Abstract
Burkholderia xenovorans LB400 (LB400), a well studied, effective polychlorinated biphenyl-degrader, has one of the two largest known bacterial genomes and is the first nonpathogenic Burkholderia isolate sequenced. From an evolutionary perspective, we find significant differences in functional specialization between the three replicons of LB400, as well as a more relaxed selective pressure for genes located on the two smaller vs. the largest replicon. High genomic plasticity, diversity, and specialization within the Burkholderia genus are exemplified by the conservation of only 44% of the genes between LB400 and Burkholderia cepacia complex strain 383. Even among four B. xenovorans strains, genome size varies from 7.4 to 9.73 Mbp. The latter is largely explained by our findings that >20% of the LB400 sequence was recently acquired by means of lateral gene transfer. Although a range of genetic factors associated with in vivo survival and intercellular interactions are present, these genetic factors are likely related to niche breadth rather than determinants of pathogenicity. The presence of at least eleven “central aromatic” and twenty “peripheral aromatic” pathways in LB400, among the highest in any sequenced bacterial genome, supports this hypothesis. Finally, in addition to the experimentally observed redundancy in benzoate degradation and formaldehyde oxidation pathways, the fact that 17.6% of proteins have a better LB400 paralog than an ortholog in a different genome highlights the importance of gene duplication and repeated acquirement, which, coupled with their divergence, raises questions regarding the role of paralogs and potential functional redundancies in large-genome microbes.
Keywords: genomics, niche adaptation, evolution, biodegradation, redundancy
Burkholderia xenovorans strain LB400 (LB400) (1) (formerly Pseudomonas sp. LB400, Burkholderia sp. LB400, Burkholderia fungorum LB400) is one of the most important aerobic polychlorinated biphenyl (PCB) degraders yet discovered. It oxidizes >20 PCB congeners, with up to six chlorine substitutions on the biphenyl rings (2–5). LB400 was isolated from a PCB-containing landfill in upper New York State (6) and has since been the subject of >70 studies related to PCB degradation, making it the model organism for PCB biodegradation studies. B. xenovorans belongs to the Burkholderia graminis clade, with members most commonly found in the rhizosphere of grass plants (1, –9).
The diversity of the Burkholderia genus is exemplified by the high diversity of ecological niches occupied by the different species, ranging from soil to aqueous environments, associated with plants, fungi, amoeba, animals, and humans, from saprophytes to endosymbionts, from biocontrol agents to pathogens (10–15). Historically, most of the attention toward this genus was directed toward species with malign properties, including plant pathogens and species causing animal and human disease, such as Burkholderia mallei and Burkholderia pseudomallei (16, 17). A subgroup of at least ten closely related species, the Burkholderia cepacia complex (Bcc), are secondary colonizers of the lungs of cystic fibrosis patients, often causing rapid decline in lung function (12). Similar to most other Burkholderia species, Bcc species are commonly found in the environment, often associated with the plant rhizosphere (18).
Common to most Burkholderia species are a large, multireplicon genome and the presence of multiple insertion sequences that confer high genome plasticity, which could help explain the versatility of the genus (19). Twenty-two Burkholderia strains are being, or have been recently, sequenced [Joint Genome Institute (JGI)/Integrated Microbial Genomes (IMG); http://img.jgi.doe.gov]. The complete genomes of the closely related B. pseudomallei K96243 (20) and B. mallei ATCC23344 (21) became available in 2004, whereas Burkholderia cenocepacia J2315 (http://www.sanger.ac.uk), B. cenocepacia HI2424 (JGI), and the environmental isolate Bcc strain 383 (JGI) are the Bcc strains thus far fully sequenced. This series of genome projects aims to determine the factors differentiating environmental from pathogenic strains, as well as to unravel the origin and evolution of the niche versatility of this genus. Because of the narrow focus (PCB degradation) of past studies on LB400, most of its general physiology and lifestyle remain to be discovered. Since the initiation of the genome project, the interest in LB400 has broadened and has highlighted it as a versatile biodegrading soil organism metabolizing compounds containing single-carbon (C1) groups (22, 23), isoflavonoids (24), diterpenoids (25), and sulfonates (26).
Results and Discussion
General Genome Description.
General overview.
The LB400 genome has a size of 9.73 Mbp, harboring ≈9,000 coding sequences distributed over three circular replicons that have been designated chromosome 1 (4.90 Mbp), chromosome 2 (3.36 Mbp), and megaplasmid (1.47 Mbp) (Table 1, which is published as supporting information on the PNAS web site). Origins of replication were determined based on G+C skew analysis (Fig. 1) and the presence of genes involved in DNA replication close to the predicted origin for each of the replicons. Homologs of dnaA and dnaN are present on chromosome 1, whereas parA and parB, which are responsible for plasmid partitioning and replication, are present on chromosome 2. Similarly, genes encoding ParA and ParB are also found on the small chromosome of other Burkholderia species (20, 21), and these proteins are >80% identical to those of LB400. We defined the 3.36-Mbp replicon as chromosome 2 and not a megaplasmid based on the presence of three ribosomal operons and several tRNAs (although none are unique), as well as the unique occurrence of core cellular functions involved in DNA replication [DNA primase (dnaG), DNA polymerase I (polA)] and associated elongation factor (polB) and amino acid metabolism. Moreover, there are several functions crucial to this organism's adaptation to its niche uniquely located on this replicon (see Genetic Factors Indicating the Ecological Niche of B. xenovorans). The 1.42-Mbp megaplasmid, on the other hand, contains neither RNA genes nor any essential functions and is mostly absent in other strains of the same species (see Comparison with other Burkholderia species). The overall G+C% of the megaplasmid is 1% lower than the two chromosomes, and, together with the G+C% patterns and the high fraction of inserted sequences, this replicon appears to be a mosaic of foreign genomic material. Nevertheless, the megaplasmid's G+C skew is well defined, although asymmetrical (Fig. 1C). ParAB share only ≈20% amino acid sequence identity with ParAB on chromosome 2 whereas they are >80% identical to ParAB on the 0.64-Mbp replicon of Cupriavidus necator (Ralstonia eutropha) JMP134.
Fig. 1.
Schematic representation of the large chromosome (chromosome 1) (A), small chromosome (chromosome 2) (B), and megaplasmid (C) of B. xenovorans LB400. Radii are scaled based on replicon size, except for the megaplasmid, an extra ring for which was added to indicate true size relative to the chromosomes. The outer two rings (1 and 2) represent the LB400 genes on the forward and reverse strands, respectively, colored by functional class. The next two sets of rings represent the CGH data of LB400 vs. LMG 16224 (rings 3 and 4) and LMG 21720 (rings 5 and 6), respectively (blue, gene absent; yellow, gene present). Ring 7 shows the locations of the genomic islands (identified based on bias in Karlin score, G+C% and G+C skew; Table 4). The next four sets of rings are based on reciprocal best BLAST hit analysis (cutoffs: 30% amino acid identity, alignment over at least 70% of the length) of a test genome vs. the LB400 genome, which was displayed by using GenomeViz (66) with bar height relative to the % amino acid identity: B. cenocepacia J2315 (red, rings 8 and 9) [the J2315 sequence data were produced by the Pathogen Sequencing Group at the Sanger Institute (http://www.sanger.ac.uk/Projects/B_cenocepacia/)]; B. pseudomallei (blue, rings 10 and 11); Bcc strain 383 (green, rings 12 and 13); and Ralstonia solanacearum (magenta, rings 14 and 15). Ring 16 (black) represents G+C content, and ring 17 represents the G+C skew.
Gross functional content.
The genome's functional content and its distribution over the replicons were analyzed based on the replicons' bias toward particular clusters of orthologous groups of proteins (COG) (Fig. 2A). The large chromosome carries the core cellular function (e.g., translational machinery, DNA replication, cell division, and nucleotide metabolism) and can be considered the “core” chromosome. The small chromosome and the megaplasmid have a functional bias toward energy metabolism, secondary metabolism, inorganic ion transport and metabolism, and amino acid metabolism and transport (COG E). The bias toward COG E can be explained by the large number of amino acid transporters on these replicons: 58% of the megaplasmid-located and 42% of small chromosome-located COG E proteins compared with only 32% of the large chromosome-located COG E proteins. On the megaplasmid, the high number of proteins in the subclass of branched amino acid transporters is remarkable: 49 (28% of its COG E proteins) vs. 25 (8%) on the small and 40 (12%) on the large chromosome. The role of this subclass of transporters is unclear but is most likely not limited to amino acid transport. The bias toward inorganic ion transport/metabolism is caused by an uneven presence of oxygenase enzymes on these replicons, which are classified in this COG because they contain metal ions. Compared with the other replicons, the protein content of chromosome 2 is biased toward transcription, carbohydrate transport and metabolism, and signal transduction mechanisms, whereas the megaplasmid has a bias toward lipid metabolism. The latter observation is explained by the presence of a large number of CoA ligases as well as other enzymes similar to fatty acid metabolic pathways. This distribution of function leads us to define the small chromosome as the “lifestyle-determining” replicon whereas the megaplasmid encodes highly specialized strain-specific functionality. A similar separation of core and secondary functions was also observed in other multireplicon genomes, such as the other sequenced Burkholderia species and the legume symbiont Sinorhizobium meliloti (27).
Fig. 2.
Functional distribution over the three replicons based on the COG classifications. Presented is the percent of genes on each replicon belonging to each COG.
Paralogs and functional redundancy.
Gene and functional redundancy seems to be an important theme in large bacterial genomes. Based on our analysis, 1,581 coding sequences (17.6% of all genes) were found to have a better match within the LB400 genome than in the database of 260 completed genomes. To date, this percentage represents the highest extent of gene and potential functional redundancy among the genomes of free-living bacteria, where the average is 7.6% (±4.0%) of their genes. The list of paralogs in the LB400 genome is enriched in genes related to transport (230 paralogs), signal transduction (164 paralogs), mobile elements (112 paralogs), membrane proteins (66 paralogs), and secondary metabolism, including 120 dehydrogenases, 32 di-oxygenases, and 13 mono-oxygenases. Although the aim of our analysis was to uncover potential functional redundancy, it is important to recognize that, for most paralogs, divergent evolution can lead to changes in substrate range, kinetic properties, or even function, thus further extending LB400's great metabolic versatility. Interestingly, these 1,581 paralogs are numerically more or less equally distributed among the three replicons (≈500 genes per replicon), and, consequently, the relative contribution of the smaller replicons to gene redundancy is substantially greater than chromosome 1.
The high number of paralogs among transport proteins correlates with the large fraction of LB400 genes (≈1,400, encoding ≈610 transport systems; Table 2, which is published as supporting information on the PNAS web site) dedicated to this function. There are >180 efflux systems, including 89 drug efflux pumps, 18 protein secretion systems, 21 heavy metal efflux pumps, and 18 amino acid/amino acid lactone efflux pumps (LivE, RhtB, and LysE). The latter suggests a role beyond amino acid transport. The other 430 systems are involved in uptake of organic compounds as well as all necessary inorganic cations and several anions. The most extensive redundancy is observed for the abundant branched and polar amino acid transporter families, which can indicate functional diversification to transport-related compounds.
A second class of proteins for which a high number of paralogs is observed is signal transduction. In light of this trait and the fact that it has been previously shown that the signal transduction network size increases disproportionately with genome size (28), we analyzed in more detail this class of proteins (Table 3, which is published as supporting information on the PNAS web site). Among currently published bacterial genomes, LB400 dedicates the second-largest share (9%) of its genome to signal transduction (second to Streptomyces coelicolor) and utilizes almost the entire spectrum of input (sensory) and output (regulatory) domains found in prokaryotic signal transduction (29). Whereas regulation of gene expression (vs. enzymatic domains) on average constitutes ≈75% of the output activity in prokaryotic signal transduction (29), it constitutes almost 90% in LB400. One-component signal transduction (1CST) proteins (29) that combine a sensory and a regulatory module are the dominant signal transduction mechanism in LB400 (629 proteins). The LysR and AraC families (transcription activation in response to a small ligand) constitute the two largest classes (176 and 74 proteins, respectively) among 1CST systems. Most are encoded within metabolic gene clusters. In contrast, we found fewer GntR-type (71 proteins) and TetR-type transcriptional repressors (57 proteins), indicating that most metabolic pathways in LB400 require induction. The majority of 1CST systems in LB400 are predicted to be soluble, cytoplasmic sensors, indicating that it detects most environmental signals intracellularly after transport into the cell. Two-component signal transduction (2CST) systems are less abundant. Apart from the chemotaxis system (see Metabolic traits and Chemotaxis), the genome contains 76 sensor histidine kinases and 73 response regulators. Thirty histidine kinases are predicted to be soluble cytoplasmic sensors whereas many membrane-bound sensor histidine kinases contain intracellular sensory domains, such as PAS (30). The extraordinary sensing capacity of LB400 is further emphasized by the fact that Rhodococcus sp. RHA1, an organism with similar niche and genome size, has 25% fewer transporter systems and less than half the number of 2CST systems (31).
What is the added value of gene and functional redundancy for organisms with such large genomes? In LB400, functional redundancy of formaldehyde oxidation (23) and benzoate catabolism (32), for each process which LB400 has three potential pathways, has been studied, and, in both cases, the functionality of each of the three pathways present in the genome was confirmed. Redundancy is not entirely unexpected because these organisms presumably have to contend with varying conditions in nutrient source types and concentrations. The latter is exemplified by the detailed study of benzoate catabolism, which suggests that each of these pathways is conditionally regulated depending on benzoate and oxygen levels presented to the cell (33). Further studies of LB400 metabolic and other functional redundancies, such as the presence of multiple Type IV secretion systems (see Burkholderia virulence-associated genes), will be important to gain a better understanding of their evolutionary origin, their role, and whether or not there is a selective benefit of functional redundancy in large-genome bacteria.
Genome Evolution.
Comparison with other Burkholderia species.
We compared the gene complement of LB400 with that of B. pseudomallei and Bcc strain 383, two strains about equally distant to LB400 (77.5% and 76.8% average nucleotide identity, respectively). The comparisons gave very similar results, and we summarize the findings for the comparison with strain 383 only for simplicity. Less than half of the genes in the LB400 genome (3,961) are conserved between LB400 and strain 383, revealing the great genetic diversity within the Burkholderia genus. These 3,961 conserved genes are unevenly distributed in the three replicons of LB400: 2,602 conserved genes on chromosome 1 (57.7% of the replicon's genes), 1,150 on chromosome 2 (37.8%), and 209 on the megaplasmid (15%). Therefore, the smaller replicons seem to contribute more to the phenotypic uniqueness of the species and strains, whereas the largest replicon possesses most of the core functions of the Burkholderia genus. This uneven distribution of conserved genes over the replicons is consistent with comparisons of B. pseudomallei and B. mallei genomes (20, 21) and reveals an important, universal property of the Burkholderia genus. Furthermore, extending our analysis to other fully sequenced Burkholderia strains confirms the large chromosome as the core and most conserved chromosome (Fig. 1). Similar to the comparison with Bcc strain 383 alone, the small chromosome displays a significantly larger variation between the various species as compared with the large chromosome (Fig. 1B), whereas the megaplasmid is virtually unique to LB400 (Fig. 1C). The large number of shared genes on the small chromosomes of the different Bcc strains (data not shown) and the high sequence similarity of the small chromosome's parAB genes between all Burkholderia species (see General Genome Description) suggest a shared ancestry.
Interestingly, our best reciprocal BLAST comparison of LB400 with Bcc strain 383 showed that the orthologous genes between the LB400 and strain 383 genomes are generally found in the same replicon for the genes in the largest replicon whereas this is not always the case for the smaller replicons. For instance, 40, 125, and 44 of the orthologs on the LB400 megaplasmid are located on the largest, medium, and smallest replicon of strain 383, respectively. The sequence identity of the orthologs is also noticeably different between the replicons: the orthologs share 74.9% average amino acid identity for chromosome 1, 66.3% for chromosome 2, and 55.9% for the megaplasmid. Together, these results reveal that the smaller replicons are more “plastic” and show higher sequence evolution rates. Although we cannot exclude that some “false” orthologies may confound some of our findings, when we repeated the analysis with genomes of more closely related Burkholderia strains, or more stringent cut-offs (which are less error-prone), we found a very similar pattern.
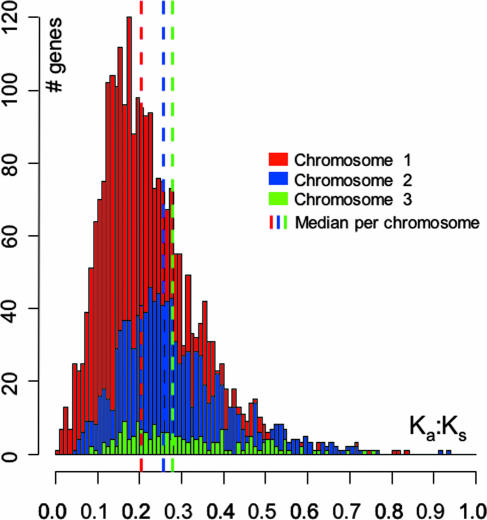
To further test for differences in selective pressure acting on the replicons, we used the ratio of nonsynonymous vs. synonymous substitutions (Ka:Ks) between orthologous pairs in LB400 and Bcc strain 383, with smaller Ka/Ks ratios representing higher negative, purifying selection. The Ka/Ks ratio differs substantially between replicons: the megaplasmid's genes have an average Ka/Ks ratio of 0.32 (±0.14), vs. 0.28 (±0.13) for chromosome 2 and 0.22 (±0.11) for chromosome 1 (Fig. 3). Also, a significantly larger share of the Ka/Ks values was significantly <1 (Z tests, P < 0.05) on the large chromosome as compared with the two smaller replicons (Fig. 5, which is published as supporting information on the PNAS web site). These results further support the notion that the smaller replicons may be under more relaxed selective pressure, which probably accommodates and drives their plasticity. It is interesting to note that, in most of the previous comparisons, chromosome 2 behaves more like the megaplasmid than like chromosome 1. Consistent with these interpretations, whole genome alignments (34) of LB400 with B. pseudomallei and Bcc strain 383 indicate that the degree of recombination/rearrangement is significantly higher for the small chromosome (Fig. 6, which is published as supporting information on the PNAS web site). Interestingly however, and similar to B. pseudomallei (20), chromosome 2 carries a smaller fraction of recently acquired functions (6 genomic islands, containing ≈7% of the small chromosome) compared with chromosome 1 (15 genomic islands containing ≈17% of the large chromosome) (Table 1 and Table 4, which is published as supporting information on the PNAS web site). Considering that 7 of 15 genomic islands on the large chromosome (8/21 in total) are adjacent to a tRNA gene and contain integrases or transposases, this preferential insertion into the large chromosome is probably due to the much larger number of tRNA genes on the large chromosome (57 of 64). The lower number of insertions into the small chromosome could also indicate that the specific functions encoded by genes located on the small chromosome, which presumably help differentiate the various Burkholderia species, were acquired long ago and have been maintained by selection.
Fig. 3.
The ratio of asynonymous vs. synonymous nucleotide substitutions (Ka:Ks) between orthologous pairs in LB400 and Bcc strain 383, plotted conjointly for the three chromosomes. There is a significant trend toward more spread ratios (flattening of the distribution) going from the large chromosome to the smaller ones. Although no positive selection could be evidenced overall for any pair of genes compared (i.e., Ka > Ks), this trend can be interpreted as a progressive relaxation of the selection pressure for amino acid substitution (pressure Chr1>Chr2>MP) (cf. Fig. 5).
Comparison with other B. xenovorans strains.
By using the LB400 genomic microarray, comparative genome hybridizations (CGHs) were performed for B. xenovorans LMG 21720 (plant rhizosphere origin) and LMG 16224 (blood sample origin). Sixty-nine percent of LB400's genes are conserved in both strains. Pairwise comparisons correlate well with previously determined DNA:DNA hybridization ratios (1): 78% are conserved in LMG 21720 and 73% in LMG 16224 as compared with DNA:DNA hybridization ratios of 83% and 73%, respectively. Based on pulsed-field gel electrophoresis (PFGE), LMG 21720 and LMG 16224 were estimated to be 7.83 and 8.87 Mbp, respectively. A fourth B. xenovorans strain, LMG 22943 (plant rhizosphere origin), was sized at 7.42 Mbp. These three strains harbor two chromosomes, whereas only LMG 22943 and LMG 16224 contain a plasmid, roughly half the size of the one in LB400 (Fig. 7, which is published as supporting information on the PNAS web site). These data support the CGH results, indicating that most of the megaplasmid is indeed missing from the other members of the species. Based on these data ≈1.75 Mbp of additional genomic material is found in LMG 16224 compared with only 0.30 Mbp in LMG 21720.
The most evident CGH result was that 70% of the genes located on the megaplasmid were uniquely present in LB400. Consequently, the functional distribution of the genes unique to LB400 correlates well with the functional bias of the megaplasmid (Fig. 8, which is published as supporting information on the PNAS web site). Apart from five islands on the megaplasmid (≈12%) that are conserved in the other B. xenovorans strains (Fig. 1C), conserved genes are scattered throughout the megaplasmid and are mostly involved in transport and regulation. The conserved islands include genes encoding carbon monoxide dehydrogenase (cox), diterpenoid metabolism (dit), anthranilate dioxygenase (andA), and formaldehyde oxidation. We also verified the presence of dit genes experimentally by growth of both LMG strains on abietic and dehydroabietic acid. Important megaplasmid-derived functions, unique to LB400, include the periplasmic nitrate reductase (nap), the biphenyl pathway for PCB degradation, and a copy of the aerobic CoA ligation-dependent benzoate pathway (boxM). Further differences between B. xenovorans strains were mostly attributed to the unique presence of genomic islands on the chromosomes (Fig. 1 A and B and Table 4). This observation suggests that most genes unique to LB400 were recently acquired by lateral gene transfer mediated by mobile elements, which is supported by their absence in other Burkholderia and Ralstonia species (Fig. 1). Except for the acquisition of several aromatic degradation capabilities (Table 4), the functional impact of the extensive import of foreign genes in LB400 remains largely unknown.
The CGH results also reveal that 6.2% of LB400's genes are uniquely shared with LMG 16227 (and not LMG 21720) whereas 9.4% are uniquely shared with LMG 21720. Whereas some of these are located within the identified genomic islands (Table 4), other differences between strains are presumably due to differential gene loss. Interestingly, whereas chromosome 2 contains significantly fewer genomic islands than chromosome 1, most of this differential gene loss appears to have occurred on the small chromosome (Fig. 1 and Table 4). The more extensive shuffling and differential gene loss on the small chromosome can be seen as genomic fine-tuning, where selective pressure for niche-specific environmental factors plays a large role.
We have also found in LB400, but not in the other B. xenovorans strains used for CGH (confirmed by PCR, data not shown), the presence of ISPpu12, a 3,372-bp insertion sequence first described on the toluene-xylene TOL catabolic plasmid pWWO of Pseudomonas putida mt-2 (35). In LB400, ISPpu12 was found in its entirety in seven different locations (three copies on the large chromosome, two on the small chromosome, and two on the megaplasmid). Furthermore, because its sequence was found in a portion of the sequencing reads at four additional locations (one on chromosome 1, two on chromosome 2, and one on the megaplasmid), we believe this IS element is actively copying itself within the genome, which resulted in distinct genotypic (and potentially phenotypic) subpopulations within the original culture. Similar observations of strain variation within clonal populations grown for sequencing have been previously reported for large-scale genomic rearrangements in Yersinia pestis (36) and for phase variable regions in the Bacteroides fragilis genome (37). It is interesting to note that ISPpu12 occurs in several other bacteria, generally associated with plasmids and xenobiotic degradation genes, and that it has been suggested to be involved in gene silencing and activation (38).
Genetic Factors Indicating the Ecological Niche of B. xenovorans.
Metabolic traits.
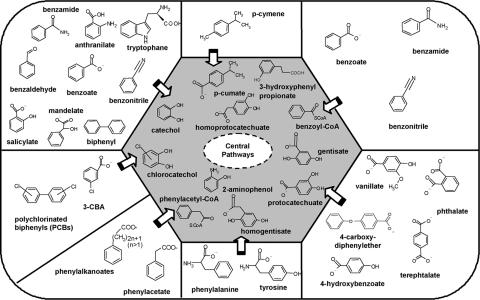
Based on the origin of its isolates, B. xenovorans presumably has a versatile environmental niche (soil and plant rhizosphere), which could imply an ability to degrade aromatic compounds that originate from root exudates and root turnover. CGH with other B. xenovorans strains showed that LB400 is enriched in aromatic catabolic pathways contained in genomic islands, providing biphenyl, 3-chlorocatechol, 2-aminophenol, and other catabolic capacities (Table 4) to this strain. Based on in silico analysis, we found eleven “central aromatic” catabolic pathways in the LB400 genome (Fig. 4 and Table 5, which is published as supporting information on the PNAS web site), indicating an unusually high metabolic versatility. Other sequenced aromatic compound-degrading bacteria, such as the aerobic P. putida KT2440 (39), Rhodococcus RHA1 (31), and anaerobic strain EbN1 (40), have six, eight, and five central aromatic pathways, respectively. Nine such pathways were found in the combined genomes of all fully or partially sequenced Pseudomonas strains (41). In addition, the LB400 genome contains twenty “peripheral aromatic” catabolic pathways (Fig. 4). Seven and 13 such pathways were described in P. putida KT2440 (39) and strain EbN1, respectively (40), whereas Rhodococcus RHA1 (31) contains at least 25. LB400 has >170 genes encoding these aromatic catabolic pathways, spread over the three replicons (Fig. 9, which is published as supporting information on the PNAS web site, and Table 4). The genes of the central aromatic pathways are generally organized in operon-like structures, and genes encoding transcriptional regulators are adjacent to 10 of 11 central aromatic pathway operons, which suggests integration into the signal transduction network. Several operons contain transporter genes as well (Table 4). There are at least 23 aromatic acid transporters, and some of the ≈100 (undefined) major facilitator superfamily and amino acid family transporters could be involved as well, e.g., branched amino acid transporters, two systems of which are within aromatic degradation operons (Table 4). The high abundance of transporters in LB400 suggests that its diverse metabolism is complemented by the same diversity in transport capacity.
Fig. 4.
Schematic representation of all central aromatic pathways (gray background), based on their main substrate, present in LB400. Peripheral aromatic pathways are indicated in the outer sectors.
Primary reactions.
To be able to metabolize aromatic, nonhydroxylated carbon sources, initial ring activation is generally required. This can occur by di- or monohydroxylation through the activity of a primary oxygenase. In other aerobic aromatic degradation pathways, CoA ligation is the initial step mediated by an AMP-dependent CoA ligase. In the latter case, ring hydroxylation is still required, but the initial substrate selectivity is determined by the CoA ligase. The LB400 genome contains 12 Rieske-type primary dioxygenases (Fig. 10A, which is published as supporting information on the PNAS web site), 6 cytochrome P450 monooxygenases, and 45 AMP-dependent synthetases/ligases (Fig. 10B). The latter group are mainly studied for their role in fatty acid metabolism, but the same mechanism is involved in degradation of benzoate, phenylacetate, and diterpenoids (see Aerobic hybrid pathways). The functions of most of these divergent CoA ligases and associated β-oxidation-like pathways are unclear and require experimental verification.
The β-ketoadipate pathway and peripheral aromatic pathways.
Degradation pathways for many aromatic compounds eventually funnel into the β-ketoadipate pathway via the catechol branch or the protocatechuate branch, which converge at β-ketoadipate enol-lactone. The cat genes, which encode the three enzymes of the catechol pathway and participate in the transformation of catechol to β-ketoadipate enol-lactone, are arranged as a catRBAC cluster located on chromosome 1. The genes of the complete protocatechuate pathway, arranged as pcaIJBDC and pcaHG clusters and a single pcaF gene, are located on chromosome 2. The cat and pca gene arrangement is similar to other β-proteobacteria such as B. pseudomallei and Ralstonia metallidurans (39). Fifteen peripheral aromatic pathways that lead to the β-ketoadipate pathway were identified. Nine substrates are degraded via catechol: (i) (chloro)biphenyl (bph, megaplasmid); (ii) benzoate, (ben, chromosome 2); (iii) benzonitrile (nit, chromosome 1), which is metabolized to benzoate via (iv) benzamide (ami, megaplasmid); (v) mandelate (mdl, chromosome 2), which is transformed to benzoate via (vi) benzaldehyde (xyl, megaplasmid); (vii) tryptophan (kyn, chromosome 1 and 2), which is converted to benzoate via (viii) anthranilate, (and, two paralogous copies on chromosome 2, megaplasmid); and (ix) salicylate, which is transformed to catechol by salicylate hydroxylase (nahW, megaplasmid). Five substrates are degraded via protocatechuate: (i) vanilline (van, chromosome 1); (ii) 4-hydroxybenzoate (pob, chromosome 1) (41); (iii) phtalate (oph, chromosome 1 and 2); (iv) terephthalate (tph, two copies on chromosome 2); and (v) 4-carboxydiphenyl ether (pob, copies on chromosome 1 and 2). Finally, 3-chlorocatechol is degraded to β-ketoadipate by the so called modified ortho pathway (clc, chromosome 1), organized similar to Pseudomonas sp. B13 (42).
Aerobic hybrid pathways.
Recently, a new aerobic hybrid pathway for benzoate degradation has been described in various bacteria including LB400, which has two homologous and functional copies of this pathway (chromosome 1, megaplasmid) (32, 33, 43, 44). Phenylacetate is transformed by a similar hybrid pathway via phenylacetyl-CoA (45) because of activity of the paa pathway (chromosome 1, megaplasmid). Also, a particular cluster of genes, which includes the characterized Pseudomonas abietaniphila BKME-9 dit cluster, encodes proteins required for abietane diterpenoid catabolism. Abietane diterpenoids are tricyclic compounds commonly found in the oleoresin of coniferous trees. The dit cluster includes genes coding for two cytochromes P450 (ditQ, ditU), a Rieske-type dioxygenase (ditA1) (Fig. 10A), an extradiol ring-cleavage dioxygenase (ditC), a CoA ligase (ditJ) (Fig. 10B), and associated β-oxidation homologs (D.S., J. Park, J.M.T., and W. Mohn, unpublished data). Based on the abundance of CoA ligase/β-oxidation homologs, more of such auxiliary, aerobic hybrid pathways are probably present in LB400 but are yet to be recognized.
Other metabolic traits.
Similar to some other Burkholderia species, B. xenovorans contains the nif nitrogen fixation genes (chromosome 2). Regarding nitrogen metabolism, the presence of nap genes (megaplasmid) for aerobic nitrate reduction is interesting as well. Furthermore, C1 oxidation and assimilation may play an important role in this species because we observed in the LB400 genome (i) three formaldehyde oxidation pathways, (ii) a methanol dehydrogenase-like enzyme (XoxF) and associated PQQ cofactor synthesis genes, (iii) three carbon monoxide dehydrogenases (acetogenesis), and (iv) the entire Calvin–Benson–Bassham cycle for CO2-fixation in autotrophic organisms. Importantly, several of these pathways also seem to be present in the B. xenovorans strains tested by CGH, but not in other sequenced Burkholderia species (23). There is no evidence of a recent acquisition event for most C1 genes, indicating that these genes were presumably acquired early on in the evolution of the B. xenovorans lineage. No growth of LB400 on C1 compounds has yet been reported; however, growth was observed on the methylated-glycine derivatives betaine and choline (23). Because many plant aromatic compounds are methylated or methoxylated, it is possible that these C1 pathways are involved in plant aromatics metabolism, which is a possible explanation for their coexpression in biphenyl-grown LB400 cells at transition to stationary phase (33, 44).
Chemotaxis.
The signal transduction network of LB400 contains several chemotaxis genes, some of which might be involved in taxis toward aromatic compounds. Only one chemoreceptor (BxeA0121, methyl-accepting chemotaxis protein, MCP) is located within the chemotaxis operon, whereas 31 other chemoreceptors are scattered throughout the genome. Twenty-one chemoreceptors belong to Class I MCPs (46), i.e., are transmembrane proteins with a periplasmic sensory domain and are therefore predicted to detect extracellular signals. One MCP (BxeA2405) is an ortholog of the Escherichia coli Aer aerotaxis transducer, and all amino acid residues implicated in the FAD binding and the aerotactic response (47) are conserved in this protein, suggesting that it governs energy taxis (48) in LB400.
Presence/absence of pathogenicity traits.
Burkholderia virulence-associated genes.
Although the genetic factors required for animal and human pathogenicity by Burkholderia are largely unknown, multiple traits have been experimentally implicated in the virulence of B. pseudomallei (20) and the Bcc (12) (Table 6, which is published as supporting information on the PNAS web site). Resistance to cationic antimicrobial agents such as polymixin B, presence of a flagellum (49), and iron acquisition by siderophores (50) (pyoverdin, pyochelin biosynthesis, chromosome 1) are virulence traits but also important in their soil habitat. Also, several genes are present that may mediate resistance to reactive oxygen species and enable LB400 to survive within intracellular environments. Although LB400 encodes three phospholipase homologs and an LasA-type protease, its repertoire of phospholipases, proteases, and collagenases, which have been implicated in Burkholderia virulence (20, 51), is limited. LB400 also lacks homologs of the B. cenocepacia cable pilus, a defining pathogenic feature of one of the most problematic strains in cystic fibrosis infection (12). However, in common with other Burkholderia genomes (20), LB400 possesses multiple hemagglutinin-like adhesin genes (n = 9), suggesting that binding to microbial and eukaryotic cells is important to its lifestyle. LB400 also possesses one type III secretion system (20, 52, 53). We found potential virulence-related functions only present on islands 9, 11, and 4 (type IV secretion and adhesion functions; Table 4), suggesting that laterally acquired DNA has been retained by LB400 by selection in the natural environment rather than during primary infection. A plasmid-encoded type IV secretion system in B. cenocepacia has been implicated in the secretion of plant cytotoxic proteins (54); however, neither of the LB400 systems shares significant sequence similarity with the latter plant pathogenic gene cluster.
The Cenocepacia island (12) is not present in LB400. However, components of the amino acid metabolism gene cluster within the B. cenocepacia pathogenicity-related island show remarkable synteny with LB400 megaplasmid segments (Fig. 11, which is published as supporting information on the PNAS web site). Six of the eight branched-chain amino acid metabolism genes, including the porin implicated in inflammation during infection (55) (BxeC0438), are highly conserved between the two species. The LB400 megaplasmid also has a cluster of type IV secretion genes immediately adjacent to the latter amino acid gene cluster, which shares considerable sequence similarity with a B. cenocepacia type IV secretion system island encoded ≈46 kb downstream of the Cenocepacia island on the J2315 second chromosome (54). These corresponding LB400 megaplasmid regions do not possess the hallmarks of genomic islands, suggesting that the megaplasmid may represent an ancestral replicon from which the B. cenocepacia laterally acquired the Cenocepacia and type IV secretion islands. The fact that the quorum-sensing system on the Cenocepacia island is most closely related to that encoded on the LB400 megaplasmid supports this hypothesis. Quorum-sensing systems regulate the expression of a number of traits involved in Burkholderia pathogenicity (20, 56, 57). LB400 possesses two gene sets with homology to autoinducer synthesis and response regulator systems, one on the megaplasmid (BxeC0415, BxeC0416) and a classical luxIR-like system (BxeB0608, BxeB0610 on chromosome 2), which is phylogenetically most closely related to the MupIR, PpuIR, and LasIR systems of Pseudomonas species (57). Their regulatory roles are uncharacterized.
Genes associated with in vivo survival.
Hunt et al. (58) reported the isolation of 102 transposon mutants of B. cenocepacia strain K56-2 that were unable to survive in a rat model of chronic lung infection. Of the 69 mutants that were mapped to orthologs within the finished B. cenocepacia J2315 genome (58), direct orthologs were found for 46 genes in LB400 (Table 7, which is published as supporting information on the PNAS web site). Although these findings suggest that LB400 encodes a large proportion of the Burkholderia genes required for in vivo survival, the lack of LB400 homologs for several of the virulence factors for pathogenic Burkholderia species indicates that LB400 has little potential to cause infection.
Concluding Remarks.
The LB400 genome and its comparison to closely related B. xenovorans strains, as well as to more distantly related Burkholderia species, underscore major themes in prokaryotic niche adaptation and genome evolution. Our analysis highlights that the niche and phenotypic diversity and versatility of the Burkholderia genus are reflected in the genomic composition of this group. This genus exemplifies the importance to niche versatility of lateral gene transfer and plasmid acquisition, as well as extensive gene and functional redundancy, because of both gene duplication and independent acquisition events. Based on (i) the distribution of gene function among the replicons, (ii) the dominance of intra- over interreplicon rearrangements when compared with other Burkholderia, and (iii) the differential gene conservation and evolutionary rates observed for the replicons, we (i) define the large chromosome as the core chromosome representing the major phenotypic characteristics of the Burkholderia genus, (ii) define the small chromosome as the lifestyle-determining replicon representing the adaptation of the species to its niche, and (iii) assert that the megaplasmid, the individuality replicon, provides the highly specialized, unique metabolic capabilities to LB400. The ecologic, phenotypic, and genomic features of B. xenovorans suggest that it and probably many environmental Burkholderia are versaphiles, i.e., adapted to complex or diverse niches.
Materials and Methods
Bacterial Strains.
The sequenced strain B. xenovorans LB400 was isolated from PCB-contaminated landfill soil in Moreau, New York (6). The sequenced clone is the original strain, which is deposited in the U.S. Department of Agriculture–Agricultural Research Service (USDA-ARS) Culture Collection (Peoria, IL) (NRRL B-18064). Three additional B. xenovorans strains were used in this study: LMG 16224 was retrieved from a blood culture specimen from a 31-year-old woman in Göteborg, Sweden; LMG 21720 is a coffee plant rhizosphere isolate from Coatepec, Veracruz State, Mexico (8); and LMG 22943 was isolated from a grass rhizosphere in Wageningen, The Netherlands (9).
Genome Sequencing.
Whole-genome shotgun sequencing was performed on 3-kb, 8-kb, and 40-kb insert DNA libraries by the Production Genomics Facility of the Department of Energy (DOE)-Joint Genome Institute (DOE-JGI, Walnut Creek, CA). Paired-end sequencing generated >270,000 reads and resulted in approximately 16× depth of coverage. The reads were assembled to produce the primary high-quality draft assembly of 715 contigs, which were linked into larger scaffolds (55 total) by paired-end information. Genome finishing, including gap closure, repeat resolution, and polishing, was performed as described (59).
Genome Annotation and Analysis.
Automated gene prediction was performed by using the output of Critica (60) complemented with the output of Generation and Glimmer (61), and is available at http://genome.ornl.gov/microbial/bfun/. The tRNAScanSE tool (62) was used to find tRNA genes, whereas ribosomal RNAs were found by using BLASTn vs. the 16S and 23S ribosomal RNA databases. Other “standard” structural RNAs (e.g., 5S rRNA, rnpB, tmRNA, SRP RNA) were found by using covariance models with the Infernal search tool (63). The assignment of product descriptions was made by using search results of the following curated databases in this order: TIGRFam; PRIAM (e−30 cutoff); Pfam; Smart; COGs; Swissprot/TrEMBL (SPTR); and KEGG. If there was no significant similarity to any protein in another organism, it was described as “hypothetical protein.” “Conserved hypothetical protein” was used if at least one match was found to a hypothetical protein in another organism. EC numbering was based on searches in PRIAM at an e−10 cutoff; COG and KEGG functional classifications were based on homology searches in the respective databases.
The automatic annotation was manually curated. The start codons were refined, and pseudogenes were identified. The product description was then curated for all genes. Those genes that had a >70% identity match in B. pseudomallei or B. mallei or a >70% identity match to a protein with an experimentally verified function were designated with the same product description unless other evidence overruled this annotation. All other product descriptions, except (conserved) hypothetical proteins, were preceded by “putative.” Next, previously studied functions in LB400 were manually curated (taurine metabolism; C1 metabolism; diterpenoid metabolism; and biphenyl and benzoate metabolism). Additionally, a detailed annotation of all aromatic catabolic pathways in LB400 was performed.
Functional redundancy in the LB400 genome was evaluated in the following way: every protein-coding gene in the LB400 genome was searched against the protein database of 260 fully sequenced genomes, including the LB400 genome itself, by using the BLASTp algorithm and a minimum cut-off for a match of at least 30% amino acid identity and an alignable region at least 70% of the length of the query sequence. The cut-off is above the twilight zone of similarity searches where inference of homology is error-prone because of low similarity between aligned sequences (64, 65). Therefore, query proteins were presumably homologous to their match and share at least the same general biochemical function. The genes that had the best match within the LB400 genome (with the exclusion of the self-match) presumably represent duplicated genes (paralogs) and/or genes of similar function that have been laterally acquired (these two processes cannot easily be differentiated) after the emergence of the LB400 lineage and, thus, represent a measure of the functional redundancy within the genome.
We also compared the gene complement of strain LB400 to that of the published B. pseudomallei and the environmental Bcc strain 383, which has been fully sequenced by our group, using a reciprocal best BLASTP approach and the same cut-off for a match as above.
Comparative DNA Microarray Hybridizations.
LB400, LMG 16224, and LMG 21720 DNA were fragmented to 1–5 kb by sonication. Aminoallyl labeling was performed by using the BioPrime labeling kit, based on the manufacturer's protocol (Invitrogen, Carlsbad, CA), except for the dNTP mixture (final concentrations of 0.5 mM dATP, dGTP, and dCTP; 0.1 mM dTTP; and 0.4 mM aa-dUTP). Sheared DNA (250 ng) was mixed with random hexamers, denatured, snap-cooled, mixed with the dNTP mix and Klenow polymerase, and incubated for 2 h at 37°C. Reaction cleanup, coupling of monoreactive fluorescent Cy dye, and hybridization to the LB400 microarray were performed as described (32, 44). Data normalization was performed in Genespring (Agilent Technologies, Palo Alto, CA) by using “division by control channel,” and data of two biological replicates with dye-swapping were averaged. If for a particular gene the signal produced by the labeled test strain DNA was less than half the signal produced by the LB400 control DNA (threshold) and displayed less than a difference of 2 between the ratios of the biological replicates (data quality filter), that gene was considered absent in the test genome. The current LB400 microarray lacks probes for 556 genes (6.2%). Data for 8,049 genes passed the used data quality filter for both strains.
Supplementary Material
Acknowledgments
P.S.G.C. thanks W. Marrs for assistance during sequencing. V.J.D. thanks B. Chai (Michigan State University) for bioinformatics support and A. M. Cook (University of Konstanz, Konstanz, Germany) and the Integrated Microbial Genomes group (JGI) for assistance during annotation. M.S. thanks R. de la Iglesia and B. González (Pontificia Universidad Católica de Chile, Santiago, Chile) for technical advice. We thank the reviewers for their useful comments. This work was supported by the Genomics:GTL program of the U.S. Department of Energy (DOE) and by Superfund Basic Research Program Grant P42 ES 04911-12 from the U.S. National Institute of Environmental Health Sciences. Work at Lawrence Livermore National Laboratory was performed under the auspices of the U.S. DOE by the University of California, under Contract No. W-7405-Eng-48. This work was supported by project grants Fondecyt and USM 130522 (to M.S.). E.M. acknowledges funding for bioinformatics support from the Natural Environment Research Council Environmental Genomics Program (Grant NER/T/S/2001/00299).
Abbreviations
- CGH
comparative genome hybridization
- LB400
Burkholderia xenovorans LB400
- Bcc
Burkholderia cepacia complex
- PCB
polychlorinated biphenyl
- COG
clusters of orthologous groups of proteins.
Footnotes
References
- 1.Goris J, De Vos P, Caballero-Mellado J, Park J, Falsen E, Quensen JF III, Tiedje JM, Vandamme P. Int J Syst Evol Microbiol. 2004;54:1677–1681. doi: 10.1099/ijs.0.63101-0. [DOI] [PubMed] [Google Scholar]
- 2.Seeger M, Zielinski M, Timmis KN, Hofer B. Appl Environ Microbiol. 1999;65:3614–3621. doi: 10.1128/aem.65.8.3614-3621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeger M, Timmis KN, Hofer B. Appl Environ Microbiol. 1995;61:2654–2658. doi: 10.1128/aem.61.7.2654-2658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maltseva OV, Tsoi TV, Quensen JF III, Fukuda M, Tiedje JM. Biodegradation. 1999;10:363–371. doi: 10.1023/a:1008319306757. [DOI] [PubMed] [Google Scholar]
- 5.Bedard DL, Unterman R, Bopp LH, Brennan MJ, Haberl ML, Johnson C. Appl Environ Microbiol. 1986;4:761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bopp LH. J Ind Microbiol. 1986;1:23–29. [Google Scholar]
- 7.Viallard V, Poirier I, Cournoyer B, Haurat J, Wiebkin S, Ophel-Keller K, Balandreau J. Int J Syst Bacteriol. 1998;48:549–563. doi: 10.1099/00207713-48-2-549. [DOI] [PubMed] [Google Scholar]
- 8.Estrada-De Los Santos P, Bustillos-Cristales R, Caballero-Mellado J. Appl Environ Microbiol. 2001;67:2790–2798. doi: 10.1128/AEM.67.6.2790-2798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salles JF, Samyn E, Vandamme P, van Veen JA, van Elsas JD. Soil Biol Biochem. 2005 in press. [Google Scholar]
- 10.Coenye T, Vandamme P. Environ Microbiol. 2003;5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 11.Partida-Martinez LP, Hertweck C. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 12.Mahenthiralingam E, Urban TA, Goldberg JB. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 13.Moulin L, Munive A, Dreyfus B, Boivin-Masson C. Nature. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- 14.Chen W-M, Moulin L, Bontemps C, Vandamme P, Bena G, Boivin-Masson C. J Bacteriol. 2003;185:7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landers P, Kerr KG, Rowbotham TJ, Tipper JL, Keig PM, Ingham E, Denton M. Eur J Clin Microbiol Infect Dis. 2000;19:121–123. doi: 10.1007/s100960050442. [DOI] [PubMed] [Google Scholar]
- 16.Lopez J, Copps J, Wilhelmsen C, Moore R, Kubay J, St-Jacques M, Halayko S, Kranendonk C, Toback S, DeShazer D. Microb Inf. 2003;5:1125–1131. doi: 10.1016/j.micinf.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Dance DA. Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramette A, LiPuma JJ, Tiedje JM. Appl Environ Microbiol. 2005;71:1193–1201. doi: 10.1128/AEM.71.3.1193-1201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessie TG, Hendrickson W, Manning BD, Devereux R. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 20.Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, et al. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, et al. Proc Natl Acad Sci USA. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King GM. Appl Environ Microbiol. 2003;69:7257–7265. doi: 10.1128/AEM.69.12.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marx CJ, Miller JA, Chistoserdova L, Lidstrom ME. J Bacteriol. 2004;186:2173–2178. doi: 10.1128/JB.186.7.2173-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeger M, Gonzalez M, Camara B, Munoz L, Ponce E, Mejias L, Mascayano C, Vasquez Y, Sepulveda-Boza S. Appl Environ Microbiol. 2003;69:5045–5050. doi: 10.1128/AEM.69.9.5045-5050.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DJ, Martin VJ, Mohn WW. J Bacteriol. 2004;186:3631–3639. doi: 10.1128/JB.186.11.3631-3639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruff J, Denger K, Cook AM. Biochem J. 2003;369:275–285. doi: 10.1042/BJ20021455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, et al. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 28.Konstantinidis KT, Tiedje JM. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulrich LE, Koonin EV, Zhulin IB. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor BL, Zhulin IB. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, et al. Proc Natl Acad Sci USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denef VJ, Klappenbach JA, Patrauchan MA, Florizone C, Rodrigues JLM, Tsoi TV, Verstraete W, Eltis LD, Tiedje JM. Appl Environ Microbiol. 2006;72:585–595. doi: 10.1128/AEM.72.1.585-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denef VJ, Patrauchan MA, Florizone C, Park J, Tsoi TV, Verstraete W, Tiedje JM, Eltis LD. J Bacteriol. 2005;187:7996–8005. doi: 10.1128/JB.187.23.7996-8005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 35.Williams PA, Jones RM, Shaw LE. J Bacteriol. 2002;184:6572–6580. doi: 10.1128/JB.184.23.6572-6580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, et al. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 37.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, et al. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 38.Weightman AJ, Topping AW, Hill KE, Lee LL, Sakai K, Slater JH, Thomas AW. J Bacteriol. 2002;184:6581–6591. doi: 10.1128/JB.184.23.6581-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez JI, Minambres B, Garcia JL, Diaz E. Environ Microbiol. 2002;4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- 40.Rabus R, Kube M, Heider J, Beck A, Heitmann K, Widdel F, Reinhardt R. Arch Microbiol. 2005;183:27–36. doi: 10.1007/s00203-004-0742-9. [DOI] [PubMed] [Google Scholar]
- 41.Jimenez JI, Miñambres B, García J, Díaz E. Pseudomonas. Vol 3. New York: Kluwer; 2004. pp. 425–462. [Google Scholar]
- 42.Kasberg T, Seibert V, Schlomann M, Reineke W. J Bacteriol. 1997;179:3801–3803. doi: 10.1128/jb.179.11.3801-3803.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gescher J, Zaar A, Mohamed M, Schägger H, Fuchs G. J Bacteriol. 2002;184:6301–6315. doi: 10.1128/JB.184.22.6301-6315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denef VJ, Park J, Tsoi TV, Rouillard JM, Zhang H, Wibbenmeyer JA, Verstraete W, Gulari E, Hashsham SA, Tiedje JM. Appl Environ Microbiol. 2004;70:4961–4970. doi: 10.1128/AEM.70.8.4961-4970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luengo JM, Garcia JL, Olivera ER. Mol Microbiol. 2001;39:1434–1442. doi: 10.1046/j.1365-2958.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhulin IB. Adv Microb Physiol. 2001;45:157–198. doi: 10.1016/s0065-2911(01)45004-1. [DOI] [PubMed] [Google Scholar]
- 47.Repik A, Rebbapragada A, Johnson MS, Haznedar JO, Zhulin IB, Taylor BL. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor BL, Zhulin IB, Johnson MS. Annu Rev Microbiol. 1999;53:103–128. doi: 10.1146/annurev.micro.53.1.103. [DOI] [PubMed] [Google Scholar]
- 49.Urban TA, Griffith A, Torok AM, Smolkin ME, Burns JL, Goldberg JB. Infect Immun. 2004;72:5126–5134. doi: 10.1128/IAI.72.9.5126-5134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokol PA, Darling P, Woods DE, Mahenthiralingam E, Kooi C. Infect Immun. 1999;67:4443–4455. doi: 10.1128/iai.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbett CR, Burtnick MN, Kooi C, Woods DE, Sokol PA. Microbiology. 2003;149:2263–2271. doi: 10.1099/mic.0.26243-0. [DOI] [PubMed] [Google Scholar]
- 52.Warawa J, Woods DE. FEMS Microbiol Lett. 2005;242:101–108. doi: 10.1016/j.femsle.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 53.Tomich M, Griffith A, Herfst CA, Burns JL, Mohr CD. Infect Immun. 2003;71:1405–1415. doi: 10.1128/IAI.71.3.1405-1415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engledow AS, Medrano EG, Mahenthiralingam E, LiPuma JJ, Gonzalez CF. J Bacteriol. 2004;186:6015–6024. doi: 10.1128/JB.186.18.6015-6024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MC, Govan JR, Speert DP, Lipuma JJ, Vandamme P, Dowson CG. J Clin Microbiol. 2005;43:4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J, Kim JG, Kang Y, Jang JY, Jog GJ, Lim JY, Kim S, Suga H, Nagamatsu T, Hwang I. Mol Microbiol. 2004;54:921–934. doi: 10.1111/j.1365-2958.2004.04338.x. [DOI] [PubMed] [Google Scholar]
- 57.Malott RJ, Baldwin A, Mahenthiralingam E, Sokol PA. Infect Immun. 2005;73:4982–4992. doi: 10.1128/IAI.73.8.4982-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunt TA, Kooi C, Sokol PA, Valvano MA. Infect Immun. 2004;72:4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, et al. J Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Badger J, Olsen G. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 61.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowe T, Eddy S. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eddy S. BMC Bioinformatics. 2002;3:18. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rost B. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 65.Rasko DA, Myers GS, Ravel J. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghai R, Hain T, Chakraborty T. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.