Abstract
Mismatch repair (MMR) is critical to maintaining the integrity of the genome, and deficiencies in MMR are correlated with cancerous transformations. Bulky rhodium intercalators target DNA base mismatches with high specificity. Here we describe the application of bulky rhodium intercalators to inhibit cellular proliferation differentially in MMR-deficient cells compared with cells that are MMR-proficient. Preferential inhibition by the rhodium complexes associated with MMR deficiency is seen both in a human colon cancer cell line and in normal mouse fibroblast cells; the inhibition of cellular proliferation depends strictly on the MMR deficiency of the cell. Furthermore, our assay of cellular proliferation is found to correlate with DNA mismatch targeting by the bulky metallointercalators. It is the Δ-isomer that is active both in targeting base mismatches and in inhibiting DNA synthesis. Additionally, the rhodium intercalators promote strand cleavage at the mismatch site with photoactivation, and we observe that the cellular response is enhanced with photoactivation. Targeting DNA mismatches may therefore provide a cell-selective strategy for chemotherapeutic design.
Keywords: DNA base mismatch, metallointercalator
The DNA mismatch repair (MMR) machinery is responsible for the recognition and repair of mispaired or damaged DNA bases (1, 2). With a deficiency in MMR, cellular mutation rates are enhanced (3, 4), and individuals who have inherited a defective copy of MMR genes are at risk of developing cancers, in particular, hereditary nonpolyposis colon cancer (5, 6). Also noteworthy, ≈18% of solid tumors and 10% of leukemias tested are deficient in MMR (7–9). Although MMR deficiencies are not a constant in cancer, they can greatly accelerate the rate at which mismatches and, thus, mutations accumulate.
Deficiencies in MMR have been particularly difficult to target therapeutically, because, in addition to being a predisposing factor for the formation of tumors, MMR deficiency is associated with a tolerance to many common chemotherapeutics, including alkylating agents, platinum compounds, topoisomerase inhibitors, and metabolic analogues (10). MMR-deficient cell lines have, however, recently been shown to be hypersensitive to a small family of drugs that target DNA (11–13). It has also been suggested that some recurrent tumors that are drug-resistant obtain this resistance through MMR deficiency (14).
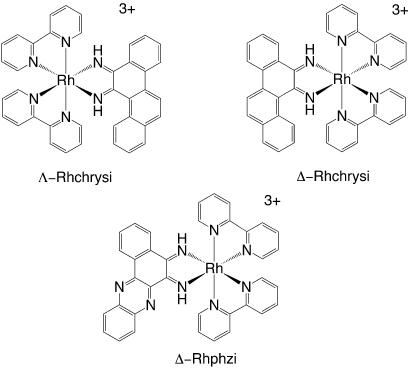
We have developed bulky rhodium intercalators (Fig. 1) that bind single base mismatches in DNA selectively and irrespective of sequence context (15–18). These compounds bind the mismatch by intercalation and, upon photoexcitation, promote strand breaks at the mismatched site. [Bis(2,2′-bipyridine)(chrysene-5,6-quinone-diimine)Rh(III)] (Rhchrysi) binds mismatches with moderate affinity and high specificity. The site-specificity depends on the thermodynamic instability associated with a base pair mismatch; the bulky intercalator cannot stack easily within well matched DNA but can more easily insert neighboring a destabilized mismatched site. The high specificity of Rhchrysi is well demonstrated in its ability to cleave a single mismatched site in a 2,700-bp duplex (16). This specificity has been used advantageously in a site-specific assay for single-nucleotide polymorphisms in pooled genomic DNA (19). Our second generation complex [bis(2,2′-bipyridine)(benzo[a]phenazine-5,6-quinonediimine)Rh(III)] (Rhphzi) utilizes heterocyclic nitrogen atoms within the bulky intercalating ligand to boost the affinity of the complex for mismatched sites (18). Rhphzi is capable of binding mismatched sites with 100 nM affinity and with specificity similar to Rhchrysi.
Fig. 1.
Structures of Λ-Rhchrysi (Upper Left), Δ-Rhchrysi (Upper Right), and Δ-Rhphzi (Lower).
Importantly, both of these rhodium complexes recently have been applied in probing whether MMR-deficient cells accumulate mismatches (18). In photocleavage assays of genomic DNA from a series of cell lines, some proficient in MMR and others wholly or partially deficient in repair, a clear correlation between photocleavage by Rhchrysi and Rhphzi and MMR deficiency was evident.
Here we explore the efficacy of these rhodium complexes in inhibiting the growth of MMR-deficient versus MMR-proficient cell lines in the presence and absence of photoactivation. The biological effects are assessed in two cell lines, both of which are derivatives of HCT116. These cell lines have had an extra copy of either chromosome 2, HCT116O, or chromosome 3, HCT116N, inserted in the cell; placing a copy of chromosome 3 within this cell line corrects the MMR deficiency by introducing a functional copy of the MLH1 gene with a normal promoter (20). Additionally, mouse fibroblast cells derived from litter mates that are Msh2− or Msh2+ were studied. These cells and cell lines provide a convenient method for testing whether a particular compound will differentially target MMR-deficient cells, because these cells are essentially identical genetically, except with regard to MMR.
Results
Preferential Inhibition of Cellular Proliferation with MMR Deficiency by the Rhodium Complexes.
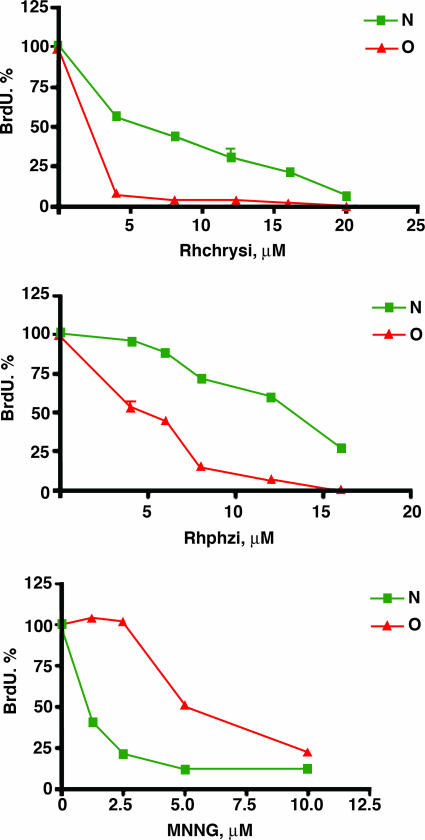
The HCT116-derived cells were treated with either Rhchrysi or Rhphzi for 96 h at various concentrations in the absence of irradiation. As is evident for both complexes at micromolar concentrations (Fig. 2), we see a significant inhibition of cell proliferation. Importantly, for both complexes we also see a preferential inhibition in the MMR-deficient strain. This selectivity seen with the rhodium complexes for the MMR-deficient HCT116O contrasts the action, also shown, of N-methyl, N′-nitro-nitrosoguanidine (MNNG), a common DNA-methylating agent that is found to be more toxic to the MMR-proficient cell line, HCT116N (20). MNNG is typical of most chemotherapeutics that target genomic DNA.
Fig. 2.
Differential antiproliferative effect of rac-Rhchrysi, rac-Rhphzi, and MNNG on MMR-deficient (red) and MMR-proficient (green) cells. MMR-deficient, O, and MMR-proficient, N, cell lines derived from the HCT116 cell line were treated with the methylating agent MNNG, Rhchrysi, or Rhphzi as described in Materials and Methods. Complexes and MNNG were incubated with cells for 96 h with the addition of BrdU after 72 h. Note that the MMR-deficient population HCT116O is resistant to the action of MNNG but is sensitive preferentially to Rhchrysi and Rhphzi.
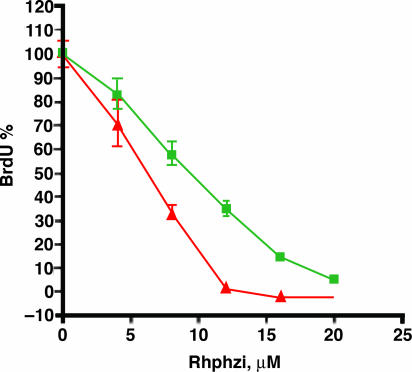
Experiments using normal mouse fibroblasts that are deficient for MMR, Msh2−, or proficient for MMR, Msh2+, were also performed. These experiments complement those conducted with HCT116, because these cells also are genetically identical except for the Msh2 gene. Experiments probing these fibroblast cells, which are not cancerous, therefore provide a rigorous test that targets MMR deficiency. The data given in Fig. 3 show that the MMR-defective mouse embryo fibroblasts that are Msh2− yield decreased DNA synthesis and are therefore more sensitive to Rhphzi compared with the control littermate cells that are Msh2+. These results parallel those obtained with the HCT116 model system. These results furthermore provide strong support for the idea that the inhibition of cell proliferation is related to the MMR deficiency of the cell, regardless of which MMR gene is absent.
Fig. 3.
Antiproliferative effect of rac-Rhphzi on MMR-deficient, Msh2− (red), and MMR-proficient, Msh2+ (green), mouse embryonic fibroblasts. Inhibition of DNA synthesis in the MMR-deficient mouse cells parallels the effects seen with the HCT116 cell lines.
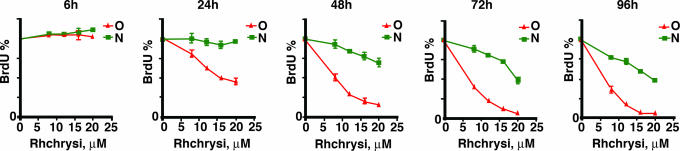
The effect of incubation time was also investigated, and the results are shown in Fig. 4. HCT116 cells were exposed to various concentrations of Rhchrysi for various amounts of time, from 6 to 96 h, before testing the effects on DNA synthesis. In this series, it is clear that the maximum effect is obtained after 48 h of incubation. Based on cellular uptake studies with closely related ruthenium complexes that are fluorescent (J.K.B. and C. Puckett, unpublished results), we expect cellular uptake to be rate-limiting; indeed, with more hydrophobic chrysi analogues, we find differential biological effects with much shorter incubation times.
Fig. 4.
Effect of varying drug incubation time on cell proliferation. HCT116 cells that are deficient (red) and proficient (green) in MMR were exposed to various concentrations of rac-Rhchrysi for different incubation times as shown. Antiproliferative effects increase with longer incubation times up to 48 h.
Significantly, the results seen here with the bulky rhodium complexes are distinctive, in contrast to common therapeutics and MNNG. MMR-deficient cell lines are generally resistant to the majority of alkylating drugs, platinum compounds, and metabolic analogues, because the antiproliferative effects these therapeutic agents have on cancerous cells are due in part to recognition of the drug-induced genomic DNA damage by the MMR system (10, 21). In contrast, we find with the rhodium complexes the preferential inhibition of cellular proliferation in cells that depends strictly on their deficiency in MMR.
Correlations Between Biological Effects and DNA Mismatch Targeting.
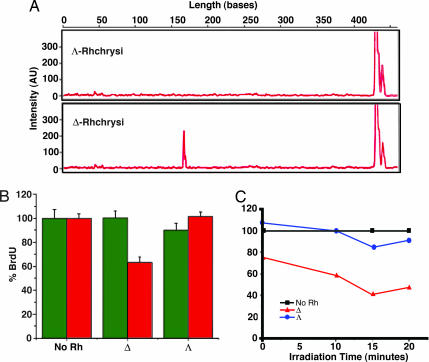
In an effort to correlate the biological effects we observed by using these bulky intercalators with their DNA-binding characteristics, experiments also were conducted with enantiomers of Rhchrysi. Only the Δ-isomer of Rhchrysi binds and cleaves, with photoactivation, a neighboring mismatched DNA site (15). Shown in Fig. 5 is an illustration of the enantiospecific reaction of Δ-Rhchrysi at a single CC mismatch on a 436-mer DNA duplex PCR product containing a single CC mismatch. As illustrated in the capillary gel electrophoresis results, upon photolysis of the complex bound to DNA, the bulky Δ-isomer cleaves the DNA neighboring the destabilized mismatch. No cleavage is seen without metal complex (light control) or after photolysis using the Λ-isomer. In general, by surveying different mismatched sites in a variety of sequence contexts, we have found the recognition to be enantioselective; it is Δ-Rhchrysi that binds and cleaves mismatched DNA preferentially (17).
Fig. 5.
DNA cleavage and biological activities with stereoisomers and light. (A) Effect of stereoisomers on photocleavage. Shown are capillary electropherograms after photolysis of 436-mer PCR products containing a single CC mismatch with 500 nM Λ-Rhchrysi (Upper) or Δ-Rhchrysi (Lower), as described earlier (12). Mismatch-specific photocleavage to produce a 170-bp fragment is evident only with the Δ-isomer. (B) Antiproliferative effects of different stereoisomers of Rhchrysi. Shown are the effects on BrdU incorporation in HCT116O (red) and HCT116N (green) cells after 48 h of incubation without rhodium or with 5 μM Λ- or Δ-Rhchrysi. The selective inhibition of the MMR-deficient HCT116O cell line is seen only with the Δ-isomer. (C) Antiproliferative effect of different stereoisomers of Rhchrysi with irradiation. Shown are the effects of various amounts of irradiation of HCT116O cells in the absence of Rhchrysi (black), in the presence of 5 μM Δ-Rhchrysi (red), or in the presence of 5 μM Λ-Rhchrysi (blue) after 48 h of incubation.
We therefore also compared the antiproliferative effects of the separated isomers (Fig. 5). Consistent with DNA targeting studies, it is Δ-Rhchrysi that shows selective inhibition of cellular proliferation in the MMR-deficient HCT116O cell line. At 5 μM rhodium, incubation with Λ-Rhchrysi shows no effect on DNA synthesis in either HCT116O or HCT116N cells, whereas the Δ-isomer selectively inhibits DNA synthesis in the MMR-deficient HCT116O cells. Although these rhodium complexes are coordinatively saturated and generally inert to substitution, we had considered the possibility that the biological activity of the complexes might be associated with complex decomposition and ligand release. The finding of high enantioselectivity associated with the biological effect argues against complex decomposition as being responsible for the inhibition of cellular proliferation, given that a similar decomposition would be likely for both isomers.
Because the complexes bind mismatched DNA only noncovalently in the absence of light but, with photolysis, promote DNA strand breaks, we also explored cell proliferation assays after exposing the cells to compound and light, conditions under which the rhodium complex might be expected to be more potent. We therefore irradiated cells for 10, 15, or 20 min after a 48-h incubation with rhodium. These results also are shown in Fig. 5. Here it should be noted that longer periods of irradiation even without metal complex lead to some inhibitory effects in both MMR deficient and proficient cell lines; evaporation of the medium is also somewhat of a problem. Nonetheless, light activation over short periods does indeed lead to greater inhibition of cell proliferation and only for Δ-Rhchrysi in the MMR deficient HCT116O cells. Again, parallel experiments with Λ-Rhchrysi provide a useful control. Under the conditions used, light activation only appears to enhance the inhibition of DNA synthesis by Δ-Rhchrysi and only in the MMR deficient HCT116O cells. These data are consistent with DNA serving as target for the rhodium complex inside the cell. Certainly these results demonstrate a clear correlation between mismatch targeting by rhodium photocleavage and biological potency in a cell-selective manner.
Discussion
The results reported here establish that the bulky rhodium intercalators we have designed serve as inhibitors of cellular proliferation and, significantly, that the complexes do so with cell selectivity. Biological effects are seen preferentially in MMR-deficient cells. These results do not establish DNA mismatch targeting as the basis for the cell selectivity. However, these data are consistent with mismatched DNA being the target of these complexes and, hence, the basis of the preferential reaction in MMR-deficient cell lines, in which mismatches are more abundant.
It is noteworthy that in the absence of light the complexes bind DNA mismatches noncovalently, albeit with high affinity and specificity. Because a covalent adduct is not formed and, in the absence of light, no DNA damage is directly generated, any biological effect would be expected to be more modest; irradiation, which does cause DNA strand breaks, should enhance the effect, as we observe. Likely, then, without light the metal-mismatch complex provides a secondary signal for protein binding or activation. It should also be noted that light activation of the metal complex generates only a short-lived ligand radical; thus, intimate association of the rhodium complex with its target is needed for a light-dependent reaction (22).
Significantly, the biological effects we observed in MMR-deficient cells also are enantiospecific, just as binding of the bulky rhodium intercalators at mismatched DNA sites are enantiospecific. It is the Δ-isomer of Rhchrysi that binds and selectively cleaves a neighboring base pair mismatch, and it is the Δ-isomer that shows the biological effect preferentially in MMR-deficient cells. There is therefore a clear correlation if not an established causality between DNA mismatch targeting by the rhodium complexes and the observed inhibition of cellular proliferation in MMR-deficient cells.
These results highlight a class of compounds for possible application in cancer therapeutics. The work furthermore underscores a cell-selective strategy in chemotherapeutic design by chemically and site-specifically targeting DNA mismatches.
Materials and Methods
Materials.
Rac-Rhchrysi and rac-Rhphzi were synthesized by using established procedures (15, 18). Rac-Rhchrysi was resolved into Δ- and Λ-isomers by using potassium antimonyl tartrate as previously described (23). Salts were exchanged to chloride before use with Sephadex QAE ion exchange resin. Media and supplements were purchased from Invitrogen (Carlsbad, CA). BrdU, antibodies, buffers, and peroxidase substrate were purchased in kit format from Roche Molecular Biochemicals (Mannheim, Germany). Irradiations were performed with an Oriel (Darmstadt, Germany) 1,000-W Hg/Xe solar simulator using a UVB/UVC-blocking filter.
Cell Lines, Media, and Culture.
The HCT116N and HCT116O cell lines were obtained from Bert Vogelstein (The Johns Hopkins University, Baltimore, MD) and were originally derived from the laboratory of C. R. Boland (University of California at San Diego, La Jolla, CA) (20). Cells were grown in RPMI medium1640 supplemented with 10% FBS/2 mM l-glutamine/0.1 mM nonessential amino acids/1 mM sodium pyruvate/100 units/ml penicillin/100 μg/ml streptomycin/400 μg/ml geneticin (G418). Cells were grown in tissue culture flasks and dishes (Corning Costar, Acton, MA) at 37°C under 5% CO2 atmosphere. Mouse fibroblasts were obtained as previously reported (24) and established for heterozygous and homozygous littermates and used at passages 5 and 6. Cells were grown in DMEM with media as for HCT116 cells.
Cell Proliferation Assays.
Cell proliferation was measured using the BrdU incorporation assay (25). Cells were plated in 96-well plates at 2,000 cells per well and grown for 24 h. At this point, variable concentrations of rhodium complexes were added. The cell cultures were then allowed to grow for 6–96 h as indicated. Cells were then either irradiated or not followed by the addition of BrdU. Irradiations of cells were performed by using a solar simulator adapted for irradiation from the bottom of the well plates. The cells were grown for an additional 24 h. The BrdU incorporation was quantified by antibody assay with established procedures (24, 25). For the experiments with mouse fibroblasts, cells were plated and after 24 h Rhphzi was added; 12 h later BrdU was added, and the assays were carried out as established.
Photocleavage of Mismatched DNA with Rhodium Isomers.
Photocleavage experiments were conducted on the 436-mer PCR product as previously described in detail (19) but using 500 nM of the purified isomers, Δ- or Λ-Rhchrysi.
Acknowledgments
We thank Drs. R. Fishel and M. McIlhatton (Thomas Jefferson University, Philadelphia, PA) for providing the mouse embryo fibroblast cells from the relevant null, heterozygote, and wild-type Msh2 animals. This work was supported by National Institutes of Health Grant GM33309 (to J.K.B.).
Abbreviations
- MMR
mismatch repair
- Rhchrysi
[bis(2,2′-bipyridine)(chrysene-5,6-quinone-diimine)Rh(III)]
- Rhphzi
[bis(2,2′-bipyridine)(benzo[a]phenazine-5,6-quinonediimine)Rh(III)]
- MNNG
N-methyl, N′-nitro-nitrosoguanidine.
Footnotes
The authors declare no conflict of interest.
References
- 1.Kunkel TA, Erie DA. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner RD, Marsischky GT. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauss BS. Mutat Res. 1999;437:195–203. doi: 10.1016/s1383-5742(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos N, Lindblom A. Hum Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Peltomaki P. Hum Mol Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 7.Arzimanoglou II, Gilbert F, Barber HRK. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 8.Loeb LA, Loeb KR, Anderson JP. Proc Natl Acad Sci USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb LA, Springgate CF, Battula N. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 10.Fink D, Aebi S, Howell SB. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 11.Jacob S, Aguado M, Fallik D, Praz F. Cancer Res. 2001;61:6555–6562. [PubMed] [Google Scholar]
- 12.Li HR, Shagisultanova EI, Yamashita K, Piao Z, Perucho M, Malkhosyan SR. Cancer Res. 2004;64:4760–4767. doi: 10.1158/0008-5472.CAN-04-0975. [DOI] [PubMed] [Google Scholar]
- 13.Liu LL, Nakatsuru Y, Gerson SL. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 14.Karran P, Offman J, Bignami M. Biochimie. 2003;85:1149–1160. doi: 10.1016/j.biochi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Jackson BA, Barton JK. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 16.Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 17.Jackson BA, Barton JK. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 18.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. Proc Natl Acad Sci USA. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart JR, Johnson MD, Barton JK. Proc Natl Acad Sci USA. 2004;126:14040–14044. doi: 10.1073/pnas.0406169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 21.O'Brien V, Brown R. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 22.Sitlani A, Pyle AM, Barton JK. J Am Chem Soc. 1992;114:2303–2312. [Google Scholar]
- 23.Murner H, Jackson BA, Barton JK. Inorg Chem. 1998;37:3007–3012. [Google Scholar]
- 24.Reitmar , Armin H, Risley R, Bristow RG, Wilson T, Ganesh A, Jang A, Peacock J, Benchimol S, Hill RP, et al. Cancer Res. 1997;57:3765–3771. [PubMed] [Google Scholar]
- 25.Gratzner HG. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]