Abstract
Glycerophosphocholine (GPC) is an osmoprotective compatible and counteracting organic osmolyte that accumulates in renal inner medullary cells in response to high NaCl and urea. We previously found that high NaCl increases GPC in renal [Madin–Darby canine kidney (MDCK)] cells. The GPC is derived from phosphatidylcholine, catalyzed by a phospholipase that was not identified at that time. Neuropathy target esterase (NTE) was recently shown to be a phospholipase B that catalyzes production of GPC from phosphatidylcholine. The purpose of the present study was to test whether NTE contributes to the high NaCl-induced increase of GPC synthesis in renal cells. We find that in mouse inner medullary collecting duct cells, high NaCl increases NTE mRNA within 8 h and NTE protein within 16 h. Diisopropyl fluorophosphate, which inhibits NTE esterase activity, reduces GPC accumulation, as does an siRNA that specifically reduces NTE protein abundance. The 20-h half-life of NTE mRNA is unaffected by high NaCl. TonEBP/OREBP is a transcription factor that is activated by high NaCl. Knockdown of TonEBP/OREBP by a specific siRNA inhibits the high NaCl-induced increase of NTE mRNA. Further, the lower renal inner medullary interstitial NaCl concentration that occurs chronically in ClCK1−/− mice and acutely in normal mice given furosemide is associated with lower NTE mRNA and protein. We conclude that high NaCl increases transcription of NTE, likely mediated by TonEBP/OREBP, and that the resultant increase of NTE expression contributes to increased production and accumulation of GPC in mammalian renal cells in tissue culture and in vivo.
Keywords: diisopropyl fluorophosphate, phospholipase B
Neuropathy target esterase (NTE) was originally identified as a target of organophosphates that is involved in their induction of peripheral neuropathy in humans (1). It is an endoplasmic reticulum-associated protein, mostly exposed on the cytoplasmic face of the membranes (2). NTE is highly conserved in eukaryotes and even has homologues in bacteria (3). The mechanism of organophosphate neurotoxicity is that certain organophosphates act as pseudosubstrates for serine hydrolases, and the covalent organophosphorylated intermediate formed by this reaction is hydrolyzed extremely slowly, thereby inhibiting the enzyme. Loss of activity disrupts phosphatidylcholine (PC) homeostasis, resulting in neuronal and glial death (4). The normal role of NTE in nerves is not completely understood.
NTE is also expressed outside of the nervous system in numerous organs, including the kidney (5, 6), but its role in those tissues also is not well understood. A possible role was revealed recently when NTE was shown to be a phospholipase B that deacylates PC sequentially at positions sn1 and sn2 in Saccharomyces cerevisiae, yielding glycerophosphocholine (GPC). Yeast mutants that lack NTE activity are not able to produce intracellular GPC (7).
The interstitial concentration of NaCl normally is very high in the renal inner medulla, associated with its role in concentrating the urine. High NaCl is hypertonic. It shrinks cells by osmosis, increasing the concentration of all cellular components, including inorganic salts. This can cause cell cycle delay and apoptosis (8, 9). Cells generally protect themselves from hypertonicity by accumulating compatible organic osmolytes that serve to restore cell volume and normalize intracellular salt concentration (10). GPC is one of the organic osmolytes normally accumulated by renal cells in response to high NaCl, both in vivo and in cell culture (11). Osmotic regulation of GPC by high NaCl has been studied in isolated renal inner medullary collecting duct cells (12, 13), in cell cultures (14–18), and in vivo (18). It involves changes in both GPC synthesis and degradation. On the one hand, high NaCl inhibits the activity of GPC–choline phosphodiesterase, an enzyme that degrades GPC to choline and α-glycerophosphate (12, 15, 16); the slower degradation of GPC increases its concentration. On the other hand, high NaCl also raises phospholipase activity, increasing the rate of synthesis of GPC from PC in renal cells (17, 18). High NaCl also increases production of GPC from PC in yeast, which helps maintain their growth during osmotic stress (19). Despite this biochemical understanding, the molecular identities of the renal phosphodiesterase and phospholipase that are involved have remained unknown.
We previously found that high NaCl increases phospholipase A2 or phospholipase B but not lysophospholipase activity in Madin–Darby canine kidney (MDCK) cells (17). After the discovery that NTE is a phospholipase B that catalyzes production of GPC (7), we were interested in testing whether it might contribute to high NaCl-induced accumulation of GPC in renal cells. In support of this hypothesis, we find in the present study that high salt increases NTE mRNA and protein expression in mouse inner medullary collecting duct (mIMCD3) cells in culture and in mouse inner medulla in vivo. Further, inhibiting NTE with an organophosphate or reducing its expression by RNA interference reduces the high NaCl-induced increase of GPC in mIMCD3 cells.
Results
High NaCl Increases GPC Content of mIMCD3 Cells.
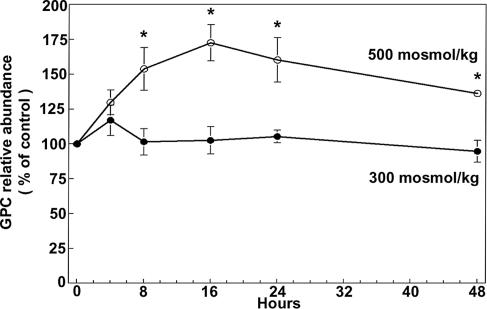
mIMCD3 cells accumulate GPC when they are chronically adapted to elevation of NaCl and urea in combination (20). MDCK (14, 16, 21) and PAP-HT25 (21) cells accumulate GPC not only when NaCl and urea are raised in combination but also when either is raised alone. Because mIMCD3 cells, which are murine, promised to be more useful for our studies than either MDCK (dog) or PAP-HT25 (rabbit), we began by testing whether high NaCl alone increases GPC in these cells. We found that raising osmolality from 300 to 500 milliosmol (mosmol)/kg by adding NaCl increases mIMCD3 cell GPC within 8 h (Fig. 1), which provides a model for testing the mechanism involved.
Fig. 1.
Effect of high NaCl on mIMCD3 cell GPC. Cells, initially grown at 300 mosmol/kg were either maintained at 300 mosmol/kg or increased to 500 mosmol/kg by addition of NaCl. GPC abundance is expressed as percent of the initial value at 300 mosmol/kg, which is 17.6 ± 0.6 nmol/mg protein (mean ± SEM, n = 3; ∗, P < 0.05).
High NaCl Increases Expression of NTE in mIMCD3 Cells.
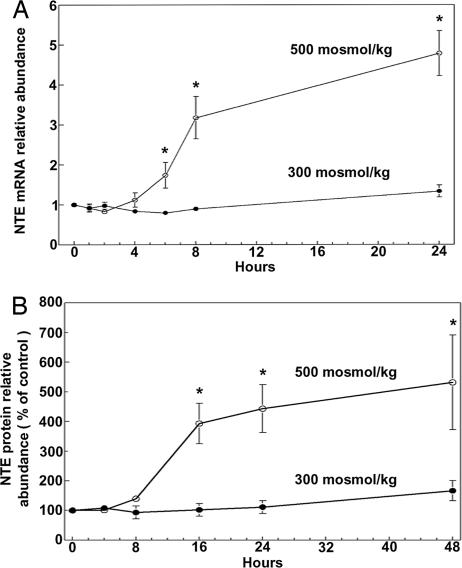
We previously found that high NaCl increases phospholipase activity in MDCK cells, which contributes to the elevation of GPC by increasing the rate of synthesis of GPC from PC (17). NTE is a phospholipase that catalyzes production of GPC from PC in COS7 and HeLa cells (7), making it a candidate for the phospholipase that is activated by high NaCl. Therefore, we determined the effect of high NaCl on NTE expression. Raising osmolality from 300 to 500 mosmol/kg by adding NaCl increases NTE mRNA abundance in mIMCD3 cells within 6–8 h (Fig. 2A) and increases NTE protein within 16 h (Fig. 2B). In the same extracts, neither cyclophilin mRNA nor β-actin protein abundance changed significantly (data not shown).
Fig. 2.
Effect of high salt on mNTE mRNA (A) and protein (B) abundance in mIMCD3. Cells, initially grown at 300 mosmol/kg, were either maintained at 300 mosmol/kg or increased to 500 mosmol/kg by addition of NaCl. (A) mRNA abundance is expressed as fold change relative to the initial value at 300 mosmol/kg. (B) Protein abundance is expressed as percent of the initial value at 300 mosmol/kg (mean ± SEM, n = 3; ∗, P < 0.05).
Expression of NTE in Renal Inner Medullary Cells in Vivo Depends on Interstitial Osmolality.
Interstitial osmolality normally is high in the renal medulla, because of high levels of NaCl and urea, resulting in intracellular accumulation of GPC (11). The high osmolality depends on NaCl transport by the thick ascending limbs of Henle's loop and countercurrent multiplication. To test whether expression of NTE might depend on interstitial osmolality in vivo, we measured NTE under conditions known to reduce renal inner medullary interstitial NaCl concentration.
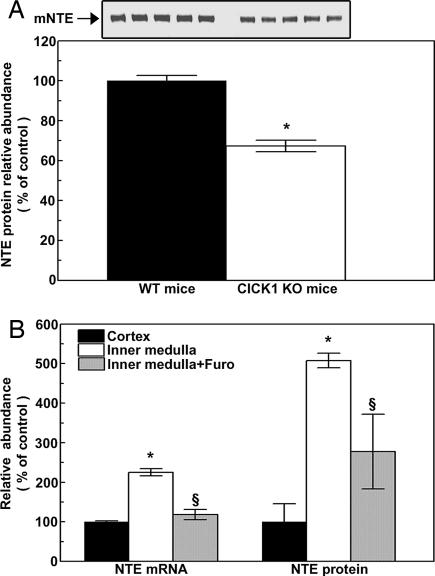
The ClC-K1 chloride channel, which normally is expressed in medullary thin ascending limbs, is necessary for renal medullary countercurrent multiplication, and mice lacking it (22) have lower urine concentration because of reduced interstitial osmolality. Therefore, we compared NTE expression in renal inner medullas of ClC-K1−/− mice to that of ClC-K1+/+ mice. NTE protein expression is 35% less in the renal medullas of ClC-K1−/− mice (Fig. 3A). NTE protein abundance is greater in the inner medulla of wild-type mice than in the cortex (Fig. 3B). Treatment of mice with furosemide reduces interstitial osmolality. When mice are treated with furosemide for 4 h, NTE mRNA and protein abundances are reduced in the inner medulla (Fig. 3B). We conclude that NTE expression is regulated by osmolality in vivo, like it is in cell culture.
Fig. 3.
Renal NTE expression in vivo. (A) (Lower) NTE protein expression in renal inner medullas of wild-type (WT) and Clcnk1−/− (CLC KO) mice. (Upper) Western blot results. (B) Effects of furosemide on NTE mRNA and protein. Kidneys were removed for analysis 4 h after i.p. injection of 1.5 mg of furosemide per 20 g of body weight (mean ± SEM, n = 3–5, ∗, P < 0.05 vs. control; §, P < 0.05 vs. inner medulla, no furosemide).
Having found that NTE expression depends on the level of extracellular NaCl, the following studies were directed at determining whether NTE is necessary for the high NaCl-induced increase in GPC.
Diisopropyl Fluorophosphate (DFP) Reduces High NaCl-Induced Increase of GPC.
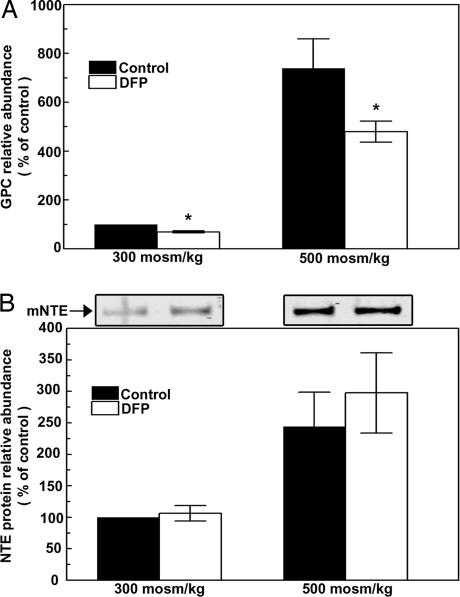
DFP is a neurotoxic organophosphate (1) that inhibits the phospholipase B activity of NTE (7). DFP (1.0 M) inhibits the increase of GPC that occurs 24 h after osmolality is increased from 300 to 500 mosmol/kg by adding NaCl (Fig. 4A). The inhibition by DFP is not accompanied by any significant change in NTE protein abundance (Fig. 4B). We conclude that this supports participation of NTE in the high NaCl-induced increase of GPC.
Fig. 4.
Effect of DFP. Cells initially grown at 300 mosmol/kg were treated with 1 μM DFP or with vehicle (isopropanol) for 2 h, then were either maintained at 300 or increased to 500 mosmol/kg for 24 h while continuing DFP or vehicle. (A) Relative GPC abundance. (B) (Lower) NTE protein abundance. (Upper) A representative Western blot is shown (mean ± SEM, n = 3–5; ∗, P < 0.05).
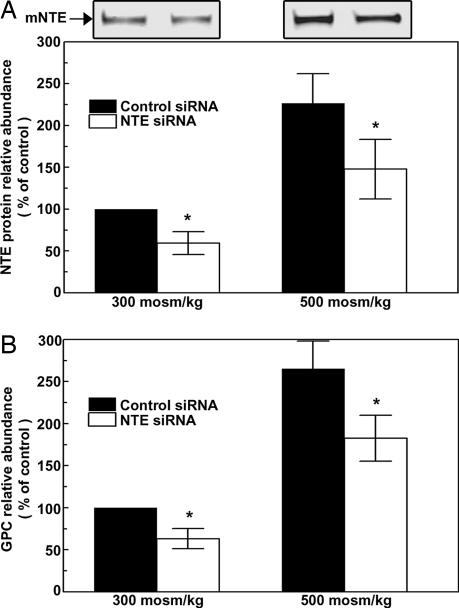
siRNA-Mediated Knockdown of NTE Reduces High NaCl-Induced Accumulation of GPC.
siRNA-mediated knockdown of NTE was previously demonstrated in COS7 cells (7). Based on the sequence previously used, we designed a “Dicer substrate RNAi duplex” that effectively knocks down NTE protein expression in mIMCD3 cells (Fig. 5A), despite the generally low transfection efficiency of these cells. We transfected the mIMCD3 cells with either nonspecific siRNA (as a control) or with NTE-specific siRNA for 24 h, then increased osmolality from 300 to 500 mosmol/kg for an additional 24 h by adding NaCl. High NaCl increases NTE protein abundance by only 47% in cells transfected with NTE siRNA, compared with a 120% increase with the control siRNA. In the same experiments (Fig. 5B), the specific siRNA reduces GPC by 30% at 300 mosmol/kg (compared with the nonspecific siRNA) and by 40% at 500 mosmol/kg. We conclude that NTE contributes to high NaCl-induced GPC accumulation.
Fig. 5.
Effect of siRNA-mediated reduction of NTE. mIMCD3 cells were transfected with either control siRNA or NTE-specific siRNA at 300 mosmol/kg. Twenty-four hours later, the media were replaced by media without the siRNAs and at either 300 or 500 mosmol/kg (NaCl added). (A) (Lower) Relative NTE protein abundance. (Upper) A representative Western blot. (B) Relative GPC abundance (mean ± SEM, n = 3; ∗, P < 0.05).
Transcription Factor TonEBP/OREBP Is Necessary for High NaCl-Induced Increase in NTE Protein.
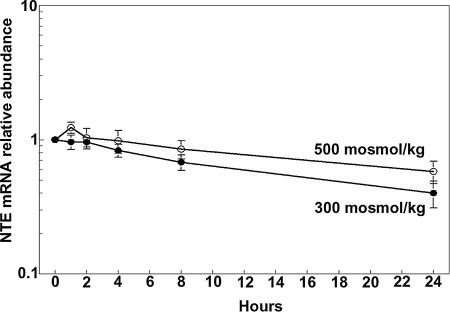
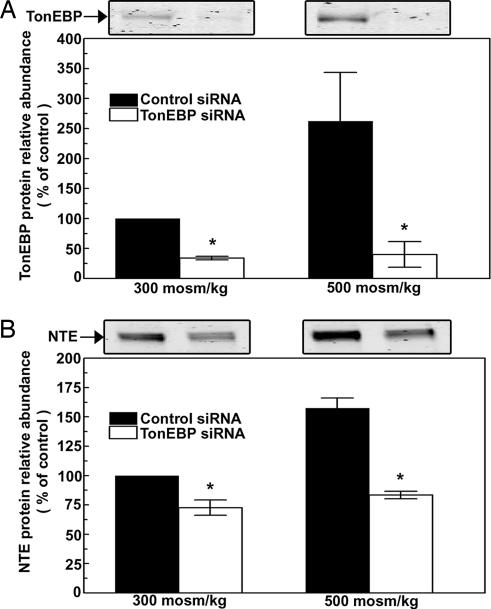
The high NaCl-induced increase in NTE mRNA (Fig. 2) could result either from increased transcription or from reduced degradation of the mRNA. We measured the stability of NTE mRNA from the rate of its decrease when transcription is inhibited by Actinomycin B. The half-life of NTE mRNA in mIMCD3 cells is ≈24 h both at 300 mosmol/kg and when NaCl is added, increasing osmolality to 500 mosmol/kg (Fig. 6). The implication is that high NaCl does not stabilize NTE mRNA but rather increases its transcription. The transcription factor TonEBP/OREBP is activated by high NaCl, resulting in increased transcription of multiple osmoprotective genes (23, 24). To determine whether TonEBP/OREBP is involved in high NaCl-induced increase of NTE mRNA, we reduced TonEBP/OREBP expression by RNA interference in HEK293 cells. The reason for using HEK293 cells here is that they can be more efficiently transfected than mIMCD3 cells. High NaCl increases TonEBP/OREBP protein abundance (23, 24), which is seen here in the presence of nonspecific siRNA (Fig. 7A). In contrast, the specific siRNA greatly decreases TonEBP/OREBP protein abundance to the same low level both at 300 and 500 mosmol/kg (Fig. 7A). At 300 mosmol/kg, reducing TonEBP/OREBP expression significantly decreases NTE protein abundance (Fig. 7B). In the same extracts, β-actin protein abundance did not change significantly (data not shown). At 500 mosmol/kg, the 70% increase of NTE protein that occurs in the presence of control nonspecific siRNA is totally inhibited. We conclude that the high NaCl-induced increase of NTE expression is mediated by the transcription factor, TonEBP/OREBP.
Fig. 6.
NTE mRNA stability. Medium bathing subconfluent mIMCD3 cells was maintained at 300 mosmol/kg or increased to 500 mosmol/kg (NaCl added) for 2 h, then 5 μg/ml actinomycinD was added. NTE mRNA was measured by real-time RT-PCR (n = 5).
Fig. 7.
High NaCl-induced increase of NTE protein abundance depends on TonEBP/OREBP. HEK293 cells were transfected with either control or TonEBP/OREBP-specific siRNA. Twenty-four hours later, the media were replaced by media without the siRNAs at either 300 or 500 mosmol/kg (NaCl added). (A) Relative TonEBP/OREBP protein abundance. (B) Relative NTE protein abundance. (A and B Upper) Representative Western blots are shown (mean ± SEM, n = 3,∗, P < 0.05).
Discussion
High NaCl Raises Phospholipase Activity in Renal Medullary Cells, Increasing Synthesis of GPC from PC.
NaCl is high in renal inner medullary interstitial fluid, and high NaCl causes renal cells to accumulate large amounts of GPC (14). The biochemical basis for high NaCl-induced accumulation of GPC was elucidated in MDCK cells (17). In those cells, the GPC is synthesized from PC, catalyzed by phospholipase activity. High NaCl increases phospholipase activity. Phospholipase activity also increases in inner medullas of rats when they are thirsted (18), water deprivation being associated with rises of urine concentration and inner medullary interstitial NaCl concentration.
High NaCl Increases NTE Expression.
NTE was originally identified as an esterase involved in the neuropathy produced by organophosphate toxicity (1, 5). Knocking out NTE in transgenic mice revealed that its expression is necessary for viability and that reduced expression impairs neural development (25). NTE is expressed not only within but also outside of the nervous system (5, 6). Although NTE was found to have esterase activity (5, 6), its normal cellular functions remained obscure until it was shown to be a phospholipase B that degrades PC to GPC (7). Given that increased phospholipase activity contributes to high NaCl-induced increase of GPC, NTE seemed to be a promising candidate for the phospholipase that is involved. That possibility is confirmed in the present studies. Thus, in tissue culture, when mIMCD3 cells are exposed to high NaCl, the abundance of NTE mRNA increases within 6–8 h and NTE protein within 16 h (Fig. 2). In vivo, NTE protein expression is higher in the renal inner medulla, where interstitial NaCl concentration is higher than in the renal cortex, where it is not (Fig. 3). Further, when the diuretic furosemide reduces renal inner medulla interstitial salt concentration, renal medullary NTE mRNA and protein decrease. Thus, high NaCl increases NTE in renal cells both in tissue culture and in vivo.
NTE Contributes to High NaCl-Induced Increase in GPC.
We tested for a role of NTE in high NaCl-induced accumulation of GPC by inhibiting its activity and by knocking it out. Organophosphates, including DFP, inhibit NTE activity by occupying its serine esterase site (1). DFP decreases NTE phospholipase activity and reduces formation of GPC in COS7 cells and in yeast (7). We find that DFP decreases high NaCl-induced GPC accumulation of mIMCD3 (Fig. 4), supporting the role of NTE in osmoregulatory accumulation of GPC. In addition, we used a specific siRNA to decrease NTE expression in mIMCD3 cells. This reduces GPC accumulation in response to high NaCl (Fig. 5). Neither way of reducing NTE activity completely eliminated the high NaCl-induced increase of GPC. However, this is not surprising, considering that high NaCl not only increases synthesis of GPC but also reduces its degradation by inhibiting GPC–choline phosphodiesterase activity (12, 15, 16). Further, the reduced degradation occurs within 2 h (16), which is faster than the increase in NTE expression (Fig. 2) and accounts for why the rise of GPC (Fig. 1) precedes that of NTE (Fig. 2). We conclude that NTE is a phospholipase that is activated by high NaCl, contributing to the resultant accumulation of GPC.
Osmotically Regulated Transcription Factor TonEBP/OREBP Is Necessary for High NaCl-Induced NTE Expression.
Hypertonicity activates TonEBP/OREBP, increasing transcription of numerous osmoprotective genes, including those involved in accumulation of the compatible organic osmolytes sorbitol, glycine betaine, myo-inositol, and taurine (23, 24). Hypertonicity apparently also increases transcription of NTE, because the high NaCl-induced increase of NTE mRNA (Fig. 2) occurs without any change in its half-life (Fig. 6), implicating increased transcription rather than reduction of degradation. Further, the increase in NTE requires expression of TonEBP/OREBP (Fig. 7), implicating transcriptional regulation by TonEBP/OREBP. Thus, high NaCl-induced activation of TonEBP/OREBP apparently increases transcription of NTE.
Because transcriptional activity of TonEBP/OREBP requires binding to a specific DNA element, ORE/TonE (26, 27), we searched for ORE/TonEs in the NTE gene. The ORE/TonE consensus sequence is NGGAAAWDHMC(N) (26). Between base pairs −900 and −1 in the 5′ flanking region of NTE, there are four DNA elements that fit this consensus. However, we do not yet know which ones are involved in its regulation by tonicity.
Effect of Hypertonicity on PC Abundance.
PC is the major membrane lipid of eukaryotic cells. Maintenance of the level of PC in cell membranes is necessary for their integrity (28). Its level generally is tightly regulated by the balance between synthesis and degradation. PC is synthesized by the CDP choline pathway in both yeast (29) and mammalia (30). PC is metabolized to GPC in yeast and catalyzed by NTE1, the yeast homologue of NTE (7). Knockdown of NTE1 reduces synthesis of GPC, but this does not affect the abundance of PC, because the rate of synthesis of PC is also decreased (7). On the other hand, when high NaCl increases synthesis of GPC from PC in yeast, PC decreases (19). Nevertheless, the yeast survive well without evident deterioration of their plasma membranes. The proposed explanation is that the source of the GPC is a pool of PC outside of the plasma membranes (19).
In contrast to yeast, when mammalian renal cells (MDCK) increase GPC in response to high NaCl, the level of PC does not change (17). At least part of the explanation is that the increase of GPC begins with inhibition of GPC–choline phosphodiesterase, which catalyzes degradation of GPC (15, 16). The reduced degradation of GPC elevates its level without a change in synthesis. Because this part of the response does not involve increased degradation of PC, it should not affect its level. Increased degradation of PC, catalyzed by NTE, does not occur until later (17). It remains to be seen whether increased synthesis of PC also occurs, which could contribute to maintaining its level in mammalian cells stressed by high NaCl.
Materials
Low-glucose DMEM was from Irvine Scientific (Irvine, CA), and Coon's improved medium F-12 was from BioSource (Camarillo, CA). QiaShredder column, RNeasy mini kit, RNase-free DNase, and Quantitect SYBR Green PCR kit were from Qiagen (Valencia, CA). Lipofectamine 2000 was from Invitrogen (Carlsbad, CA). ABI Prism 7900 sequence detection system and TaqMan reverse-transcription reagents kit were from Applied Biosystems (Foster City, CA). Mammalian protein extraction reagent was from Pierce (Rockford, IL), and complete miniprotease inhibitor tablets were from Roche (Indianapolis, IN). Double-stranded siRNA (Dicer substrate RNAi duplex) against mNTE and nonspecific control were designed following the Integrated DNA Technologies (Coralville, IA) instructions and recommendations (31). The siRNAs were synthesized by Integrated DNA Technologies. The sequences of mNTE siRNA were as follows: sense 5′-AAGAUUAUGCGGAAGGUGAGUUCCA-3′ and antisense, 5′-UGGAACUCACCUUCCGCAUAAUCUUCC-3′; for TonEBP/OREBP, sense, 5′-UAAAGGAGCUGCAAGCACCGCCCAUGG-3′ (based on ref. 32), and antisense, 5′-AUGGGCGGUGCUUGCAGCUCCUUTA-3′; and for nonspecific control, sense, 5′AACUCCCUCCCUGGGUCAGGUUCAUU-3′, and antisense, 5′-UGAACCUGACCCAGGGGAGGGAGTT-3′.
Cell Culture.
mIMCD3 cells (20) (a gift from S. Gullans, Harvard University, Boston, MA) were grown in 45% low-glucose DMEM plus 45% Coon's improved medium F-12 plus 10% FBS. HEK293 cells (American Type Culture Collection) were grown in Eagle's minimal essential medium plus 10% FBS. Osmolality of the control medium was 300 mosmol/kg H2O. All cells were cultured at 37°C in 5% CO2 and were used while subconfluent.
Animal Experiments.
Two- to 3-month-old male black six-strain mice (Taconic Farms, Germantown, NY) were injected i.p. with 1.5 mg/20 g of furosemide or the same volume of saline solution (controls). Kidneys were removed 4 h later, and inner medullas and cortices were dissected for RNA and protein analysis.
siRNAs and Inhibitors.
mIMCD3 cells (passages 14–20) were grown at 300 mosmol/kg and were split into 12-well plates 1 day before transfection. Transfection was at 300 mosmol/kg and 30–50% confluency, using 1 μg/ml mNTE specific or control siRNA or 0.1 μg/ml TonEBP/OREBP specific or control siRNA. Twenty-four hours later, fresh medium was substituted that had the same osmolality or was increased to 500 mosmol/kg by adding NaCl. mIMCD3 cells were exposed to 1 μM DFP (Sigma, St. Louis, MO) in isotonic medium for 2 h before altering NaCl concentration.
Immunoblotting.
Cells were lysed with mammalian protein extraction reagent (Pierce, Rockford, IL) according to the manufacturer's instructions with added protease inhibitors. Western blotting was performed as described (33). Briefly, 10 μg of protein was loaded onto each lane of 4–12% gradient acrylamide-Tris-glycine gels, and the separated proteins were transferred electrophoretically to Nitrocellulose membranes (Invitrogen). Membranes were blocked with blocking buffer (Odyssey, LICOR Biosciences, Lincoln, NE) plus Tween (0.1%), then incubated overnight at 4°C with the rabbit anti-NTE polyclonal antibody (diluted 1:1,000) provided by Moses V. Chao (25) or anti-TonEBP/OREBP polyclonal antibody from rabbit (diluted 1:1,000; PA1–023, anti-NFAT5; Affinity BioReagents, Golden, CO), followed by Alexa Fluor 680 goat anti-rabbit secondary antibody (diluted 1:5,000) for 1 h at room temperature. Blots were visualized and quantitated by using a LICOR (Lincoln, NE) Odyssey Infrared Imager.
RNA Isolation and cDNA Preparation.
Total RNA from mIMCD3 cells was isolated by using QiaShredder columns, followed by Qiagen RNeasy columns, according to the manufacturer's directions. RNA was treated with DNase while bound to the RNeasy column. Total RNA concentration was measured by spectrophotometry, and the RNA was run on agarose gels to assess its quality. Then 2.0 μg of total RNA were reverse-transcribed with random hexamers using TaqMan reverse transcription reagent kits (Applied Biosystems), following the manufacturer's recommendations.
Real-Time PCR.
For real-time PCR, specific primers were designed for mouse NTE mRNA by using Primer Express software (Applied Biosystems). The forward primer was 5′-CTCGGGCAGGGGGACTATGTTTTT-3′, and the reverse primer was 5′-CCGCTGGGGATGCTGGTGAC-3′. Multiplex PCR was performed with an ABI Prism 7900 sequence detection system, by using a Quantitect SYBR Green PCR kit. Triplicates of each sample at each of two levels of added cDNA (8 and 80 ng) were included in each PCR run. We analyzed the results using ABI Prism 7900 system software.
Analysis of mRNA Stability.
Osmolality was increased from 300 to 500 mosmol/kg for 2 h by addition of NaCl or was kept constant. Then, 5 μg/ml actinomycin D was added, and cells were harvested at indicated times for measurement of NTE mRNA abundance by real-time PCR.
Measurement of GPC.
Cell GPC was measured by chemiluminescence as described (16). Briefly, perchloric acid (PCA, 7%) was added to attached cells for 10 min on ice after removal of the experimental medium. Then, the PCA extract was kept at −80°C until analyzed. NaOH (0.5 M) was added to the cell remnants for protein determination. Fifty microliters of the PCA extract was diluted 1:1 with 7% PCA in duplicate. One aliquot was heated at 95°C for 1 h to hydrolyze GPC to choline. The other aliquot was kept on ice for 1 h to prevent hydrolysis. Then, 36 μl of 2 M K2CO3 was added to each aliquot, followed by centrifugation at 16,000 × g for 15 min. Twenty microliters of each supernatant were diluted to 1 ml with 67 mM glycine buffer (pH 8.6). One hundred microliters of this dilution plus 300 μl of glycine buffer were analyzed for choline in a luminometer (model 2010; Monolight, San Diego, CA) by rapidly adding 100 μl of reagent containing 67 mM glycine buffer (pH 8.6), 20 μg of horseradish peroxidase, 0.4 units of choline oxidase, and 6 nmol luminol.
Statistical Analysis.
Data were compared with an ANOVA test (followed by a Student–Newman–Keuls posttest). Normalized data were log-transformed before analysis by ANOVA. Results are expressed as means ± SE (n = number of independent experiments). Differences were considered significant for P < 0.05.
Abbreviations
- NTE
neuropathy target esterase
- PC
phosphatidylcholine
- GPC
glycerophosphocholine
- mIMCD3
mouse inner medullary collecting duct
- MDCK
Madin–Darby canine kidney
- DFP
diisopropyl fluorophosphate; mosmol, milliosmol.
Footnotes
The authors declare no conflict of interest.
References
- 1.Johnson MK. Biochem J. 1969;114:711–717. doi: 10.1042/bj1140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Dinsdale D, Glynn P. J Biol Chem. 2003;278:8820–8825. doi: 10.1074/jbc.M210743200. [DOI] [PubMed] [Google Scholar]
- 3.Lush MJ, Li Y, Read DJ, Willis AC, Glynn P. Biochem J. 1998;332:1–4. doi: 10.1042/bj3320001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muhlig-Versen M, da Cruz AB, Tschape JA, Moser M, Buttner R, Athenstaedt K, Glynn P, Kretzschmar D. J Neurosci. 2005;25:2865–2873. doi: 10.1523/JNEUROSCI.5097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson MK. In: The Target for Delayed Neurotoxicity by Organophosphorus Esters: Biochemical Studies and Toxicological Applications. Hodgson E, Bend JR, Philpot RM, editors. New York: Elsevier; 1982. [Google Scholar]
- 6.Glynn P. Biochem J. 1999;344:625–631. [PMC free article] [PubMed] [Google Scholar]
- 7.Zaccheo O, Dinsdale D, Meacock PA, Glynn P. J Biol Chem. 2004;279:24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]
- 8.Michea L, Ferguson DR, Peters EM, Andrews PM, Kirby MR, Burg MB. Am J Physiol. 2000;278:F209–F218. doi: 10.1152/ajprenal.2000.278.2.F209. [DOI] [PubMed] [Google Scholar]
- 9.Santos BC, Chevaile A, Hebert MJ, Zagajeski J, Gullans SR. Am J Physiol. 1998;274:F1167–F1173. doi: 10.1152/ajprenal.1998.274.6.F1167. [DOI] [PubMed] [Google Scholar]
- 10.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Perez A, Burg MB. Physiol Rev. 1991;71:1081–1115. doi: 10.1152/physrev.1991.71.4.1081. [DOI] [PubMed] [Google Scholar]
- 12.Bauernschmitt HG, Kinne RK. Biochim Biophys Acta. 1993;1150:25–34. doi: 10.1016/0005-2736(93)90117-i. [DOI] [PubMed] [Google Scholar]
- 13.Bauernschmitt HG, Kinne RK. Biochim Biophys Acta. 1993;1148:331–341. doi: 10.1016/0005-2736(93)90147-r. [DOI] [PubMed] [Google Scholar]
- 14.Nakanishi T, Burg MB. Am J Physiol. 1989;257:C795–C801. doi: 10.1152/ajpcell.1989.257.4.C795. [DOI] [PubMed] [Google Scholar]
- 15.Zablocki K, Miller SP, Garcia-Perez A, Burg MB. Proc Natl Acad Sci USA. 1991;88:7820–7824. doi: 10.1073/pnas.88.17.7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon ED, Zablocki K, Jung KY, Peters EM, Garcia-Perez A, Burg MB. Am J Physiol. 1995;269:C35–C41. doi: 10.1152/ajpcell.1995.269.1.C35. [DOI] [PubMed] [Google Scholar]
- 17.Kwon ED, Jung KY, Edsall LC, Kim HY, Garcia-Perez A, Burg MB. Am J Physiol. 1995;268:C402–C412. doi: 10.1152/ajpcell.1995.268.2.C402. [DOI] [PubMed] [Google Scholar]
- 18.Zablocki K. Int J Biochem Cell Biol. 1995;27:1055–1063. doi: 10.1016/1357-2725(95)90938-a. [DOI] [PubMed] [Google Scholar]
- 19.Kiewietdejonge A, Pitts M, Cabuhat L, Sherman C, Kladwang W, Miramontes G, Floresvillar J, Chan J, Ramirez RM. FEMS Yeast Res. 2006;6:205–217. doi: 10.1111/j.1567-1364.2006.00030.x. [DOI] [PubMed] [Google Scholar]
- 20.Rauchman MI, Nigam SK, Delpire E, Gullans SR. Am J Physiol. 1993;265:F416–F424. doi: 10.1152/ajprenal.1993.265.3.F416. [DOI] [PubMed] [Google Scholar]
- 21.Moriyama T, Garcia-Perez A, Burg MB. Am J Physiol. 1990;259:F847–F858. doi: 10.1152/ajprenal.1990.259.5.F847. [DOI] [PubMed] [Google Scholar]
- 22.Matsumura Y, Uchida S, Kondo Y, Miyazaki H, Ko SB, Hayama A, Morimoto T, Liu W, Arisawa M, Sasaki S, Marumo F. Nat Genet. 1999;21:95–98. doi: 10.1038/5036. [DOI] [PubMed] [Google Scholar]
- 23.Ko BC, Turck CW, Lee KW, Yang Y, Chung SS. Biochem Biophys Res Commun. 2000;270:52–61. doi: 10.1006/bbrc.2000.2376. [DOI] [PubMed] [Google Scholar]
- 24.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Proc Natl Acad Sci USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. Proc Natl Acad Sci USA. 2004;101:5075–5080. doi: 10.1073/pnas.0401030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferraris JD, Williams CK, Jung KY, Bedford JJ, Burg MB, Garcia-Perez A. J Biol Chem. 1996;271:18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- 27.Takenaka M, Preston AS, Kwon HM, Handler JS. J Biol Chem. 1994;269:29379–29381. [PubMed] [Google Scholar]
- 28.McMaster CR, Jackson TR. In: Lipid Metabolism and Membrane Biogenesis. Daum G, editor. Vol. 6. Berlin: Springer; 2004. pp. 5–88. [Google Scholar]
- 29.Dowd SR, Bier ME, Patton-Vogt JL. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- 30.Baburina I, Jackowski S. J Biol Chem. 1999;274:9400–9408. doi: 10.1074/jbc.274.14.9400. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 32.Na KY, Woo SK, Lee SD, Kwon HM. J Am Soc Nephrol. 2003;14:283–288. doi: 10.1097/01.asn.0000045050.19544.b2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Ferraris J, Irarrazabal CE, Dmitireva NI, Park JH, Burg MB. Am J Physiol. 2005;289:F506–F511. doi: 10.1152/ajprenal.00417.2004. [DOI] [PubMed] [Google Scholar]