Abstract
Activating signal cointegrator-2 (ASC-2), a coactivator of multiple transcription factors that include retinoic acid receptor (RAR), associates with histone H3-K4 methyltranferases (H3K4MTs) MLL3 and MLL4 in mixed-lineage leukemia. Here, we show that mice expressing a SET domain mutant of MLL3 share phenotypes with isogenic ASC2+/− mice and that expression and H3-K4 trimethylation of RAR target gene RAR-β2 are impaired in ASC-2-null mouse embryo fibroblasts (MEFs) or in MEFs expressing siRNAs against both MLL3 and MLL4. We also show that MLL3 and MLL4 are found in distinct ASC-2-containing complexes rather than in a common ASC-2 complex, and they are recruited to RAR-β2 by ASC-2. In contrast, RAR-β2 expression is intact in MEFs devoid of menin, a component of MLL1 and MLL2 H3K4MT complexes. These results suggest that ASC-2 confers target gene specificity to MLL3 and MLL4 H3K4MT complexes and that recruitment of H3K4MTs to their target genes generally involves interactions between integral components of H3K4MT complexes and transcription factors.
Keywords: mixed-lineage leukemia (MLL), retinoic acid receptor, transcription
Nuclear receptors (NRs) bind hormone response elements in target genes and regulate transcriptional initiation in a ligand-dependent manner (1). During ligand binding, the conserved C-terminal activation function 2 domain undergoes a structural change (1) that is recognized by an α-helical LXXLL motif (NR box) in transcriptional coactivators (2). Activating signal cointegrator-2 (ASC-2; also named AIB3, TRBP, TRAP250, NRC, and PRIP), a coactivator of many NRs and other transcription factors, contains two NR boxes (3). NR box 1 binds multiple NRs, including retinoic acid receptor (RAR), whereas NR box 2 interacts with liver X receptors. The physiological importance of ASC-2 as a key coactivator of these NRs and the pivotal roles of both NR boxes in this context have been proposed from recent studies with various ASC-2 mouse models (3).
In HeLa nuclei, ASC-2 resides in a steady-state complex [ASC-2 complex (ASCOM)] (4) that contains retinoblastoma-binding protein RbBP5, α/β-tubulins, and trithorax group proteins Ash2L, MLL4-1/ALR-1, MLL4-2/ALR-2, and MLL3/HALR (the paralog of MLL4/ARL).†† MLL4-1 and MLL4-2 (collectively MLL4s) are encoded by the same gene, and they differ only at their N termini (4). The C termini of MLL3 and MLL4s contain a SET domain (5) with an intrinsic histone lysine-specific methyltransferase activity. Indeed, recombinant MLL3 and MLL4 SET domains and partially immunopurified ASCOM exhibit weak but specific histone H3-K4 methyltransferase (H3K4MT) activity in vitro (4).
H3-K4 methylation, an evolutionarily conserved mark linked to transcriptionally active chromatin, has been proposed to counter the generally repressive chromatin environment imposed by H3-K9/K27 methylation in higher eukaryotes (6). In particular, H3-K4 trimethylation is associated with promoters and early transcribed regions of active genes (7, 8). H3K4MTs include yeast Set1 (ySet1), hSet1, MLL1, MLL2, and MLL3 and MLL4s, as well as Ash1 and Set7/9 (4, 6, 9–12). Mixed-lineage leukemias (MLLs), hSet1, and ySet1 form similar complexes, called Set1-like complexes (6). Interestingly, H3-K4 methylation has been linked to other chromatin-modifiers such as histone acetyltransferases and chromatin remodelers (13–17). In yeast, H3-K4 methylation by ySet1 is downstream from histone H2B ubiquitination, and it requires Paf1 and other transcription elongation factors (6, 18).
Transactivation of RAR-β2, a well characterized RAR target gene, involves ligand-dependent recruitment of ASCOM to the RAR-β2 promoter in vivo, likely through the NR box 1 of ASC-2 (4). Thus, an ASC-2 fragment with an intact NR box 1 (DN1), but not one with a mutated LXXLL that disables NR interactions (DN1/m), blocks RAR transactivation in transient cotransfections, abolishes the ligand-dependent recruitment of ASC-2 and other ASCOM components (Ash2L and MLL4s) to the RAR-β2 promoter, and inhibits retinoid-induced H3-K4 trimethylation at RAR-β2 (4). These results suggest that direct ligand-dependent interactions between ASC-2 NR boxes and NRs (3) allow MLL3 and MLL4s in ASCOM to affect H3-K4 trimethylation and expression of NR target genes. However, this model has two prominent issues that remain to be addressed. First, the NR box 1-containing DN1 fragment could block not only ASC-2 but also the function of other essential NR box-containing coactivators (19, 20). Second, the weak in vitro H3K4MT activity of ASCOM (4) and the presence of multiple H3K4MTs in mammalian cells (4, 6, 9–12) question the direct role of MLL3 or MLL4s in retinoid-induced RAR-β2 H3-K4 trimethylation (4).
Our original purification of ASCOM revealed the presence of multiple H3K4MTs (4). Here, we show that ASCOM represents a pool of similar complexes and that each complex contains a single H3K4MT (i.e., MLL4-1, MLL4-2, or MLL3) and an evolutionarily conserved WDR5·RbBP5·Ash2L core complex (21). We also demonstrate that ASC-2 is a key adaptor for RAR-dependent recruitment of MLL3 and MLL4s and their H3K4MT activities to RAR-β2, whereas RAR-β2 expression is intact in mouse embryo fibroblasts (MEFs) devoid of menin, an integral component of MLL1 and MLL2 H3K4MT complexes. Thus, ASC-2 confers target gene specificity to ASCOM. Recruitment of other Set1-like H3K4MTs to their target genes may also involve direct interactions between bona fide components of Set1-like complexes and DNA-binding transcription factors.
Results
Similar Phenotypes of MLL3 and ASC-2 Mutant Mice.
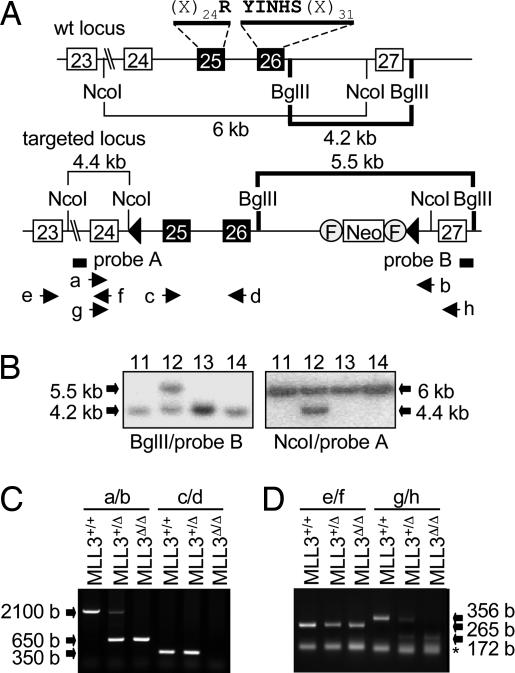
To elucidate the physiological role of ASCOM, we decided to establish mouse models for MLL3 and MLL4. MLL3 contains 4,025 aa, and, in an effort to preserve the overall structural integrity of its interactions with other components in ASCOM, we designed a targeting vector for an in-frame deletion of two exons that encode a 61-aa catalytic core region in the MLL3 SET domain (Fig. 1A). 129SVJ embryonic stem cell clones containing the correctly integrated construct were identified by Southern blot assays (Fig. 1B), and they were used to generate chimeric mice. Heterozygous MLL3+/Δ mice, created by crossing with protamine-Cre animals (22), were mated to generate MLL3Δ/Δ mice, as confirmed by genomic DNA (Fig. 1C) and mRNA (Fig. 1D) analyses.
Fig. 1.
Generation of MLL3Δ/Δ mice. (A) Wild-type (wt) and targeted MLL3 loci are shown schematically with loxP sites (filled triangles) and the neomycin resistance gene flanked by two Frt sites. Exons 25 and 26 encode 61 aa that include the RYINHS motif, which contributes to the catalytic core region of the SET domain. The genomic positions of the Southern blotting probes and PCR primers are indicated. (B) Southern blot analyses of recombined MLL3 locus. A representative clone (clone 12) generates targeted locus-specific bands of 5.5 and 4.4 kb from BglII and NcoI restriction digestions, respectively. (C) In genomic PCR, primers a/b generate a 650-bp band for the targeted MLL3 locus and a 2,100-bp band for the wild-type MLL3 locus. Primers c/d generate a 350-bp band only from the wild-type MLL3 locus. (D) In RT-PCR, primers e/f generate a 265-bp band for both wild-type and targeted MLL3 mRNAs, whereas primers g/h generate a 356-bp band for wild-type MLL3 mRNAs and a 172-bp band for targeted MLL3 mRNAs. The asterisk indicates primers.
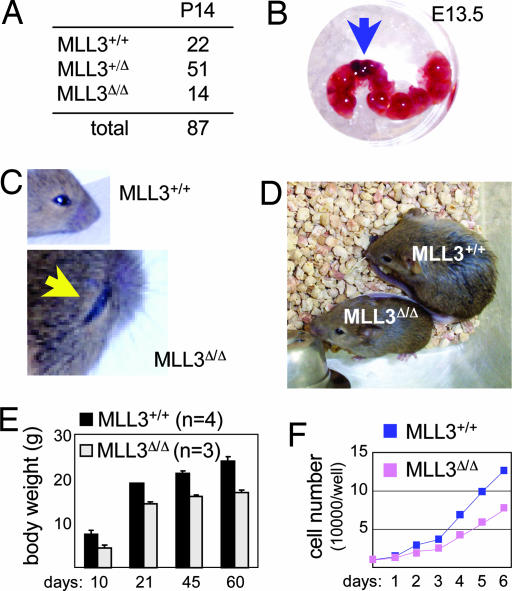
Genotyping of 87 P14 pups from intercrosses of MLL3+/Δ mice revealed that the number of MLL3Δ/Δ mice is significantly lower than the expected Mendelian ratio (Fig. 2A), suggesting a reduced survival of homozygous mice. Indeed, we observed occasional MLL3Δ/Δ embryos undergoing necrosis (Fig. 2B). Thus, a homozygous deletion of exons 25 and 26 appears to result in partial embryonic lethality.
Fig. 2.
Similarity of MLL3 and ASC-2 at the genetic level. (A) Genotypes of 87 P14. (B) Representative picture of a dying E13.5 MLL3Δ/Δ embryo (arrow). (C) Representative 2-week-old MLL3Δ/Δ mouse, retarded in eye opening (arrow), and its wild-type littermate. (D) Representative 4-week-old MLL3Δ/Δ mouse, with stunted growth, shown with its wild-type littermate. (E) Body weights of MLL3Δ/Δ (n = 3) mice and their wild-type littermates (n = 4) over a 60-day period. (F) Ten thousand wild-type and MLL3Δ/Δ MEFs were seeded in 9-cm2 wells, and the cells were harvested and counted each day for 6 days. A representative result is shown.
ASC-2−/− mice die at embryonic day (E) 9.5–13.5 (3). Although MLL3+/Δ mice exhibited no apparent phenotypic defects, MLL3Δ/Δ mice showed at least three phenotypes strikingly similar to those recently described for isogenic ASC-2+/− mice (23). First, MLL3Δ/Δ mice were retarded in eye opening (Fig. 2C) and stunted in their overall growth (Fig. 2D) relative to wild-type littermates. They weighed ≈30–40% less at birth, and although they gained on wild-type littermates with age, they remained ≈20% smaller through adulthood (Fig. 2E). Second, in a comparison of isolated E12.5 MEFs, MLL3Δ/Δ MEFs doubled at approximately half the rate of wild-type MEFs (Fig. 2F). Similarly, HEK293 cells expressing siRNA for ASC-2, MLL3, or MLL4s also showed decreased cell proliferation (data not shown). Finally, MLL3Δ/Δ females were infertile to hypofertile, whereas MLL3Δ/Δ males were slightly hypofertile (data not shown). These similarities with isogenic ASC-2+/− mice (23) provide genetic evidence for complex formation between MLL3 and ASC-2. The detailed analyses of fertility, fat, and renal phenotypes of MLL3Δ/Δ mice will be reported elsewhere.
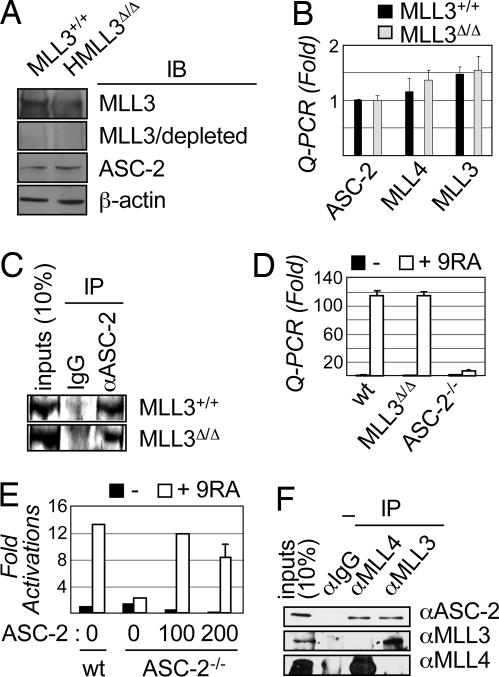
The phenotypes of MLL3Δ/Δ mice are the result of deletion of the catalytic region of the MLL3 SET domain because this mutation does not significantly affect the stability of mutant MLL3, the mRNA levels of mutant MLL3 and wild-type ASC-2 and MLL4s, or the incorporation of mutant MLL3 protein into ASCOM. Thus, immunoblots from three independent experiments revealed comparable amounts of MLL3 and ASC-2 proteins in wild-type and MLL3Δ/Δ MEFs (Fig. 3A and data not shown). Comparable amounts of ASC-2, MLL3, and MLL4 proteins were found at organism levels in wild-type and MLL3Δ/Δ mice (data not shown). Consistently, quantitative PCR (Q-PCR) analyses revealed no significant differences in the mRNA levels of ASC-2, MLL3, and MLL4s between wild-type and MLL3Δ/Δ MEFs (Fig. 3B). Mutant MLL3 protein is incorporated into the ASC-2 complex because ASC-2 antibody coimmunoprecipitated mutant MLL3 protein from MLL3Δ/Δ MEFs as readily as wild-type MLL3 from wild-type MEFs (Fig. 3C).
Fig. 3.
MLL3Δ/Δ MEFs support RAR transactivation. (A) Nuclear extracts of E12.5 MEFs from wild-type and MLL3Δ/Δ mice were immunoblotted (IB) with MLL3, ASC-2, and β-actin antibodies as well as MLL3 antibody preincubated with recombinant MLL3 protein or BSA (data not shown). (B) In Q-PCR, the transcript levels of ASC-2, MLL3, and MLL4s were measured in E12.5 wild-type and MLL3Δ/Δ MEFs. (C) Immunoprecipitation (IP) of nuclear extracts of E12.5 wild-type and MLL3Δ/Δ MEFs by IgG and ASC-2 antibody was followed by immunoblotting with MLL3 antibody. (D) E9.5 wild-type (wt), MLL3Δ/Δ, and ASC-2−/− MEFs were treated with vehicle or 0.1 μM 9-cis-retinoic acid (RA) for 12 h, and RAR-β2 transcript levels were determined by Q-PCR. (E) E9.5 wild-type and ASC-2−/− MEFs were transfected with β2-RARE-Luc reporter and an increasing amount of ASC-2 expression vector; then they were treated with vehicle or 0.1 μM 9-cis-RA for 12 h and tested for luciferase activity. (F) Nuclear extracts of HeLa cells were subjected to immunoprecipitations with IgG and antibodies against MLL4s and MLL3 followed by immunoblotting with the indicated antibodies (4).
ASC-2 as a Key Adaptor for RAR-Mediated Recruitment of MLL3 and MLL4s.
To explore the roles of MLL3 and its SET domain in NR transactivation, we focused on RAR and its target gene RAR-β2. We isolated E9.5 MEFs from wild-type, MLL3Δ/Δ, and ASC-2−/− mice. Surprisingly, MLL3Δ/Δ MEFs fully supported RAR transactivation, as evidenced by Q-PCR analysis of 9-cis-RA-induced levels of RAR-β2 mRNAs (Fig. 3D). In contrast, ASC-2−/− MEFs failed to support RAR transactivation (Fig. 3D). Supporting the specificity of ASC-2−/− MEFs, reexpression of ASC-2 in these cells restored RAR transactivation (Fig. 3E). These results demonstrate that ASC-2, but not MLL3, is essential for RAR transactivation in MEFs.
Although several H3K4MTs were originally shown to interact with ASC-2 (4), a further analysis with antibodies specific for MLL3 and MLL4s (4) indicated that these H3K4MTs form distinct ASC2-containing complexes. Thus, MLL3 antibody coimmunoprecipitated ASC-2 and MLL3 but not MLL4, whereas MLL4 antibody coimmunoprecipitated ASC-2 and MLL4 but not MLL3 from HeLa nuclear extracts (Fig. 3F). Similarly, MLL4-1-specific antibody coimmunoprecipitated ASC-2 and MLL4-1 but not MLL3; and MLL3 antibody coimmunoprecipitated no MLL4-1 (data not shown). These results, along with the presence of a single H3K4MT in all of the known Set1-like complexes, suggest that ASCOM is likely a pool of three complexes, namely, ASCOM(MLL4-1), ASCOM(MLL4-2), and ASCOM(MLL3).
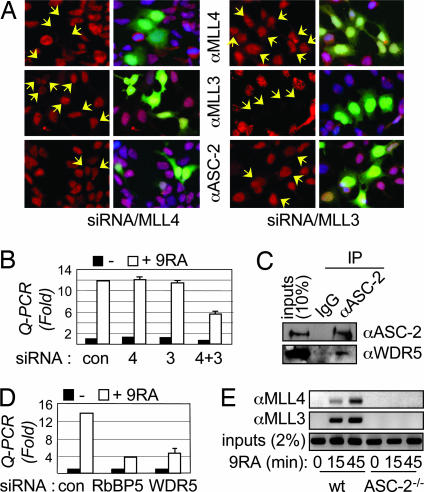
The presence of several distinct ASCOMs and the essentiality of ASC-2 but not MLL3 for RAR transactivation in MEFs (Fig. 3D) suggested the possibility of a functional redundancy of ASCOMs for RAR transactivation. To test this possibility, we examined a series of different siRNAs against MLL3 and MLL4s. The selected siRNAs were highly specific when transiently expressed in HEK293 cells, as demonstrated by the observation that a given siRNA affected its own expression but neither the mRNA levels nor the protein levels of other key components of ASCOM (Fig. 4A and data not shown). Consistent with the results from MLL3Δ/Δ MEFs (Fig. 3D), each siRNA alone did not affect 9-cis-RA-dependent expression of RAR-β2 mRNAs in HEK293 cells (Fig. 4B). However, cotransfection of both siRNAs resulted in an ≈50% reduction in the 9-cis-RA-induced level of RAR-β2 mRNAs (Fig. 4B). Transfection efficiencies for single siRNA transfections were 70–85%, and, as measured by Q-PCR, both MLL3 and MLL4 mRNA levels were simultaneously down-regulated in doubly transfected cells (data not shown). These results suggest that MLL3 and MLL4s both function with RAR and that the intact RAR transactivation in MLL3Δ/Δ MEFs (Fig. 3D) likely results from the function of ASCOM(MLL4s).
Fig. 4.
ASC-2 is a key adaptor for RAR-dependent recruitment of MLL3 and MLL4s. (A) HEK293 cells were transfected with siRNA against MLL3 or MLL4s, along with CMV-GFP. These cells were immunostained for MLL3, MLL4s, and ASC-2, 3 days posttransfection. GFP-positive cells (i.e., transfected cells; arrows) were monitored for the expression of MLL3, MLL4s, and ASC-2. (B) HEK293 cells were transfected with control siRNA, siRNA against MLL3 or MLL4s, or siRNAs for both MLL3 and MLL4s. Two days posttransfection, cells were treated with vehicle or 0.1 μM 9-cis-RA (9-RA) for 12 h, and they were then tested for RAR-β2 transcript levels by Q-PCR. (C) Nuclear extracts of HeLa cells were subjected to immunoprecipitations (IP) with IgG and ASC-2 antibody followed by immunoblotting with ASC-2 and WDR5 antibodies. (D) HEK293 cells were transfected with control (con) siRNA or siRNA against WDR5 or RbBP5. Two days posttransfection, cells were treated with vehicle or 0.1 μM 9-cis-RA for 12 h and tested for RAR-β2 and GAPDH (control; data not shown) transcripts by Q-PCR. (E) Immortalized cell lines from wild-type (wt) and ASC-2−/− MEFs were treated with 0.1 μM 9-cis-RA for 0–45 min and subjected to ChIPs with antibodies against MLL3 and MLL4s.
To demonstrate further the functional redundancy of MLL3 and MLL4 complexes, we tested siRNAs directed against other shared components of ASCOMs. WDR5 is a common component of ySet1, MLL1, MLL2, and hSet1 complexes; it associates with histone H3-K4-dimethylated nucleosomes (24), and it plays a key role in presenting the H3 substrate to the MLL1 H3K4MT complex (21). Because we failed to identify all ASCOM components in our original purification (4), we tested whether WDR5 is included in ASCOM. Immunoprecipitation of HeLa nuclear extracts by ASC-2 antibody, followed by immunoblotting with WDR5 antibody, revealed that WDR5 is indeed associated with ASC-2 (Fig. 4C). Thus, the core components (WDR5, RbBP5, and Ash2L) of H3K4MT complexes are conserved in ASCOM. Taking advantage of the key regulatory roles of these core components in H3K4MT activity, we performed experiments with siRNA directed against WDR5 or RbBP5. By using Q-PCR, we found that RAR-β2 mRNAs are significantly decreased after 9-cis-RA induction in HEK293 cells treated with RbBP5 or WDR5 siRNA compared with control siRNA-treated HEK293 cells (Fig. 4D). These siRNAs did not affect the level of GAPDH, whose expression is RA-independent (data not shown). Thus, RAR transactivation of RAR-β2 should involve H3K4MT complex(es) containing the core WDR5·RbBP5·Ash2L subcomplex but not other complexes that lack this subcomplex (9–12). These results, along with those obtained with ASC-2−/− MEFs (Fig. 3D) and cells expressing MLL3 and MLL4 siRNAs (Fig. 4B), suggest that ASCOMs provide the primary H3K4MTs responsible for RAR transactivation of RAR-β2.
Next, we directly tested the proposed adaptor role of ASC-2 in recruiting MLL3 and MLL4s to promoter-bound RAR. Chromatin immunoprecipitation (ChIP) assays revealed that both MLL3 and MLL4s are recruited in a ligand-dependent manner to the RAR-β2 promoter in wild-type MEFs (Fig. 4E). Remarkably, their recruitment to RAR-β2 was abolished in immortalized cell lines derived from ASC-2−/− MEFs (Fig. 4E). These results demonstrate that ASC-2 is an essential component for recruiting MLL3 and MLL4s to RAR, validating our previous results with DN1 (4). This experimental evidence shows that a bona fide subunit of H3K4MT complexes acts as a direct adaptor to link these complexes to a DNA-binding transcription factor.
H3-K4 Trimethylation of RAR-β2 Requires ASC-2 and MLL3 or MLL4s.
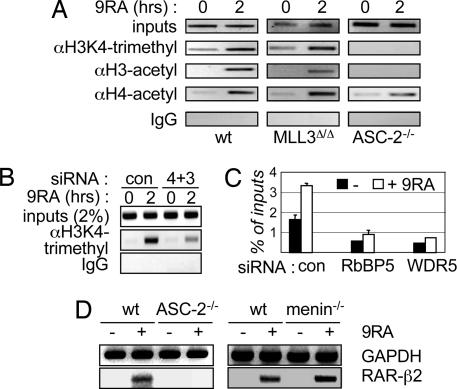
The above results lead to a model in which RAR transactivation requires MLL3 or MLL4s and ASC-2 functions as a key adaptor for RAR-mediated recruitment of MLL3 and MLL4s. The requirement for MLL3 or MLL4s likely reflects their essential function to direct H3-K4 trimethylation of RAR target genes. To test this model, we carried out ChIP assays using E9.5 MEFs from wild-type, MLL3Δ/Δ, and ASC-2−/− mice, and we specifically examined the histone-modification status of the RAR-β2 promoter. In wild-type MEFs, histone H3-K4 trimethylation as well as H3 and H4 acetylations were induced during 9-cis-RA treatment (Fig. 5A). Similar modifications were also observed in MLL3Δ/Δ MEFs (Fig. 5A). In contrast, H3-K4 trimethylation was abolished in ASC-2−/− MEFs compared with wild-type and MLL3Δ/Δ MEFs (Fig. 5A). These results argue that ASC-2 targets H3-K4 trimethylation activity mediated by both MLL3 and MLL4s to RAR-β2 (Fig. 4E). To test this idea directly, we cotransfected HepG2 cells with siRNAs against MLL3 and MLL4s (Fig. 4A). Indeed, 9-cis-RA-induced H3-K4 trimethylation of RAR-β2 was significantly impaired in these cells (Fig. 5B). Similar results were obtained with HEK293 cells (data not shown). These results suggest that ASC-2 is a key adaptor for RAR in mediating recruitment of MLL3 and MLL4s to RAR-β2 and that MLL3 and MLL4s in turn methylate RAR-β2 H3-K4 residues. Interestingly, H3 acetylation was also ablated, and H4 acetylation decreased in ASC-2−/− MEFs (Fig. 5A), suggesting cooperativity between ASCOM and H3/H4 acetylation during RAR transactivation.
Fig. 5.
RAR-β2 H3-K4 trimethylation requires ASC-2 and MLL3 or MLL4. (A) E9.5 wild-type (wt), MLL3Δ/Δ, and ASC-2−/− MEFs were subjected to ChIP assays for H3-K4 trimethylation and H3/H4 acetylation during treatment with vehicle or 0.1 μM 9-cis-RA (9RA) for 2 h. (B) HepG2 cells transfected with control (con) siRNA or siRNAs for both MLL4s and MLL3 were treated with vehicle or 0.1 μM 9-cis-RA for 2 h, 2 days posttransfection, and they were then subjected to ChIPs for H3-K4 trimethylation. (C) HepG2 cells transfected with control siRNA or siRNA for WDR5 or RbBP5 were treated with vehicle or 0.1 μM 9-cis-RA for 2 h, 2 days posttransfection, and they were then subjected to ChIPs for H3-K4 trimethylation of RAR-β2. These siRNAs had no effect on H3-K4 trimethylation of GAPDH (data not shown). (D) E9.5 wild-type and ASC-2−/− MEFs, as well as immortalized cell lines derived from wild-type and menin−/− MEFs, were treated with vehicle or 0.1 μM 9-cis-RA for 12 h, and 9-cis-RA induction of RAR-β2 mRNAs was measured by RT-PCR.
We also tested whether 9-cis-RA-induced H3-K4 trimethylation of RAR-β2 involves the core WDR5·RbBP5·Ash2L subcomplex, which is important for regulating the H3K4MT activity of Set1-like complexes (21). The decreased expression level of RAR-β2 mRNAs in cells treated with siRNA against WDR5 or RbBP5 (Fig. 4D) correlated with significantly reduced H3-K4 trimethylation of RAR-β2 (Fig. 5C). These results, along with the results observed with ASC-2−/− MEFs (Fig. 5A) and cells expressing MLL3 and MLL4s siRNAs (Fig. 5B), demonstrate that ASCOM is the primary H3K4MT responsible for RA-induced H3-K4 trimethylation of RAR-β2.
ASC-2 Confers Target Gene Specificity to ASCOM.
Menin is an essential component of two mammalian Set1-like complexes, namely, MLL1 and MLL2 complexes. Menin recently was shown to function as a coactivator of estrogen receptor (ER) and to link ER transactivation to H3-K4 trimethylation (25). Thus, we tested whether menin acts as a redundant mediator of H3-K4 trimethylation of RAR-β2. Semiquantitative RT-PCR analyses revealed that 9-cis-RA induced RAR-β2 mRNAs in wild-type cells, whereas it was inert in E9.5 ASC-2−/− MEFs (Fig. 5D Left). Interestingly, 9-cis-RA fully induced RAR-β2 mRNAs in immortalized cell lines derived from menin−/− MEFs (Fig. 5D Right). These results demonstrate that RAR transactivation requires ASCOM and that highly homologous, menin-associated MLL1 and MLL2 H3K4MT complexes cannot replace ASCOM. Importantly, these results suggest that unique components of H3K4MT complexes may serve as key specificity determinants to direct different H3K4MT complexes to specific transcription factors. Thus, ASC-2 targets ASCOMs to RAR, whereas menin may possibly tether MLL1 or MLL2 complexes to ER (25).
Discussion
The present study significantly extends our earlier report (4) of a complex (ASCOM) containing MLL3, MLL4s, and ASC-2 by showing that MLL3 and MLL4s exist with ASC-2 in distinct complexes with at least partially redundant functions. We demonstrate the highly specific and essential roles of MLL3 and MLL4s in ASC-2-dependent transactivation of RAR-β2, an essential function of ASC-2 in recruiting MLL3 and MLL4s to RAR-β2 in a retinoid-dependent manner, and a key role for MLL3- and/or MLL4-dependent H3-K4 trimethylation in transcriptional activation of RAR-β2. Collectively, we propose a model for the function of ASCOM(MLL3) and ASCOM(MLL4s) in RAR transactivation that entails (i) independent recruitment of an ASCOM complex to RAR by retinoid-dependent interactions between RAR and NR box 1 of ASC-2; (ii) stabilization of the binding of ASCOMs to the H3 tail by WDR5 in the WDR5·RbBP5·Ash2L subcomplex (21, 24); and (iii) MLL3- and MLL4-mediated H3-K4 trimethylation of the promoter region bound by RAR.
The SET Domain Is Pivotal for ASCOM·MLL3 Function.
The successful establishment of MLL3Δ/Δ mice is a major advancement in understanding the physiological function of ASCOMs. Although the MLL3 SET domain deletion mutant is expressed at a normal level and incorporated into the ASC-2 complex (Fig. 3 A–C), the observation that some MLL3 functions are impaired suggests important roles for the SET domain and its H3K4MT activity in MLL3 function. The similar phenotypes of MLL3Δ/Δ and isogenic ASC-2+/− mice suggest genetic interactions between the two proteins in vivo. The more severe phenotype of ASC-2−/− (3) can be partially explained by the redundant functions of MLL3 and MLL4s. Because ASC-2 is a common component of both MLL3 and MLL4 complexes, the ASC-2-null phenotypes are likely the sum of MLL3- and MLL4-deficient phenotypes rather than just an MLL3-null phenotype. In addition, because MLL3 is a very large protein that contains multiple domains, some of its functions are likely SET domain-independent. However, although all ASC-2 proteins appeared to be associated with MLL3 and MLL4 complexes in our original purification (4), it has not yet been tested whether all MLL3 and MLL4s are associated with ASC-2. Thus, we cannot exclude the possibility that MLL3 and MLL4s have some ASC-2-independent functions.
ASC-2 Confers Specificity to ASCOMs.
The Set1-like complexes contain unique components such as menin and host cell factors in MLL1 and MLL2 (6) and ASC-2 in MLL3 and MLL4s (4). The existence of unique components in different H3K4MT complexes argues for specific functions. Indeed, several observations suggest specific roles for different H3K4MTs in transcription regulation. Thus, MLL2 appears to be important for expression of the HOX B gene cluster but not for expression of the HOX A cluster (26), whereas HOX a9 and HOX c8 are exclusive MLL1 targets relative to other Set1-like H3K4MTs (27, 28). Although the mechanisms underlying the specificity of the different Set1-like H3K4MTs remain to be understood fully, the unique components in different H3K4MT complexes (including H3K4MTs themselves; see below) may confer specificity. Thus, as demonstrated in this work, the unique integral component of MLL3 and MLL4 complexes, ASC-2, is essential for RAR-dependent transactivation, and RAR-β2 expression and H3-K4 trimethylation are impaired in ASC2−/− cells but not in menin−/− cells. These results clearly demonstrate that RAR-β2 is a specific target for MLL3 and MLL4s but not for MLL1 and MLL2. This specificity is likely achieved through the previously demonstrated physical interactions between RAR and ASC-2 (3). In further support of this notion, the MLL3 and MLL4 complexes, which share ASC-2, demonstrate redundant functions for RAR transactivation. Because ASC-2 is required for transactivation by other NRs (3), it is likely that MLL3 and/or MLL4s are directly involved in their transactivation functions.
In addition to ASC-2, other notable components of ASCOM are the catalytic subunits (MLL3 and MLL4s) themselves. MLL family H3K4MTs are consistently large proteins with a highly conserved C-terminal SET domain and highly divergent N-terminal domains. Although MLL3 and MLL4s share a series of conserved motifs that include the SET domain, their overall homology is <30%. It will be important to determine whether MLL3 and MLL4s have unique target genes in addition to common targets such as RAR-β2. Interestingly, MLL4-1, Ash2L, RbBP5, and WDR5 were recently isolated from DU4475 nuclear extracts as ER-associated proteins. This estrogen-dependent interaction involves the fifth and sixth LXXLL motifs of MLL4-1 (29). As a result, this function of MLL4-1 is not redundant with MLL3, and MLL4-1 siRNA treatment alone impairs ER-dependent transactivation (29).
Recruitment of H3K4MTs Through Interactions with DNA-Binding Transcription Factors.
We have demonstrated here that ASC-2 tethers ASCOMs to RAR-β2 in a ligand-dependent manner and that MLL3 and MLL4, in turn, carry out H3-K4 trimethylation. The recruitment of ASCOMs is likely mediated by NR box 1 of ASC-2 because a dominant-negative fragment of ASC-2 containing NR box 1 (DN1) impairs their recruitment (4), thus providing experimental evidence for recruitment of an H3K4MT complex to a natural promoter through the interaction of an integral component with a DNA-binding transcription factor. A similar recruitment mechanism has been indicated for ER with MLL4-1 (29) as well as for β-catenin with MLL1 and MLL2 (30). Direct interactions between DNA-binding transcription factors and H3K4MTs are likely a general phenomenon. Indeed, an interaction between p53 and the MLL1 complex leads to transcription activation in vitro (13), which implicates more active roles of H3K4MTs in transcription activation than is generally assumed (see below).
Notably, this recruitment mechanism is distinct from what has been proposed for the ySet1 complex (6, 18, 31, 32) and from an in vitro transcription study (33). The latter study postulated that the H3-K4 methylation is solely a consequence of transcription activation and that H3K4MT recruitment depends on transcription elongation factors such as RNA polymerase II-associated factor (PAF) and FACT. However, this model fails to explain several observations in higher eukaryotes: first, H3-K4 trimethylation is enriched in promoter regions; second, interactions of methylated H3-K4 and/or H3K4MTs with chromatin modifiers involved in transcription initiation, such as NURF (34) and p300 (35), occur at the promoter; third, although direct interactions among FACT, the PAF complex, and the H2B ubiquitination machinery have been documented, direct interactions of these components with H3K4MTs are not yet established in mammalian cells (although Paf1 interacts with Set1 in yeast; ref. 36). Finally, several recent reports demonstrate that siRNA-mediated knockdown of components of the H2B ubiquitination machinery does not affect H3-K4me1 and/or H3-K4me2 at target genes, suggesting intact recruitment of H3K4MTs (33, 37). Future studies on the regulation H3-K4 methylation, especially through an evolutionarily conserved mechanism involving the PAF complex and H2B ubiquitination will likely reveal functional interactions between different chromatin modifiers and their involvement in transcription regulation.
Materials and Methods
Generation of MLL3Δ/Δ Mice.
The targeting construct was electroporated into embryonic stem cells and G418-selected. Correctly recombined clones were used to generate chimeric animals (38). The neo-heterozygotes (MLL3+/Neo) were bred to a protamine-Cre transgenic mouse line (22) to generate MLL3+/Δ mice, which were bred to produce MLL3Δ/Δ mice. The primer sequences used for genomic PCR are available on request.
RT-/Q-PCR.
Total RNA was isolated from cells after lysis in TRIzol reagent according to the manufacturer's protocol (Invitrogen Corp., Carlsbad, CA), and RT- and SYBR Green Q-PCRs were performed as described in refs. 20 and 39. The primer sequences are available on request.
ChIPs.
Soluble chromatin was prepared and immunoprecipitated with the indicated antibodies, as described in ref. 40. The final DNA extractions were amplified by using pairs of primers that encompass the RAR-responsive element in the RAR-β2 promoter (4).
RNA Interference.
Double-stranded DNA fragments that contained the selected RNAi sequences positioned downstream from the human U6 RNA polymerase III promoter were generated by PCR by using the Silencer express kit (Ambion, Austin, TX) according to the manufacturer's instructions. The DNA fragments were cloned into the pSec vector (Ambion). Cells were transfected with pSec derivatives. Three days later, ASC-2, MLL4, and MLL3 expression levels were determined by Q-PCR (data not shown) and by immunostaining with the indicated antibodies (Fig. 4A). The siRNA sequences are available on request.
Acknowledgments
We thank Drs. Sam Pfaff, Winship Herr, Jianming Xu, and Mathew Meyerson for reagents. This work was supported by National Institutes of Health Grants DK064678 (to J.W.L.) and DK071900 (to R.G.R.).
Abbreviations
- ASC-2
activating signal cointegrator-2
- ASCOM
ASC-2 complex
- E
embryonic day
- ER
estrogen receptor
- H3K4MT
H3-K4 methyltransferase
- MEFs
mouse embryo fibroblasts
- MLL
mixed-lineage leukemia
- NR
nuclear receptor
- Q-PCR
quantitative polymerase chain reaction
- RA
retinoic acid
- RAR
retinoic acid receptor
- RbBP
retinoblastoma-binding protein.
Footnotes
The authors declare no conflict of interest
Despite the recent NCBI designation of ALR and Trx2 as MLL2 and MLL4, respectively, Trx2 was originally, and continues to be, published as MLL2. Thus, we retain the original MLL2 usage, and for convenience we designate ALR as MLL4.
References
- 1.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hermanson O, Glass CK, Rosenfeld MG. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 3.Mahajan MA, Samuels HH. Endocr Rev. 2005;26:583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- 4.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao B, Wilson JR, Gamblin SJ. Curr Opin Struct Biol. 2003;13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Shilatifard A. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 7.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 8.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 9.Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- 10.Byrd KN, Shearn A. Proc Natl Acad Sci USA. 2003;100:11535–11540. doi: 10.1073/pnas.1933593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 13.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 14.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 17.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampsey M, Reinberg D. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 19.Kim SW, Park K, Kwak E, Choi E, Lee S, Ham J, Kang H, Kim JM, Hwang SY, Kong YY, et al. Mol Cell Biol. 2003;23:3583–3592. doi: 10.1128/MCB.23.10.3583-3592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SW, Cheong C, Sohn YC, Goo YH, Oh WJ, Park JH, Joe SY, Kang HS, Kim DK, Kee C, et al. Mol Cell Biol. 2002;22:8409–8414. doi: 10.1128/MCB.22.24.8409-8414.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Nat Struct Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 22.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahajan MA, Das S, Zhu H, Tomic-Canic M, Samuels HH. Mol Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 25.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 26.Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Development (Cambridge, UK) 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 29.Mo R, Rao SM, Zhu YJ. J Biol Chem. 2006;281:15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 30.Sierra J, Yoshida T, Joazeiro CA, Jones KA. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerber M, Shilatifard A. J Biol Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 32.Ng HH, Robert F, Young RA, Struhl K. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 33.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 34.Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Mol Cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- 35.Cho H, Orphanides G, Sun X, Yang XJ, Ogryzko V, Lees E, Nakatani Y, Reinberg D. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 37.Shahbazian MD, Zhang K, Grunstein M. Mol Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 39.Ledbetter MW, Preiner JK, Louis CF, Mickelson JR. J Biol Chem. 1994;269:31544–31551. [PubMed] [Google Scholar]
- 40.Shang Y, Myers M, Brown M. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]