Abstract
The mechanism that drives the segregation of cells into tissue-specific subpopulations during development is largely attributed to differences in intercellular adhesion. This process requires the cadherin family of calcium-dependent glycoproteins. A widely held view is that protein-level discrimination between different cadherins on cell surfaces drives this sorting process. Despite this postulated molecular selectivity, adhesion selectivity has not been quantitatively verified at the protein level. In this work, molecular force measurements and bead aggregation assays tested whether differences in cadherin bond strengths could account for cell sorting in vivo and in vitro. Studies were conducted with chicken N-cadherin, canine E-cadherin, and Xenopus C-cadherin. Both qualitative bead aggregation and quantitative force measurements show that the cadherins cross-react. Furthermore, heterophilic adhesion is not substantially weaker than homophilic adhesion, and the measured differences in adhesion do not correlate with cell sorting behavior. These results suggest that the basis for cell segregation during morphogenesis does not map exclusively to protein-level differences in cadherin adhesion.
Keywords: cell adhesion, force probe, selectivity, surface forces apparatus
Both the formation of distinct tissues during morphogenesis and the maintenance of adult tissue structure are regulated by adhesive contacts between cells. One family of adhesion proteins, cadherins, is critical to several processes associated with the formation and maintenance of tissues. The genetic regulation of the spatiotemporal expression patterns of cadherins in vivo is believed to play a central role in embryogenesis, cell differentiation, and the maintenance of the multicellular structure of an organism (1–5). One prevalent hypothesis is that differences in adhesion between cells direct cell segregation during morphogenesis, analogous to liquid–liquid phase separation (6). A fundamental unresolved question is whether this sorting out is encoded by protein-specific or cell-specific differences in adhesion. A common hypothesis is that preferential binding between identical cadherins (homophilic) relative to heterophilic binding between dissimilar cadherins induces the cell segregation.
Both the adhesive and the selectivity functions of classic cadherins map to the extracellular (EC) region (7), which comprises five tandemly arranged EC domains, EC1–5, numbered from the outermost domain (Fig. 1) (8). Classical cadherins also contain a transmembrane segment and a cytoplasmic domain. Cadherins form multiple bonds that require different EC domains (9–12). A common feature of the classical cadherins is that the N-terminal domains bind by inserting the tryptophan 2 (W2) residue from one protein into a hydrophobic pocket on an opposed N-terminal domain (13–16). However, given the weak adhesion between the outer domains (11, 12), it is questionable whether the tryptophan binding accounts for both cadherin selectivity and robust adhesion.
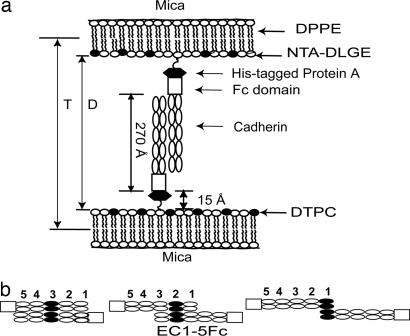
Fig. 1.
Protein configurations in the force measurements. (a) Cadherin monolayer architecture used in SFA measurements. D is the absolute separation between the bilayer surfaces, and T is the distance between the DPPE monolayers. (b) Proposed cadherin ectodomain alignments at the three different membrane separations at which the proteins adhere.
The most extensively cited evidence for protein-level control of cell sorting is the time-dependent aggregation of different cell populations that each express different cadherins (14, 17). In these assays, cells are kept in suspension by agitating the solutions. Under these conditions, the cell populations often form aggregates of a single cell type, and cells expressing different cadherins do not intermix (14, 17). Electron microscopy studies similarly suggested that epithelial E-cadherin (E-CAD) and placental P-cadherin (P-CAD) form homophilic bonds exclusively (18). However, this segregation vanishes at slower mixing speeds. Shaking cell suspensions at <10 rpm versus the standard 100 rpm generates mixed aggregates of cells, which do not intermix at the higher mixing speeds (19).
In contrast, other investigations demonstrated cadherin cross-reactivity and suggested that protein-level adhesion selectivity alone cannot explain cell segregation. Cells expressing epithelial, neural, or Xenopus cleavage-stage cadherin adhered equally well to different cadherin EC domains immobilized in a capillary flow chamber (20). Additionally, two cell populations expressing the same amounts of E- and P-CAD remained intermixed in cell clusters (19). The cells did segregate, however, when the P- and E-CAD expression levels differed by as little as 20% (3, 19). In some cases, the different cell populations formed distinct cell patterns, but the cells did not delaminate as would occur in the absence of cadherin cross-adhesion. Despite the centrality of the assumed cadherin-binding specificity, this hypothesis has not been tested at the protein level.
This work reports direct, quantitative measurements of the mechanisms and strengths of heterophilic and homophilic cadherin adhesion at the protein level. The molecular mechanisms of homophilic binding by canine E-CAD and chicken neural N-cadherin (N-CAD) are nearly identical to those of Xenopus cleavage-stage C-cadherin (C-CAD). All three cadherins cross-react by a similar mechanism. The average energies of heterophilic and homophilic bonds are similar, and there is no general preference for homophilic versus heterophilic binding. These findings are supported by semiquantitative bead aggregation measurements.
Results
In these measurements, the distance D is the absolute separation between the surfaces of opposed lipid bilayers supporting the immobilized proteins (Fig. 1a). The normalized force between the two surfaces, Fc/R, is then measured as a function of the distance D (±1 Å) during both approach and separation of the membrane-bound cadherin monolayers.
Homophilic Cadherin Interactions.
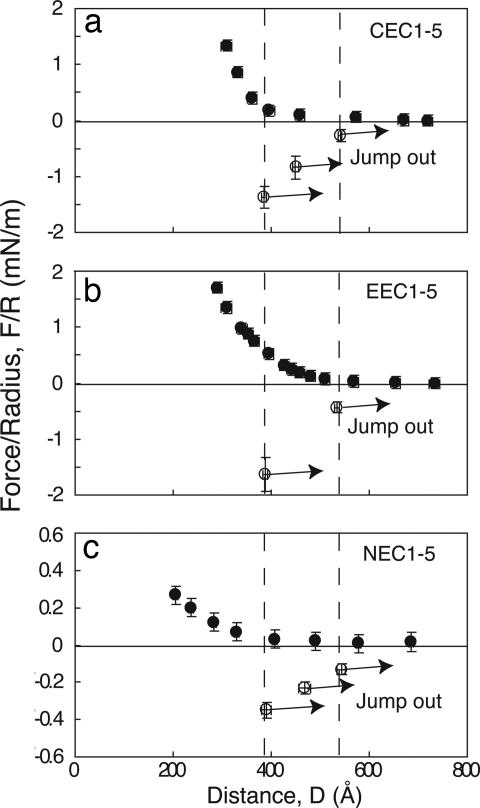
An earlier report demonstrated that opposing C-CAD (C/C) ectodomains (CEC1-5) form three spatially distinct bound states at different membrane distances (10). Fig. 2a shows the results of the normalized force profile between identical C-CAD monolayers, which are repeated here for comparison. During approach, at membrane separations of D < 570 Å, the force increases (F > 0) because of steric repulsion between opposing proteins. The maximum expected range of the interaction is 570 Å, based on the measured 15-Å thickness of the His6-tagged protein A layer (21), the 45-Å thickness of the Fc fragment, and the 225-Å cadherin ectodomain length [570 = 2(15 + 45 + 225)] (compare Fig. 1a). Upon separation, the C-CAD ectodomains adhered at each of three membrane separations: D = 383 ± 6 Å (Fpo/R = −1.9 ± 0.4 mN/m), D = 455 ± 6 Å (Fpo/R = −1.7 ± 0.4 mN/m), and D = 536 ± 3 Å (Fpo/R = −1.0 ± 0.3 mN/m). The C-CAD-Fc surface density, Γ, in this experiment was 9.5 ± 1 × 103 C-CAD-Fc per μm2. From the measured pull-off force Fpo/R, the estimated average energies per bond (Eb) were 7.8 ± 2, 6.9 ± 2, and 4.1 ± 1.6 kBT (Table 1), where kB is the Boltzmann constant and T is the absolute temperature.
Fig. 2.
Normalized force–distance profiles for homophilic interactions between C-CAD (a), E-CAD (b), and N-CAD (c). Filled circles show the normalized force versus distance during approach (decreasing D). Open circles show the positions and magnitudes of the adhesive minima (F/R < 0) measured during separation (increasing D). The arrows show distances at which the cadherin bonds rupture. The vertical dashed lines correspond to the positions of bond failure.
Table 1.
Cadherin-binding parameters
| Cadherin pair | Density, cadherins per μm2 (×103) | Distance, Å; F/R, mN/m; and bond energy, kBT |
||
|---|---|---|---|---|
| Inner bond | Middle bond | Outer bond | ||
| C-CAD/C-CAD | 9.5 ± 1 | 383 ± 6; 1.9 ± 0.4; 7.8 | 455 ± 6; 1.7 ± 0.4; 6.9 | 536 ± 3; 1.0 ± 0.3; 4.1 |
| E-CAD/E-CAD | 10 ± 1 | 388 ± 5; 1.6 ± 0.3; 6.2 | — | 525 ± 5; 0.6 ± 0.1; 2.5 |
| N-CAD/N-CAD | 7 ± 0.7 | 391 ± 10; 0.35 ± 0.04; 2.1 | 468 ± 12; 0.23 ± 0.03; 1.5 | 543 ± 9; 0.14 ± 0.03; 1.0 |
| E-CAD/C-CAD | 9/9 | 384 ± 8; 1.34 ± 0.4; 5.8 | 471 ± 5; 0.8 ± 0.2; 3.5 | 541 ± 10; 0.45 ± 0.1; 2.2 |
| N-CAD/E-CAD | 7/7 | 386 ± 8; 1.2 ± 0.2; 6.6 | 474 ± 8; 0.7 ± 0.1; 4.0 | 535 ± 4; 0.34 ± 0.04; 2.1 |
| N-CAD/C-CAD | 7/11 | 389 ± 5; 1.43 ± 0.18; 7.8 | 471 ± 6; 1.19 ± 0.07; 6.6 | 536 ± 3; 0.69 ± 0.08; 3.9 |
—, not detected.
Based on the distances, protein dimensions, and prior domain deletion studies, at D = 383 Å, the EC3 domains between opposed cadherins are aligned (Fig. 1b) (10). Adhesion at 455 Å could be due to either EC2/EC2 and/or EC1/EC3 contacts (compare Fig. 1b). Adhesion at 536 Å is due to EC1/EC1 binding (Fig. 1b) (10). The relative adhesion strengths are inner bond (383 Å) > middle bond (455 Å) > outer bond (536 Å) (9, 10, 22).
Fig. 2b shows force profiles between canine epithelial cadherin E-CAD monolayers (E/E) at 10 ± 1 × 103 E-CAD per μm2. Upon separation from 300 < D < 370 Å, the proteins adhered at D = 388 ± 5 Å. The 388-Å distance is identical to the position of the inner bond between C-CAD-Fc monolayers, which involves complete ectodomain overlap (compare Fig. 1b). When separated from distances of 390 < D < 530 Å, the proteins also bound at D = 535 ± 5 Å, which corresponds to EC1 overlap (Fig. 1b). From the measured pull-off forces (Fpo) of −1.6 ± 0.3 and −0.6 ± 0.1 mN/m at 388 and 535 Å, respectively, the average bond energies are 6.2 ± 1.5 and 2.5 ± 0.5 kBT (Table 1). The middle bond measured with C-CAD ectodomains was not measured between canine E-CAD (23).
Oriented monolayers of chicken neural cadherin (N-CAD) (N/N) at 7 ± 0.7 × 103 N-CAD per μm2 adhered at D = 391 ± 10 Å (Eb = 2.2 ± 0.3 kBT), D = 468 ± 12 Å (Eb = 1.5 ± 0.2 kBT), and at D = 543 ± 9 Å (Eb = 1.0 ± 0.2 kBT) (Fig. 2c and Table 1). Adhesion at each of the three distances was −0.35 ± 0.04, −0.23 ± 0.03, and −0.14 ± 0.03 mN/m, respectively. N-CAD binds at the same three intermembrane distances as C-CAD monolayers.
The calcium dependence of binding confirmed that the measured adhesion is due to specific cadherin binding. In calcium-free buffer, the outer two homophilic N-CAD bonds were completely abolished. The putative EC3/EC3 bond persisted, although it was substantially weaker (0.9 ± 0.3 kBT) (Table 1). C-CAD exhibited similar behavior (10, 24). Based on the similar calcium dependence of the different cadherin bonds, the calcium dependence was assumed to apply to all interactions studied.
Heterophilic Cadherin Interactions.
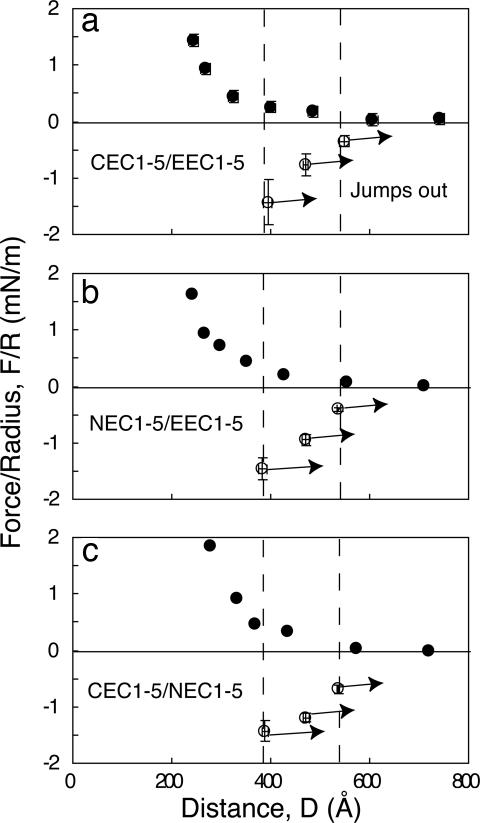
E-CAD-Fc also bound C-CAD-Fc (E/C) (Fig. 3a) at three intermembrane distances: D = 384 ± 8 Å (F/R = −1.34 ± 0.4 mN/m), D = 471 ± 5 Å (F/R = −0.8 ± 0.2 mN/m), and D = 541 ± 10 Å (F/R = −0.45 ± 0.1 mN/m) (Fig. 3a). The estimated bond energies were 5.8 ± 2, 3.5 ± 0.8, and 2.2 ± 0.4 kBT, respectively (Table 1).
Fig. 3.
Normalized force–distance profiles for heterophilic interactions between C- and E-CAD (a), N- and E-CAD (b), and C- and N-CAD (c). The filled circles indicate the normalized force versus distance during approach (decreasing D). Open circles show the positions and magnitudes of the adhesive minima (F/R < 0) measured during separation (increasing D). The arrows indicate protein–protein bond failure. The vertical dashed lines show the positions of bond failure.
Fig. 3b shows the force profile between N-CAD-Fc and E-CAD-Fc (N/E) at 7 ± 0.7 × 103 cadherins per μm2 (Table 1). These proteins also bound at D = 386 ± 8 Å (F/R = −1.2 ± 0.2 mN/m), D = 474 ± 8 Å (F/R = −0.7 ± 0.1 mN/m), and D = 535 ± 4 Å (F/R = −0.34 ± 0.04 mN/m) (Fig. 3b). The average bond energies were 6.6 ± 1.4, 4.0 ± 0.7, and 2.1 ± 0.3 kBT, respectively (Table 1).
At surface densities of 7 ± 0.7 × 103 and 11 ± 1.1 × 103 cadherins per μm2 for N- and C-CAD, respectively, N-CAD-Fc and C-CAD-Fc (N/C) (Fig. 3c) bound at D = 389 ± 5 Å (F/R = −1.43 ± 0.18 mN/m), D = 471 ± 6 Å (F/R = −1.19 ± 0.07 mN/m), and D = 536 ± 3 Å (F/R = −0.69 ± 0.08 mN/m) (Fig. 3c) (Table 1). The average bond energies at the three distances were 7.8 ± 1.1, 6.6 ± 0.4, and 3.9 ± 0.5 kBT, respectively.
Bead Aggregation Assays.
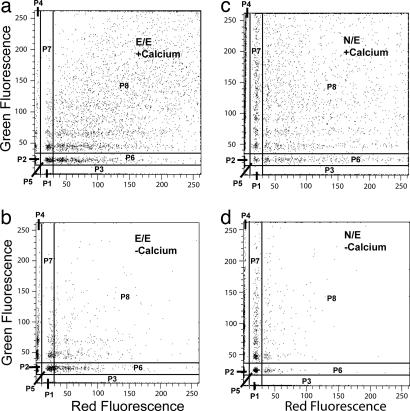
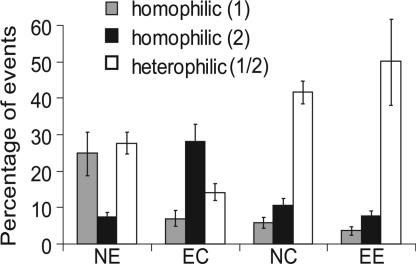
Qualitative bead aggregation assays independently verified cadherin cross-reactivity. Fig. 4a shows the 2D distribution of bead aggregates induced by homophilic E-CAD binding. In this measurement, E-CAD-Fc was immobilized on both the red and green microspheres. From the distribution of aggregates formed in the presence of calcium (Fig. 4a), we determined the percentage of beads forming mixed (red/green) aggregates relative to the entire population of events. In the analysis, we partitioned the events into different categories. Regions P1 and P2 correspond to single red and green beads. Sectors P3 and P4 correspond to homoaggregates only. Regions P6 and P7 indicate clusters with only one red or one green bead in the aggregate. We excluded P6 and P7 from the calculations to eliminate potential contributions from bead entrapment or nonspecific adhesion. The remaining regions, P8 and P5, define the population of “heteroaggregates,” which contain both red and green beads. Some “homoaggregates” also form, as expected. Some residual aggregation occurs in the absence of calcium (Fig. 4b) because of nonspecific adhesion or possibly because of residual adhesion by means of the putative EC3/EC3 bond (10, 24). However, in the absence of calcium, the majority of beads remain dispersed.
Fig. 4.
2D plots of distributions of red and green beads in aggregates quantified by flow cytometry. (a and b) Distribution of homophilic aggregates formed by mixing red and green beads coated with E-CAD in the presence (a) and absence (b) of calcium. The vertical axis is the green fluorescence intensity caused by green E-CAD-coated beads, and the horizontal axis shows the red fluorescence from red E-CAD-coated beads. The region P8 defines aggregates containing both red and green beads. Sectors P1 and P2 indicate isolated red and green beads, respectively. Sectors P3 and P4 define homoaggregates of red and green beads, respectively. Regions P6 and P7 quantify aggregates containing only one red or green bead in a larger aggregate. (c and d) Aggregates formed by mixing N-CAD and E-CAD beads in the presence (c) and absence (d) of calcium. The different sectors are defined as in a and b.
Fig. 4 c and d summarizes the aggregation of beads coated with E-CAD-Fc and with N-CAD-Fc in the presence and absence of calcium, respectively. The majority of events are due to heteroaggregates of E-CAD-Fc and N-CAD-Fc beads. Without calcium (Fig. 4d), the percentage of heteroaggregates decreases substantially, and most of the beads remain dispersed.
Fig. 5 summarizes the results obtained with N/E, E/C, N/C, and E/E bead mixtures. The histograms show the percentages of homo- and heteroaggregates in each measurement; these values are based on the total number of events rather than the total number of beads, which is difficult to quantify. In all three cases, heteroaggregates account for a substantial proportion of the measured events, confirming that these cadherins cross-react.
Fig. 5.
Percentage of homoaggregates (gray and black bars) and heteroaggregates (open bars). The cadherin combinations are indicated below the histograms. The first letter in the pair is shown by the first (gray) bar, and the second (black) bar represents the second letter of the pair.
Discussion
These results compare the homophilic and heterophilic binding of three different type I classical cadherins. In contrast to the widely held view, C-CAD, N-CAD, and E-CAD all cross-react. A surprising finding is that, although homophilic cadherin bonds are stronger than heterophilic bonds in some cases, sometimes the reverse is true. In these force measurements, the controlled, quantified protein density rules out uncertainties due to differences in protein expression levels or the amounts of adsorbed cadherin in adhesion measurements. The reported adhesion differences are due to functional differences between active proteins. The adhesion energies of the homophilic and heterophilic bonds also are similar. A recent model suggests that preferences for homophilic binding of as little as 1 kcal/mol (1.2 kBT per bond) would be sufficient to trigger cell sorting (25). Our measurements show, however, that the homophilic cadherin bonds are not always thermodynamically more favorable than heterophilic bonds.
The multistage binding mechanisms of these three cadherins as identified in the force measurements are nearly identical, and the bond energies do not differ significantly. The order of homophilic adhesion is C/C > E/E > N/N, in qualitative agreement with cell adhesion and single bond rupture measurements (11, 12, 20, 26–28).
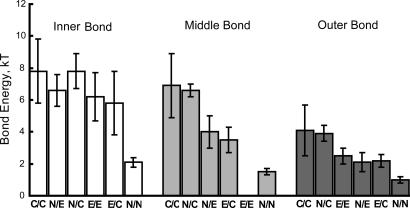
Comparisons of the bond energies for each of the three bound states reveal trends in the homophilic and heterophilic adhesion (Fig. 6). In qualitative agreement with evidence that the EC1 domains confer cadherin specificity, the outer and middle bond energies vary more than the inner bonds (Fig. 6). This observation is consistent with recent findings that the kinetics of weak E-CAD12 bonds differ ≈30-fold from C-CAD12 bonds (11) and that human N-CAD and canine E-CAD exhibit different mechanical properties (28). However, the modest absolute differences between the bond energies should result in a distribution of homophilic and heterophilic binding, as observed in the bead assays (Fig. 5).
Fig. 6.
Average energies of homophilic and heterophilic cadherin bonds. From left to right are the histograms, in decreasing order of bond energy, for the inner, middle, and outer cadherin bonds. The corresponding cadherin pairs are indicated below each bar.
The very similar (within error) average energies of the strongest inner bonds suggest that they contribute to the mechanical stability of mature junctions rather than to cadherin selectivity. Similarly, the slow dissociation rates of these bonds (11, 12) are more appropriate to stable, long-lived intercellular linkages.
These quantitative force measurements contrast with the widely held view that cadherins do not form heterophilic bonds or that heterophilic bonds are substantially weaker than homophilic linkages. Cell aggregation measurements are most often cited as evidence of preferential homophilic binding. The apparent discrepancy may be due to the aggregation assay used (19). For example, E-CAD- and P-CAD-expressing cells formed separate aggregates after 30–45 min of mixing at 100 rpm, but continuous mixing generated both uniform and mixed aggregates, albeit with cells sorted into distinct regions (29). At 1- to 3-rpm shaking, E-CAD and P-CAD cells formed mixed aggregates, whereas only uniform aggregates formed at the typical higher mixing speeds of 75–100 rpm (19). These cell aggregation assays use agitated (sheared) cell suspensions, and it is well known that particle (e.g., cell) coagulation rates under shear depend on the bond kinetics and the shear rate (30–33).
Kinetics also likely account for the differences between these surface force measurements and the apparent absence of heterophilic N-CAD/E-CAD bonding in atomic force microscopy (AFM) measurements with cells (28). In the AFM measurements, very short probe–cell contact times were used to minimize nonspecific binding. However, if heterophilic bonds form more slowly, then millisecond AFM measurements would detect fewer heterophilic bonds, even though such bonds would readily form on the much longer time scales of surface forces apparatus (SFA) and cell adhesion measurements (20).
Several other studies suggest that classical cadherins cross-react (3, 19, 20, 34). C-, E-, or N-CAD adhered similarly to immobilized C-, E-, and N-CAD ectodomains in capillary flow assays (20). L cells expressing comparable levels of P- and E-CAD intermixed in cell sorting assays and formed mixed aggregates in cell aggregation assays. The latter findings and those of Duguay et al. (19) contradict electron microscopy data that suggest that P- and E-CAD ectodomains do not form intermolecular bonds (18). Chick retina cells mixed with both heart cells (35) and liver cells (36), even though the cells express different cadherins. Although N- and E-cadherin-displaying cells do not cross-react in cell aggregation assays at high mixing speeds (100 rpm) (14), chick lens cells expressing the N-CAD homologue intermixed with liver cells expressing the chicken E-CAD homologue and formed heterotypic cell–cell junctions (35, 37). Substituting N-CAD for E-CAD in mouse hearts also resulted in normal development (38).
The proteins used in this study are from different species. Although there are some differences, the sequences across species are very similar. The chicken N-CAD sequence homology is ≈90% that of Xenopus and mammalian N-CAD. Similarly, the chicken liver cell adhesion molecule, which exhibits similar expression patterns as mammalian E-CAD, is closest in sequence identity to mammalian E-CAD. Xenopus E-CAD, Xenopus C-CAD, E-CAD, and P-CAD exhibit >55% overall sequence similarity and 75% sequence similarity in the N-terminal domains (20). Furthermore, in extensive cell sorting studies, the species origin did not affect in vitro cell segregation patterns (19, 39). The latter findings in particular suggest that our results reflect general cadherin behavior. Possible species-specific differences may exist. Nevertheless, these results show that different cadherins cross-react and that homophilic bonds are not always favored over heterophilic interactions.
Our results argue that the mechanism postulated to direct cell sorting is not solely due to differences in protein-level adhesion, which in turn raises additional interesting questions regarding the role, for example, of cadherin association rates in segregation or the force dependence of interactions. Controlled gene expression levels, cell surface receptor interactions, or intracytoplasmic associations, as reviewed in ref. 40, are also postulated to regulate cadherin function.
Inside-out signaling could also alter cadherin adhesion (40). The inability to test inside-out signaling is a limitation of using soluble, recombinant ectodomains. This same limitation applies to many reported biophysical studies, including crystallography, electron microscopy, NMR, flow cell assays, and force measurements (9, 10, 12, 13, 15, 16, 20, 22, 41–46). The importance of our findings is that they demonstrate quantitatively, at the protein level, that cadherin ectodomains do not universally favor homophilic versus heterophilic bonding. These results thus address a fundamental question of cadherin selectivity.
In summary, the C-, E-, and N-CADs investigated in this work form multiple adhesive bonds. These proteins cross-react by means of similar mechanisms and with similar adhesion energies. The homophilic adhesion strengths decreased according to C/C > E/E > N/N. The magnitude of heterophilic C/E, C/N, and N/E adhesion was also similar to homophilic adhesion. The exceptions are heterophilic N/C and N/E bonds, which were stronger than the homophilic N/N bonds. The measured average bond energies do not correlate with reported cell sorting behavior (20), suggesting that additional mechanisms contribute to cell sorting in vivo.
Materials and Methods
Chemicals.
1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and 1,2-ditridecanoyl-sn-glycero-3-phosphocholine (DTPC) were obtained from Avanti Polar Lipids (Alabaster, AL). 6-[9-[2,3-Bis(dodecyloxy)propyl]-3,6,9-trioxanonyl-1-oxycarboxylamino]-2-[di(carboxymethyl)-amino]-hexanoic acid (NTA-TRIG-DLGE) was custom-synthesized by Northern Lipids (Vancouver, BC). Tris buffer was purchased from Sigma (St. Louis, MO), and all high-purity salts were from Aldrich (St. Louis, MO).
Cell Lines.
The ectodomains of Xenopus cleavage-stage cadherin (C-CAD-Fc), canine epithelial cadherin (E-CAD-Fc), and chicken neural cadherin (N-CAD-Fc) were expressed in stably transfected cell lines. The ectodomains were expressed with the Fc domains fused to the C terminus of the cadherin ectodomain (EC1–5) just before the transmembrane sequence. The full-length ectodomain of Xenopus cleavage-stage cadherin fused with human Fc domain (C-CAD-Fc) was purified from cell culture supernatants as described in ref. 46.
The full ectodomain of canine epithelial cadherin fused to the human Fc domain (E-CAD-Fc) was purified from cultures of stably transfected 293 HEK cells (a gift from James Nelson, Stanford University, Stanford, CA) (47). The 293 HEK cells (CRL1573, American Type Culture Collection, Manassas, VA) were stably transfected with a pcDNA3.1 vector (Invitrogen, Carlsbad, CA) containing the E-CAD-Fc insert. The cells were selected by using 0.4 mg/ml geneticin, and clones with the highest cadherin expression levels were identified by Western blot (ECL Western blot analysis system, Amersham Biosciences, Uppsala, Sweden) using an anti-human IgG HRP conjugate (Promega, Madison, WI).
The plasmid pCECH (a gift from Rene-Marc Mege, Institut National de la Santé et de la Recherche Médicale, Paris, France) encoding the full-length ectodomain of chicken neural cadherin fused to the mouse Fc domain (N-CAD-Fc) is described in ref. 48. The N-CAD-Fc insert was cloned into a different vector. The pCECH plasmid was restricted with XbaI and NotI, and the DNA fragment encoding N-CAD-Fc was ligated into the pcDNA3.1(−) vector (Invitrogen) by using T4 DNA ligase (New England Biolabs, Beverly, MA). Proper insertion of the DNA fragment was verified by DNA sequencing (at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois). The resulting plasmid was used to stably transfect 293 cells (CRL1573, American Type Culture Collection). Colonies were selected by using 0.4 mg/ml geneticin (Invitrogen), and protein expression levels were determined by Western blot using the anti-mouse IgG HRP conjugate (Promega).
Protein Expression and Purification.
CHO-K1 cells expressing C-CAD-Fc (Xenopus cadherin with human Fc) were cultured in complete glutamine-free Glasgow MEM with 10% dialyzed FCS and 25 μM methionine sulfoximine (Sigma–Aldrich, St. Louis, MO) (46, 49). Cells (293 HEK) expressing E-CAD-Fc or N-CAD-Fc were cultured in DMEM containing 10% FBS and 0.4 mg/ml geneticin. During the protein collection phase, the culture medium was switched to serum-free DMEM to simplify the purification and increase protein yields.
E-CAD-Fc and N-CAD-Fc were purified from the serum-free conditioned medium on a protein A Affigel affinity column (Bio-Rad, Hercules, CA) (47, 48). C-CAD-Fc was purified with a protein A affinity column, followed by gel-filtration chromatography (46). The protein purity was assessed by SDS/PAGE.
Bead Aggregation Assay.
Cadherin-induced aggregation of fluorescent beads was quantified by flow cytometry. The beads were carboxylate-modified, 0.9-μm-diameter fluorescent microspheres (Bangs Labs, Fishers, IN). The latex beads were chemically activated with nitrilotriacetic acid as described. They were then washed twice by centrifugation and resuspension in buffer (100 mM NaNO3/50 mM Tris, pH 7.5). Then, 150 μg of the beads was incubated with a 5 μM concentration of the histidine-tagged dimer of the B domain of protein A, His6SpAB2 (21), for 1 h. The beads were again washed twice with buffer to remove unbound protein and then incubated with 1 μg of cadherin in 25 μl of the calcium-free buffer. Red fluorescent beads coated with one type of cadherin were then mixed with an identical volume of green fluorescent beads coated with the same or a second type of cadherin to give a total volume of 50 μl. The mixture was sonicated for <30 s to generate single microspheres (>80% as determined by flow cytometry). Ca2+ addition to 2 mM induced aggregation. No calcium was added to the negative controls. The extent of aggregation, as determined by the aggregate sizes and aggregate composition, was quantified with a BD LSR II flow cytometer (BD Biosciences, San Jose, CA) (at the W. M. Keck Center for Comparative and Functional Genomics). One microliter of the sample was withdrawn after 90 min, diluted 200-fold with calcium-containing buffer, and analyzed with the flow cytometer. Each aggregate was recorded as an event. A 2D density plot of the intensity of red fluorescence versus green fluorescence in each aggregate shows the size distribution and composition of the aggregates. The percentage of aggregates containing more than one red or green bead indicated the propensity for heterophilic binding. Aggregates containing only a single red or green bead were discounted, because such events could be due to nonspecific adhesion or physical entrapment.
Sample Preparation for Force Measurements.
The Fc cadherins were immobilized on lipid bilayers supported on freshly cleaved, atomically smooth mica sheets. We used a dimer of the B domain of staphylococcal protein A fused with a C-terminal hexahistidine tag (His6SpAB2) (21) to immobilize the Fc-CADs on a nitrilotriacetic acid-lipid bilayer (10). A monolayer of DPPE at 43 Å2 per lipid was first deposited onto the surface of the freshly cleaved mica surface (22, 50). A second lipid layer, containing either 75% NTA-TRIG-DLGE and 25% 1,2-ditridecanoyl-sn-glycero-3-phosphocholine (DTPC) or 100% NTA-TRIG-DLGE, was deposited on the DPPE layer at 25°C and a surface pressure of 35 mN/m (≈65 Å2 per lipid). The resulting bilayer was incubated for 1 h with 5 μM His6SpAB2 in 50 mM Tris/100 mM NaNO3/50 μM NiSO4/2 mM Ca(NO3)2 at pH 7.8 (21). The supported protein A monolayer was then incubated for 3 h with 0.05–0.2 μM Fc-CAD in the same buffer. The concentration of bulk protein determines the final cadherin coverage on the supported membranes. After removing nonspecifically bound protein, the discs were transferred to the SFA chamber. Measurements were performed at 25 ± 1°C.
Surface Force Measurements.
To establish the absolute distance D between the bilayers, we first calibrated the wavelengths of interference fringes at contact between DPPE monolayers in air. Subsequent deposition of NTA-TRIG-DLGE/DTPC lipid and protein monolayers increased the distance of closest approach T (Fig. 1a) between the DPPE monolayers. This change is quantified within ±1 Å. Between two bilayers, the absolute separation is then D = T − 2TNTA-TRIG-DLGE (50), where TNTA-TRIG-DLGE is the thickness of the NTA-TRIG-DLGE monolayer (51) (Fig. 1a).
Normalized forces between oriented Fc-tagged cadherin molecules were measured with the Mark II SFA (52). The SFA quantifies the integrated force between two macroscopic surfaces as a function of the absolute separation D (53, 54). The distances are measured by interferometry with a resolution of ±1 Å (55). A spring supporting one of the silica lenses quantifies the force between the surfaces (52). The total measured force between the curved discs (Fc) is proportional to the energy per unit area between equivalent flat plates (Ef) according to Fc = 2πREf (55, 56), where R is the geometric average radius of the two discs: R = .
The pull-off force (Fpo) to separate the adhesive layers is related to the adhesion energy per unit area EA between the surfaces by the Derjaguin–Müller–Toporov theory by EA = Fpo/2πR (57). Normalizing the adhesion energy by Γ, which is the cadherin surface density determined by plasmon resonance and radiolabeling, allows comparisons between measurements with different protein preparations. Assuming an equilibrium distribution between bound and free states, the estimated bond energy is EA = Γ(Eb/(1 + exp(−Eb/kBT))) (58). Because stresses may be unevenly distributed in the contact region, the above estimate is a lower bound.
The distance resolution (±1 Å) allows precise control of the bilayer (and protein) separation. Upon separation, at the point of adhesive failure (F < 0) or the maximum attractive force (57), the surfaces jump out of contact. Each measurement takes 5–10 min. We controlled the intersurface distances, and hence the degree of ectodomain overlap, at separations of D < 370 Å (full ectodomain overlap) and 390 Å < D < 530 Å (partial overlap). For each measurement, bond rupture occurred at discrete distances corresponding to particular alignments between opposing proteins (Fig. 1b) (9, 59, 60). Each experiment reflects more than five measurements of each bound state at two to three separate positions on the 1-cm2 sample, and each experiment was repeated at least twice.
Cadherin Surface Densities.
The adhesion energies are normalized by the measured protein coverage Γ to account for small differences (<25%) in protein densities. The lipid layer composition and bulk protein concentration used for cadherin immobilization determined the immobilized Fc-CAD coverage.
Surface plasmon resonance measurements quantified the amount of immobilized cadherin (61). A monolayer of NTA-TRIG-DLGE lipid at the same composition as that used in the surface force measurements was deposited on a dodecanethiol monolayer on gold by Langmuir–Blodgett deposition. Subsequently, His6SpAB2 and then Fc-tagged cadherins were immobilized as described in ref. 10. From measured shifts in the plasmon resonance angle, we determined the optical thickness of the adsorbed protein layer by fitting the resonance curves to the Fresnel equations for a multilayer film (61). We determined the protein surface density from the effective optical thickness by using a refractive index of 1.44 for a pure protein layer and a thickness of 270 Å. The latter is based on the 45-Å length of the Fc domain plus the 225-Å length of the cadherin ectodomain (16).
Accurate measurements were obtained with 125[I]-labeled Fc-CAD (Pierce, Rockford, IL) (62). The radiolabeling measurements were also used to calibrate the surface plasmon resonance data.
Acknowledgments
We thank Saiko Rosenburger for excellent technical assistance. This work was supported by National Institutes of Health Grant 5 R01 GM51338.
Abbreviations
- EC
extracellular
- ECn
EC domain n
- CAD
cadherin
- SFA
surface forces apparatus
- DPPE
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine
- NTA-TRIG-DLGE
6-[9-[2,3-bis(dodecyloxy)propyl]-3,6,9-trioxanonyl-1-oxycarboxylamino]-2-[di(carboxymethyl)-amino]-hexanoic acid.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gumbiner BM. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg MS. Proc Natl Acad Sci USA. 1962;48:1577–1582. doi: 10.1073/pnas.48.9.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg MS, Takeichi M. Proc Natl Acad Sci USA. 1994;91:206–209. doi: 10.1073/pnas.91.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeichi M. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 5.Takeichi Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 6.Beysens DA, Forgacs G, Glazier JA. Proc Natl Acad Sci USA. 2000;97:9467–9471. doi: 10.1073/pnas.97.17.9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap AS, Brieher WM, Gumbiner BM. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- 8.Takeichi M. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 9.Sivasankar S, Brieher W, Lavrik N, Gumbiner B, Leckband D. Proc Natl Acad Sci USA. 1999;96:11820–11824. doi: 10.1073/pnas.96.21.11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu B, Chappuis-Flament S, Wong E, Jensen I, Gumbiner BM, Leckband DE. Biophys J. 2003;41:12163–12170. doi: 10.1016/S0006-3495(03)75129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayas M, Leung A, Evans E, Leckband D. Biophys J. 2006;90:1385–1395. doi: 10.1529/biophysj.105.069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perret E, Leung A, Feracci H, Evans E. Proc Natl Acad Sci USA. 2005;101:16472–16477. doi: 10.1073/pnas.0402085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MG, Grübel G, Legrand J-F, Als-Nielsen J, Colman DR, Hendrickson WA. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Shan W-S, Hendrickson WA, Colman DR, Shapiro L. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 15.Pertz O, Bozic A, Koch W, Fauser C, Brancaccio A, Engel J. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boggon T, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 17.Nose A, Tsuji K, Takeichi M. Cell. 1990;61:147–155. doi: 10.1016/0092-8674(90)90222-z. [DOI] [PubMed] [Google Scholar]
- 18.Ahrens T, Pertz O, Häussinger D, Fauser C, Schulthess T, Engel J. J Biol Chem. 2002;277:19455–19460. doi: 10.1074/jbc.M200606200. [DOI] [PubMed] [Google Scholar]
- 19.Duguay D, Foty RA, Steinberg MS. Dev Biol. 2003;253:309–323. doi: 10.1016/s0012-1606(02)00016-7. [DOI] [PubMed] [Google Scholar]
- 20.Niessen C, Gumbiner BM. J Cell Biol. 2002;156:389–399. doi: 10.1083/jcb.200108040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CP, Jensen IE, Prakasam A, Vijayendran R, Leckband D. Bioconjug Chem. 2003;14:974–978. doi: 10.1021/bc034063t. [DOI] [PubMed] [Google Scholar]
- 22.Sivasankar S, Gumbiner B, Leckband D. Biophys J. 2001;80:1758–1768. doi: 10.1016/S0006-3495(01)76146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prakasam A, Chien Y-H, Maruthamuthu V, Leckband DE. Biochemistry. 2006;45:6930–6939. doi: 10.1021/bi060213m. [DOI] [PubMed] [Google Scholar]
- 24.Johnson CP, Fujimoto I, Rutishauser U, Leckband D. J Biol Chem. 2005;280:137–145. doi: 10.1074/jbc.M410216200. [DOI] [PubMed] [Google Scholar]
- 25.Chien PC, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Proc Natl Acad Sci USA. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Proc Natl Acad Sci USA. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumgartner W, Golenhofen N, Grundhoefer, Wiegand JH, Drenckhahn D. J Neurosci. 2003;23:11008–11014. doi: 10.1523/JNEUROSCI.23-35-11008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panorchan P, Thompson MS, Davis KJ, Tseng Y, Konstantopoulos K, Wirtz D. J Cell Sci. 2006;119:66–74. doi: 10.1242/jcs.02719. [DOI] [PubMed] [Google Scholar]
- 29.Nose A, Nagafuchi A, Takeichi M. Cell. 1988;54:147–155. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- 30.Shankaran H, Neelamegham S. Biophys J. 2004;86:576–588. doi: 10.1016/S0006-3495(04)74136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldsmith HL, Quin AT, Drury G, Spanos C, McIntosh FA, Simon SI. Biophys J. 2001;81:2020–2034. doi: 10.1016/S0006-3495(01)75852-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurenzi IJ, Diamond SL. Biophys J. 1999;77:1733–1746. doi: 10.1016/S0006-3495(99)77019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long M, Goldsmith HL, Tees DFJ, Zhu C. Biophys J. 1999;76:1112–1128. doi: 10.1016/S0006-3495(99)77276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedlander DR, Mege RM, Cunningham BA, Edelman GM. Proc Natl Acad Sci USA. 1989;86:7043–7047. doi: 10.1073/pnas.86.18.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas W, Edelman B, Lobel S, Breithart A, Steinberg M. J Supramol Struct Cell Biochem. 1981;16:15–27. doi: 10.1002/jsscb.1981.380160103. [DOI] [PubMed] [Google Scholar]
- 36.Brackenbury R, Rutishauser U, Edelman G. Proc Natl Acad Sci USA. 1981;78:387–391. doi: 10.1073/pnas.78.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volk T, Cohen O, Geiger B. Cell. 1987;50:987–994. doi: 10.1016/0092-8674(87)90525-3. [DOI] [PubMed] [Google Scholar]
- 38.Luo Y, Ferreira-Cornwall M, Baldwin H, Kostetskii I, Lenox J, Lieberman M, Radice G. Development (Cambridge, UK) 2001;128:459–469. doi: 10.1242/dev.128.4.459. [DOI] [PubMed] [Google Scholar]
- 39.Foty RA, Steinberg MS. Dev Biol. 2004;278:255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Gumbiner BM. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 41.Haussinger D, Ahrens T, Sass H-J, Pertz O, Engel J, Grzesiek S. J Mol Biol. 2002;324:823–839. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- 42.Koch A, Bozic D, Pertz O, Engel J. Curr Opin Struct Biol. 1999;9:275–281. doi: 10.1016/S0959-440X(99)80038-4. [DOI] [PubMed] [Google Scholar]
- 43.Nagar B, Overduin M, Ikura M, Rini JM. Nature. 1995;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 44.Pokutta S, Herrenknecht K, Kemler R, Engel J. Eur J Biochem. 1994;223:1019–1026. doi: 10.1111/j.1432-1033.1994.tb19080.x. [DOI] [PubMed] [Google Scholar]
- 45.Tomschy A, Charlotte F, Landwehr R, Engel J. EMBO J. 1996;15:3507–3514. [PMC free article] [PubMed] [Google Scholar]
- 46.Chappuis-Flament S, Wong E, Hicks LD, Kay CM, Gumbiner BM. J Cell Biol. 2001;154:231–243. doi: 10.1083/jcb.200103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen YT, Nelson WJ. Anal Biochem. 1996;242:276–278. doi: 10.1006/abio.1996.0466. [DOI] [PubMed] [Google Scholar]
- 48.Lambert M, Padilla F, Mege RM. J Cell Sci. 2000;113:2207–2219. doi: 10.1242/jcs.113.12.2207. [DOI] [PubMed] [Google Scholar]
- 49.Brieher WM, Yap AS, Gumbiner BM. J Cell Biol. 1996;135:487–496. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marra J, Israelachvili J. Biochemistry. 1985;24:4608–4618. doi: 10.1021/bi00338a020. [DOI] [PubMed] [Google Scholar]
- 51.Martel L, Johnson C, Boutet S, Al-Kurdi R, Konovalov O, Robinson I, Leckband D, Legrand J-F. J Phys IV. 2002;12:365–377. [Google Scholar]
- 52.Israelachvili JN, Adams GE. J Chem Soc Faraday Trans I. 1978;75:975–1001. [Google Scholar]
- 53.Israelachvili J. Surface Sci Rep. 1992;14:110–159. [Google Scholar]
- 54.Leckband D, Israelachvili JN. Q Rev Biophys. 2001;34:105–267. doi: 10.1017/s0033583501003687. [DOI] [PubMed] [Google Scholar]
- 55.Israelachvili J. J Colloid Interface Sci. 1973;44:259–272. [Google Scholar]
- 56.Hunter R. Foundations of Colloid Science. Oxford: Oxford Univ Press; 1989. [Google Scholar]
- 57.Israelachvili J. Intermolecular and Surface Forces. New York: Academic; 1992. [Google Scholar]
- 58.Tereste D, Pincet F, Brellier M, Mioskowski C, Perez E. J Am Chem Soc. 2005;127:3879–3884. doi: 10.1021/ja043525q. [DOI] [PubMed] [Google Scholar]
- 59.Zhu B, Davies EA, vanderMerwe PA, Calvert T, Leckband D. Biochemistry. 2002;41:12163–12170. doi: 10.1021/bi020296g. [DOI] [PubMed] [Google Scholar]
- 60.Johnson CP, Fujimoto I, Perrin-Tricaud C, Rutishauser U, Leckband D. Proc Natl Acad Sci USA. 2004;101:6963–6968. doi: 10.1073/pnas.0307567100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lavrik N, Leckband D. Langmuir. 2000;16:1842–1851. [Google Scholar]
- 62.Yeung C, Purves T, Kloss AA, Kuhl TL, Sligar S, Leckband D. Langmuir. 1999;15:6829–6836. [Google Scholar]