernardi G. J Neurosci. 1992;12:4224–4233. doi: 10.1523/JNEUROSCI.12-11-04224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson TE, Berridge KC. Annu Rev Psychol. 2003;54:25–53. doi: 10.1146/annurev.psych.54.101601.145237. [DOI] [PubMed] [Google Scholar]
- 33.Greenough WT, Chang FL. Brain Res. 1988;471:148–152. doi: 10.1016/0165-3806(88)90160-5. [DOI] [PubMed] [Google Scholar]
- 34.Luthi A, Schwyzer L, Mateos JM, Gahwiler BH, McKinney RA. Nat Neurosci. 2001;4:1102–1107. doi: 10.1038/nn744. [DOI] [PubMed] [Google Scholar]
- 35.Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P, Hayden MR, Raymond LA. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 36.Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, Cadigan BA, Warzecki L, Tagle DA, Reddy PH, et al. J Neurosci. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cepeda C, Ariano MA, Calvert CR, Flores-Hernandez J, Chandler SH, Leavitt BR, Hayden MR, Levine MS. J Neurosci Res. 2001;66:525–539. doi: 10.1002/jnr.1244. [DOI] [PubMed] [Google Scholar]
- 38.Hansson O, Petersen A, Leist M, Nicotera P, Castilho RF, Brundin P. Proc Natl Acad Sci USA. 1999;96:8727–8732. doi: 10.1073/pnas.96.15.8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morton AJ, Leavens W. Brain Res Bull. 2000;52:51–59. doi: 10.1016/s0361-9230(00)00238-0. [DOI] [PubMed] [Google Scholar]
- 40.Luthi-Carter R, Apostol BL, Dunah AW, DeJohn MM, Farrell LA, Bates GP, Young AB, Standaert DG, Thompson LM, Cha JH. Neurobiol Dis. 2003;14:624–636. doi: 10.1016/j.nbd.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Hahn-Barma V, Deweer B, Durr A, Dode C, Feingold J, Pillon B, Agid Y, Brice A, Dubois B. J Neurol Neurosurg Psychiatry. 1998;64:172–177. doi: 10.1136/jnnp.64.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence AD, Hodges JR, Rosser AE, Kershaw A, Ffrench-Constant C, Rubinsztein DC, Robbins TW, Sahakian BJ. Brain. 1998;121:1329–1341. doi: 10.1093/brain/121.7.1329. [DOI] [PubMed] [Google Scholar]
- 43.Snowden JS, Craufurd D, Thompson J, Neary D. J Clin Exp Neuropsychol. 2002;24:133–145. doi: 10.1076/jcen.24.2.133.998. [DOI] [PubMed] [Google Scholar]
- 44.Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, Knopfel T, Erzurumlu RS, Itohara S. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsien JZ, Huerta PT, Tonegawa S. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Erzurumlu RS, Chen S, Jhaveri S, Tonegawa S. Cell. 1994;76:427–437. doi: 10.1016/0092-8674(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 47.Bessert DA, Skoff RP. J Histochem Cytochem. 1999;47:693–702. doi: 10.1177/002215549904700511. [DOI] [PubMed] [Google Scholar]
- 48.Cheng HW, Rafols JA, Goshgarian HG, Anavi Y, Tong J, McNeill TH. Exp Neurol. 1997;147:287–298. doi: 10.1006/exnr.1997.6618. [DOI] [PubMed] [Google Scholar]
- 49.Mele A, Castellano C, Cestari V, Oliverio A. Neurobiol Learn Mem. 1995;63:143–148. doi: 10.1006/nlme.1995.1014. [DOI] [PubMed] [Google Scholar]
- 50.Yokoi F, Dang MT, Li J, Li Y. J Biochem. 2006;140:141–146. doi: 10.1093/jb/mvj138. [DOI] [PubMed] [Google Scholar]
- 51.Yin HH, Lovinger DM. Proc Natl Acad Sci USA. 2006;103:8251–8256. doi: 10.1073/pnas.0510797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerdeman GL, Ronesi J, Lovinger DM. Nat Neurosci. 2002;5:446–451. doi: 10.1038/nn832. [DOI] [PubMed] [Google Scholar]
- 53.Partridge JG, Tang K, Lovinger DM. J Neurophysiol. 2000;84:1422–1429. doi: 10.1152/jn.2000.84.3.1422. [DOI] [PubMed] [Google Scholar]
- 54.Hoffman AF, Oz M, Caulder T, Lupica CR. J Neurosci. 2003;23:4815–4820. doi: 10.1523/JNEUROSCI.23-12-04815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens CF, Tonegawa S, Wang Y. Curr Biol. 1994;4:687–693. doi: 10.1016/s0960-9822(00)00153-6. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Aghajanian GK. J Neurosci. 1990;10:3335–3343. doi: 10.1523/JNEUROSCI.10-10-03335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Xu R, Sasaoka T, Tonegawa S, Kung MP, Sankoorikal EB. J Neurosci. 2000;20:8305–8314. doi: 10.1523/JNEUROSCI.20-22-08305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovinger DM, McCool BA. J Neurophysiol. 1995;73:1076–1083. doi: 10.1152/jn.1995.73.3.1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.