Abstract
FGF-2 is an unconventionally secreted lectin that transmits proangiogenic signals through a ternary complex with high-affinity FGF receptors and heparan sulfate proteoglycans (HSPGs). Although FGF-2 signal transduction is understood in great detail, its mechanism of release from cells, which is independent of the classical secretory pathway, remains elusive. To test the hypothesis that FGF-2 secretion is linked to its cell-surface ligands, we studied FGF-2 release using mutants defective for HSPG binding and cells with impaired HSPG biosynthesis. Here, we report that a functional interaction between FGF-2 and HSPGs is required for net export of FGF-2 from mammalian cells. FGF-2 release requires extracellular, membrane-proximal HSPGs. We propose that extracellular HSPGs form a molecular trap that drives FGF-2 translocation across the plasma membrane.
Soluble proteins destined for secretion are characterized by N-terminal signal peptides that direct their translocation into the lumen of the endoplasmic reticulum (1, 2). Secretory proteins are subsequently packaged into transport vesicles and delivered to the Golgi apparatus and eventually reach the cell surface through vesicular transport (3–7). A growing number of soluble secretory proteins are being identified that lack signal peptides and do not appear to rely on vesicular transport for export to the extracellular space (8–12). These proteins exit the cell in a process termed unconventional secretion (10, 12).
A number of biomedically significant proteins are unconventionally secreted. These include the proangiogenic lectins FGF-1 and FGF-2 (13–21), the inflammatory cytokines interleukin 1β (22–24) and macrophage migration inhibitory factor (MIF) (25), and galectins, which are β-galactoside-specific lectins of the extracellular matrix that function in development and immune regulation (26–34). Consistent with their critical functions, unconventional secretory processes are, in most cases, specifically regulated (reviewed in refs. 9 and 10). For example, interleukin 1β is secreted upon activation of monocytes (22–24), MIF secretion from monocytes is triggered by bacterial lipopolysaccharides (35, 25), secretion of galectin-1 is up-regulated during differentiation (26, 36), externalization of FGF-1 is triggered by stresses such as heat shock (15, 19), and FGF-2 secretion is enhanced by shear stress (37). These and other observations suggest that there are multiple, mechanistically distinct nonclassical export routes (12). However, the molecular components involved in membrane translocation of unconventionally secreted proteins have not been identified.
Extracellular FGF-2 transmits signals to target cells as part of a ternary complex with cell-surface heparan sulfate proteoglycans (HSPGs) and high-affinity FGF receptors (FGFRs) (38–40). FGF-2 has been reported to participate in autocrine signaling under various physiological conditions (41, 14). This finding raises the possibility that FGF-2 secretion and signaling are linked. Because CHO cells lack FGFRs (42) but still secrete FGF-2 (21, 43), we chose in this study to examine the potential role of HSPGs in FGF-2 export. We engineered FGF-2 C-terminal truncation mutants that are deficient in HSPG binding. When expressed in CHO cells, these mutants were found to be export-incompetent. Conversely, we found that cells defective for heparan sulfate biosynthesis were not able to secrete wild-type FGF-2. FGF-2 secretion from HSPG-deficient cells was rescued by cocultivation with cells that present HSPGs on their surface. Importantly, recovery of FGF-2 secretion required that the two cell populations be in close proximity, because their spatial separation resulted in complete failure of FGF-2 export.
Results and Discussion
Generation of FGF-2 Mutants Deficient in Binding to HSPGs.
Two lysine-rich surface loops near the C terminus of FGF-2 have been implicated in specific binding to HSPGs (44–46). To investigate a potential role of HSPGs in FGF-2 export, we constructed C-terminally truncated FGF-2 mutants fused to GFP (see and Supporting Experimental Procedures, which are published as supporting information on the PNAS web site). The mutants were stably introduced into CHO cells by using a retroviral vector system that allows for their doxycycline-dependent expression (21). As shown in Fig. 5B, after induction with doxycycline, the FGF-2 mutant proteins were produced at levels comparable with full-length FGF-2. We prepared cell-free supernatants from each of the cell lines, normalized them for FGF-2 concentration based on GFP fluorescence, and analyzed the capacity of the FGF-2 mutants to bind heparin (Fig. 5C) and cell-surface HSPGs (Fig. 5D). Compared with wild-type FGF-2, each of the mutants was deficient in binding to both heparin and HSPGs, demonstrating that deletion of the FGF-2 C terminus causes its failure to bind to the cell surface.
FGF-2 Mutants Defective in Binding to HSPGs Are Not Exported from Mammalian Cells.
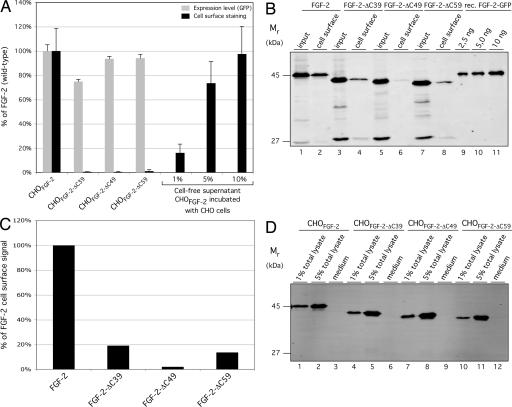
To analyze whether the inability of FGF-2 mutants to bind to HSPGs has an impact on export efficiency, secretion experiments were conducted by using the stable cell lines described above. We quantified the amount of FGF-2 bound to the cell surface and released into the medium. Under steady-state conditions, ≈10% of wild-type FGF-2-GFP was associated with the cell surface, as measured by flow cytometry and comparison with defined amounts of FGF-2-GFP from cell-free supernatants (Fig. 1A) (21). By contrast, consistent with the data shown in Fig. 5, none of the FGF-2 C-terminal truncation mutants were detected on the cell surface (Fig. 1A). These results were confirmed by cell-surface biotinylation experiments (Fig. 1B). Approximately 8.2% of wild-type FGF-2-GFP was detected at the cell surface under steady-state conditions (Fig. 1B, lanes 1 and 2). In contrast, there was a significant reduction in cell surface-associated mutant FGF-2-GFP (Fig. 1B, lanes 3–8; see Fig. 1C for quantification). These data are consistent with our flow-cytometry analysis (Fig. 1A). We also calculated the absolute amount of wild-type FGF-2-GFP associated with the cell surface by comparing its signal (Fig. 1B, lanes 1 and 2) with defined amounts of recombinant FGF-2-GFP (Fig. 1B, lanes 9–11). Under steady-state conditions (16-h expression in the presence of doxycycline), we found 14 ng of FGF-2-GFP associated with the cell surface of 150,000 cells, which corresponds to ≈1.25 × 106 FGF-2-GFP molecules per cell.
Fig. 1.
FGF-2 mutants deficient in binding to HSPGs fail to get exported from mammalian cells. (A) CHO cells (7 × 104) were induced with doxycycline for 16 h at 37°C to achieve similar expression levels of the reporter molecules indicated. Cells were decorated with anti-GFP primary and APC-coupled secondary antibodies and analyzed for both total FGF-2-GFP protein (GFP fluorescence) and FGF-2-GFP cell-surface localization (APC-derived fluorescence). To quantify the cell surface-bound fraction in relation to total FGF-2-GFP, defined amounts of cell-free supernatants (percentage of totally expressed material) prepared from 7 × 104 CHOFGF-2-GFP cells were incubated with equal numbers of nontransduced CHO cells for 30 min at 4°C (n = 3). (B) CHO cells (1.5 × 105) were cultivated as described above. Cells were incubated with a membrane-impermeable biotinylation reagent. After detergent-mediated cell lysis, biotinylated material was purified by using streptavidin beads. Aliquots from the cell lysate (10%), the cell surface-biotinylated fractions (45%), and defined amounts of recombinant FGF-2-GFP (lanes 9–11) were analyzed by Western blotting. Antigens were quantified with an Odyssey infrared imaging system (LI-COR Biosciences). (C) The biotinylation signals in B were quantified by using the Odyssey infrared imaging system (LI-COR Biosciences) and graphed as the percentage of wild-type FGF-2, which was set to 100%. (D) CHO cells were cultivated for 16 h at 37°C to a confluency of ≈80%. Growth medium was centrifuged (500 × g for 20 min at 4°C), and cells attached to the culture dish were used to prepare total lysates. Both medium and defined amounts of cell lysates were incubated with Ni-NTA-agarose for 1 h at 4°C. Bound material was analyzed by Western blotting as described in Experimental Procedures.
Because the FGF-2 truncation mutants are unable to bind to HSPGs, it is possible that these mutants are secreted but not retained on the cell surface. Because the FGF-2-GFP fusions also bear a C-terminal hexahistidine tag (Fig. 5A), we collected FGF-2 from the culture medium using Ni-affinity chromatography and analyzed affinity-purified proteins by SDS/PAGE and Western blotting. In parallel, defined amounts of total cell lysates were used to establish the lower limit of sensitivity of our detection system. As shown in Fig. 1D, 1% of the total population of each FGF-2-GFP was easily detectable, which is at least one order of magnitude more sensitive than the amount of cell-surface FGF-2 observed under steady-state conditions (Fig. 1 A and B) (21). When the media of cells expressing wild-type FGF-2 or the FGF-2 mutants deficient for HSPG binding were analyzed, none of the fusion proteins were detectable (Fig. 1D). This finding was expected for wild-type FGF-2, because it is not released into the medium of cultured cells but, rather, remains associated with cell-surface HSPGs upon secretion (20, 21). That we were unable to detect the FGF-2 truncation mutants in the medium indeed suggests a block in secretion, because they were detected neither on the cell surface (Fig. 1 A and B) nor in the medium (Fig. 1D). Because binding to HSPGs has been reported to protect FGF-2 from degradation (47, 48), we analyzed whether the C-terminally truncated mutants were simply degraded after secretion. Under the experimental conditions used, all mutants displayed a stability comparable with, if not better than, wild-type FGF-2 (see Supporting Experimental Procedures). Partial degradation of FGF-2 reporter molecules results in the accumulation of a protease-resistant GFP fragment as an end product. Because not even the GFP fragment was observed in the medium of cells that express FGF-2 mutants (Fig. 1D), we conclude that degradation does not explain their absence from the medium. In conclusion, FGF-2 mutants that cannot bind cell-surface HSPGs are not exported from mammalian cells, because they are detectable neither on the cell surface nor in the supernatant of producer cells.
Cells Deficient for Functional Cell-Surface HSPGs Do Not Export FGF-2.
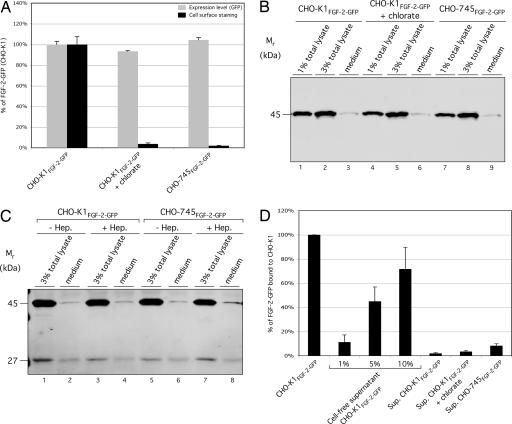
Our finding that FGF-2 mutants impaired for binding to HSPGs are not secreted (Fig. 1) does not demonstrate a direct role for HSPGs in FGF-2 secretion. Our C-terminal truncations could have deleted an export signal or folded the molecule in a way incompatible with secretion. Therefore, we conducted the opposite experiment, analyzing whether wild-type FGF-2 is exported from cells lacking HSPGs (Fig. 2). In the first set of experiments, cells were treated with chlorate, a compound that blocks heparan sulfate biosynthesis (49–51). A second approach made use of a CHO mutant cell line (CHO-745) that lacks the enzyme xylosyl transferase and, therefore, is incapable of synthesizing heparan sulfate chains on the core protein of proteoglycans (52, 53). In all experimental conditions, the cells produced similar amounts of FGF-2-GFP (Fig. 2A, gray bars). Although we were able to detect ≈10% of the total FGF-2-GFP on the surface of wild-type, untreated CHO cells (Fig. 2 A and D), FGF-2-GFP was not detectable on the surface of chlorate-treated cells or HSPG-deficient CHO-745 cells (Fig. 2A, black bars).
Fig. 2.
Full-length FGF-2 fails to get exported from cells lacking surface HSPGs. (A) CHO cells (7 × 104) were cultivated for 16 h at 37°C in the presence of varying doxycycline concentrations to achieve similar expression levels of FGF-2-GFP. Where indicated, the growth medium was supplemented with 75 mM sodium chlorate. Cells were decorated with anti-GFP primary and APC-conjugated secondary antibodies. Total fusion protein (GFP fluorescence) and cell surface-localized protein (APC-derived fluorescence) were determined by flow cytometry (n = 4). (B) CHO cells were grown to a confluency of ≈80%. The medium was centrifuged (500 ×g for 20 min at 4°C), and cells attached to the culture dish were used to prepare total lysates. Medium and defined amounts of cell lysates (as indicated) were incubated with heparin Sepharose for 1 h at 4°C. Bound material was analyzed by Western blotting as described in Materials and Methods. (C) CHO cells were grown and processed as in B. Where indicated, growth medium was supplemented with 125 μg/ml heparin (Hep.). Both medium and defined amounts of cell lysates (as indicated) were subjected to immunoprecipitation using affinity-purified anti-GFP antibodies. Bound material was eluted with SDS sample buffer, followed by Western blotting. Antigen detection was conducted as described in Materials and Methods. (D) CHO cells (7 × 104) were cultivated as described above. Cell culture supernatants were centrifuged (500 × g for 20 min at 4°C) and incubated with 7 × 104 nontransduced CHO-K1 cells for 30 min at 4°C. Cells were decorated with anti-GFP primary and APC-coupled secondary antibodies and analyzed by flow cytometry. To estimate the quantity of cell surface-bound material, defined amounts of cell-free supernatant prepared from 7 × 104 CHO-K1FGF-2-GFP cells were added as indicated to the same number of CHO-K1 cells and analyzed the same way. In addition, the APC-derived signal at the surface of CHO-K1FGF-2-GFP cells was determined (n = 4).
Because FGF-2-GFP was not found on the surface of HSPG-deficient cells, we analyzed whether it was secreted into the medium. Because the FGF-2-GFP construct expressed in CHO-745 cells does not contain a His-tag, we isolated FGF-2-GFP using heparin-affinity chromatography. We found that the supernatants of neither wild-type, untreated CHO cells nor HSPG-deficient cells contained significant amounts of FGF-2-GFP, because only a faint band representing <0.1% of total FGF-2-GFP was detected (Fig. 2B). Similar results were obtained when cells were grown in medium supplemented with heparin (Fig. 2C) to prevent potential degradation of FGF-2-GFP after secretion (see Supporting Information). Thus, even under these conditions, FGF-2-GFP was not detected in the medium of CHO-745 cells. It is important to note that, in these experiments (Fig. 2C), we used anti-GFP antibodies to immunoprecipitate FGF-2-GFP to detect the stable GFP fragment that remains after fusion protein degradation (see supporting information) (54). Because neither full-length FGF-2-GFP nor the GFP degradation product were detected to a significant extent, we conclude that degradation of FGF-2-GFP fusion proteins does not explain their absence from the medium.
We used an independent method to confirm the absence of FGF-2-GFP fusions from cell supernatants. Media obtained from FGF-2-GFP-expressing CHO wild-type and CHO-745 cells were incubated with nontransduced wild-type CHO cells (which express HSPGs). We used flow cytometry to quantify FGF-2-GFP associated with acceptor cell surfaces. As shown in Fig. 2D, and consistent with our biochemical data, none of the supernatants contained significant amounts of FGF-2-GFP. In conclusion, FGF-2-GFP fusion proteins secreted from wild-type cells are quantitatively retained on the surface. Approximately 10% of the total population is exposed on the extracellular face under steady-state conditions. By contrast, FGF-2-GFP fails to get exported from cells lacking functional HSPGs, because it is detected on neither their cell surfaces nor in their supernatants.
Rescue of FGF-2 Secretion from HSPG-Deficient Cells by Coculture with HSPG-Expressing Cells.
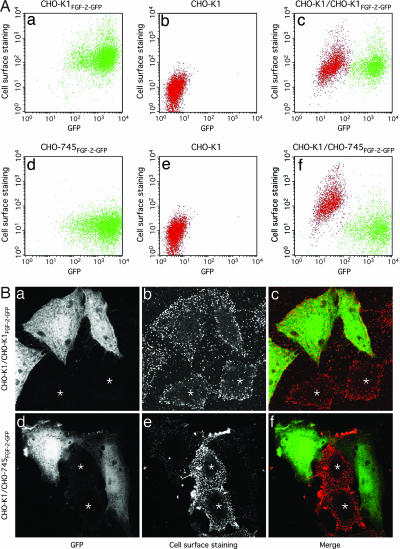
To further investigate the role of HSPGs in FGF-2 secretion, we asked whether FGF-2 export from HSPG-deficient cells can be rescued by the presence of cells expressing functional HSPGs. For this purpose, we mixed HSPG-deficient CHO-745 cells (52, 53) that express FGF-2-GFP with nontransduced, wild-type CHO cells. The cell mixture was incubated on culture plates for 16 h at 37°C in the presence of doxycycline. As shown by both flow cytometry (Fig. 3A) and confocal microscopy (Fig. 3B), FGF-2-GFP synthesized by HSPG-deficient CHO-745 cells was indeed found on the surface of cocultivated wild-type CHO cells that do not express FGF-2-GFP. By using flow cytometry, both CHOFGF-2-GFP (Fig. 3Aa, green) and CHO-745FGF-2-GFP cells (Fig. 3Ad, green) are easily recognized by GFP fluorescence (x axis). As expected, CHO-745 cells lack FGF-2-GFP cell-surface staining (Fig. 3Ad, y axis). By contrast, nontransduced CHO cells cultivated without FGF-2-GFP-expressing cells lack both GFP and cell-surface staining (Fig. 3 Ab and Ae, red). When FGF-2-GFP-containing CHO-745 cells were grown as a mixed culture with nontransduced wild-type CHO cells, the latter acquired FGF-2-GFP cell-surface staining, as indicated by a strong increase in allophycocyanin (APC)-derived fluorescence (Fig. 3Af). Similar observations were made when FGF-2-GFP-expressing wild-type CHO cells were cocultivated with nontransduced wild-type CHO cells; however, in this case, both donor and acceptor cells were positive for FGF-2-GFP cell-surface staining because both populations express cell-surface HSPGs (Fig. 3Ac). Importantly, under the experimental conditions used, neither CHO-745 nor wild-type FGF-2-GFP-expressing cells fused with nontransduced wild-type CHO cells, because the mutant cells remained negative for FGF-2 cell-surface staining, and the wild-type cells did not acquire substantial GFP fluorescence. These findings were confirmed by dual-color confocal microscopy (Fig. 3B): FGF-2-GFP-expressing wild-type CHO cells (Fig. 3B a–c) and CHO-745 cells (Fig. 3B d–f) both transferred secreted FGF-2-GFP to the surface of nontransduced wild-type CHO cells (labeled with an asterisk). Consistent with the flow-cytometry data, CHO-745 cells were not able to retain secreted FGF-2-GFP on the cell surface (Fig. 3Be). By contrast, in mixtures of transduced and nontransduced wild-type CHO cells, both populations had FGF-2-GFP cell-surface staining (Fig. 3Bb). In conclusion, the presence of HSPG-expressing acceptor cells is sufficient to rescue secretion of FGF-2-GFP from HSPG-deficient CHO-745 cells.
Fig. 3.
Reconstitution of FGF-2 secretion from HSPG-deficient cells by cocultivation with HSPG-expressing acceptor cells. (Aa–Af) CHO-K1FGF-2-GFP or CHO-745FGF-2-GFP cells (7 × 104) (both shown in green), were grown for 16 h at 37°C in 24-well plates either separately or in a mixed culture (1:1 ratio) with wild-type cells CHO (shown in red). Cells were decorated with anti-GFP primary and APC-conjugated secondary antibodies. Total FGF-2-GFP (GFP fluorescence) and cell surface-localized fusion proteins (APC-derived fluorescence) were analyzed by flow cytometry. (Ba–Bf) CHO-K1FGF-2-GFP or CHO-745FGF-2-GFP cells were mixed with CHO-K1 cells in a 1:1 ratio and grown on glass coverslips for 16 h at 37°C. After fixation with 3% paraformaldehyde, nonpermeabilized cells were processed with anti-GFP primary and Alexa Fluor 546-coupled secondary antibodies. Subsequently, specimens were embedded in Fluoromount G (Southern Biotechnology Associates, Birmingham, AL) and analyzed by confocal microscopy. Nontransduced CHO-K1 cells that express cell-surface HSPGs are labeled with an asterisk.
Spatial Requirements for HSPG Acceptor Cell-Mediated Rescue of FGF-2 Secretion from HSPG-Deficient Cells.
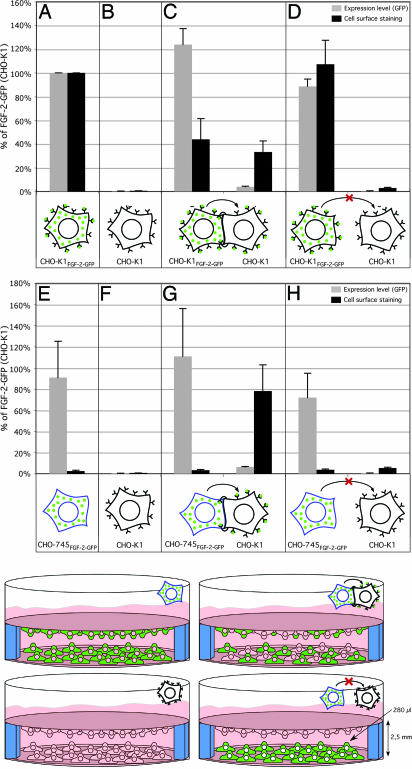
In Fig. 4A–C and E–G, we present a statistical analysis of the quantitative data (Fig. 3A) and additional controls obtained by flow cytometry. This analysis revealed that FGF-2-GFP secretion from HSPG-deficient CHO-745 cells was almost fully restored by cocultivation with HSPG-positive wild-type CHO cells (Fig. 4G). That is, nontransduced wild-type cells cultivated with FGF-2-GFP-expressing CHO-745 cells acquired an FGF-2-GFP cell-surface signal that was comparable with that of FGF-2-GFP-expressing wild-type cells (Fig. 4, compare G with A). Cell-surface fluorescence of cocultured wild-type transduced and nontransduced cells amounted to approximately half the signal obtained when FGF-2-GFP-expressing wild-type cells were grown alone (Fig. 4, compare C with A). The data from Figs. 3 and 4 emphasize the need for cell-surface HSPGs in net export of FGF-2. The data show that HSPGs act from the outside of cells rather than intracellularly to trigger FGF-2 export, because cocultured wild-type cells can complement the FGF-2 secretion defect of HSPG-deficient cells.
Fig. 4.
Spatial requirements for rescue of FGF-2 secretion from HSPG-deficient cells by wild-type acceptor cells. CHO-K1, CHO-K1FGF-2-GFP, or CHO-745FGF-2-GFP cells (8 × 104 cells per experimental condition) were seeded on glass coverslips either alone or mixed together (1:1 ratio), as indicated. Cells were cultivated at 37°C for ≈6 h. Coverslips were placed in 24-well plates, as indicated, and induced with doxycycline for 16 h at 37°C. Cells were incubated with anti-GFP primary and APC-coupled secondary antibodies and analyzed for total FGF-2-GFP (GFP fluorescence) and FGF-2-GFP cell-surface localization (APC-derived fluorescence) by using flow cytometry (n = 5).
To better understand the mechanism by which HSPGs stimulate FGF-2 export, we asked whether or not there are spatial restrictions to HSPG acceptor cell-mediated rescue of FGF-2 export from HSPG-deficient cells. FGF-2-GFP-transduced wild-type CHO cells or FGF-2-GFP-transduced CHO-745 cells were grown on coverslips. After stable attachment, they were cocultivated with nontransduced HSPG-containing acceptor cells also grown on a coverslip (see schematic in Fig. 4) and incubated for 16 h in the presence of doxycycline. Thus, FGF-2-GFP-expressing cells (wild-type or CHO-745) shared the same medium with HSPG-positive acceptor cells, but the two were unable to directly contact one other. We used flow cytometry to measure GFP fluorescence, which indicates total FGF-2-GFP expression, and APC-derived fluorescence, which indicates only cell-surface FGF-2-GFP, separately for the two cell populations. Interestingly, as opposed to the experiments shown in Fig. 4C and G, in which the two cell populations were mixed in the culture, HSPG-expressing wild-type cells were not capable of rescuing FGF-2 export from HSPG-deficient CHO-745 cells when the two populations were spatially separated (Fig. 4H). Likewise, there was almost no significant transfer of FGF-2-GFP from transduced to nontransduced wild-type CHO cells when the two populations were grown without direct cell–cell contact (Fig. 4D). These data imply that the unconventional secretion of FGF-2 depends on HSPGs in close proximity to the cell surface.
In this study, we identified HSPGs as essential components of the unconventional secretory pathway of FGF-2. This conclusion is based on our observations that (i) FGF-2 mutants deficient in binding to HSPGs are not secreted from CHO cells, and (ii) CHO cells lacking in cell-surface HSPGs do not export wild-type FGF-2. Export of FGF-2-GFP from HSPG-deficient cells can be rescued by cocultivation with HSPG-expressing acceptor cells; however, rescue is observed only when the cell populations are grown in direct contact. We had observed that FGF-2 can translocate across the membrane of affinity-purified plasma membrane inside-out vesicles (55, 12). This finding is consistent with the data presented here, in which cell-surface HSPGs from cocultivated cells can act in trans to promote net export of FGF-2 from HSPG-deficient cells. Thus, we favor a model in which FGF-2 secretion occurs by direct translocation across the plasma membrane. Our finding that FGF-2 mutants impaired for HSPG binding are also impaired for secretion might indicate that FGF-2 translocation occurs in a folded state, because the interaction between FGF-2 and HSPGs depends on the 3D structure of FGF-2 (44–46). Again, these conclusions are consistent with our earlier observation that FGF-2 export does not require its unfolding (43). In this regard, FGF-2 secretion may resemble that of double arginine-containing bacterial proteins: the plasma membrane-resident twin arginine transporters of prokaryotes and chloroplasts exclusively translocate folded substrates (56).
We propose a mechanism for FGF-2 secretion in which cell-surface HSPGs form a molecular trap that drives net export of FGF-2. In this model, FGF-2 membrane translocation from the cytoplasm to the extracellular space is made irreversible by binding of FGF-2 to the heparan sulfate chains of proteoglycans. Importantly, the binding sites must be in close proximity to the site of membrane translocation. This model resembles the mechanism by which proteins are posttranslationally translocated from the cytoplasm into the endoplasmic reticulum (ER). The molecular chaperone BiP is required in the ER lumen, where it functions as a molecular ratchet (57). This process has been reconstituted with proteoliposomes and found to require BiP-mediated ATP hydrolysis. Intriguingly, replacement of BiP with an antibody directed against the translocating protein renders the process ATP-independent (58). It is possible that FGF-2 translocation across the plasma membrane works in a similar manner: FGF-2 might be able to traverse the membrane by facilitated diffusion through a proteinaceous pore or by an as-yet-unrecognized ability to directly cross the hydrophobic core of the lipid bilayer. Heparan sulfate chains of proteoglycans may trap FGF-2 molecules that appear on the surface or help to extract FGF-2 molecules from the membrane. Both mechanisms would be compatible with our earlier finding that FGF-2 membrane translocation does not require ATP hydrolysis (55). In this regard, FGF-2 export might also resemble that of galectin-1, a component of the extracellular matrix that is unconventionally secreted (9, 10) through a mechanism thought to involve cell-surface counter receptors (34). Further work will surely establish whether lectin-like proteins such as FGF-2 and galectin-1 rely on a general mechanism for unconventional export.
Experimental Procedures
Antibodies, Cell Culture, and Generation of Stable Cell Lines.
For detection of FGF-2-GFP fusions in flow cytometry, confocal microscopy, and Western blotting, affinity-purified polyclonal rabbit anti-GFP and anti-FGF-2 (21) or monoclonal anti-GFP antibodies were used (Clontech, Mountain View, CA). As secondary antibodies (all purchased from Invitrogen, Carlsbad, CA), we used APC-conjugated anti-rabbit antibodies for flow cytometry, Alexa Fluor 546-coupled anti-rabbit antibodies for confocal microscopy, and Alexa Fluor 680-coupled anti-rabbit or anti-mouse antibodies for Western blot analyses using the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). The mammalian cell lines CHO (ECACC 85050302), CHO-K1 (ATCC CCL-61), and CHO-K1-pgsA-745 (ATCC CRL-2242; abbreviated CHO-745) were maintained according to standard protocols. FGF-2 (18-kDa isoform) and FGF-2 truncations were fused to GFP or GFP-His6, respectively. Stable CHO cell lines bearing FGF-2-GFP fusions were generated by retroviral transduction (21).
Preparation of Cell Lysates and Cell-Free Supernatants.
Expression of FGF-2-GFP fusion proteins in CHO cells was induced by the addition of doxycycline for 16 h at 37°C. For the preparation of total lysates, cells were detached with PBS containing 0.5 mM EDTA, sedimented (2,000 × g for 3 min at 4°C), and solubilized in PBS/1% Triton X-100. After a 15-min ice incubation, lysates were cleared by centrifugation (20,000 × g for 10 min at 4°C). For preparation of cell-free supernatants, cells were detached with PBS plus 0.5 mM EDTA, sedimented (2,000 × g for 3 min at 4°C), and resuspended in PBS. After homogenization using a combination of freeze/thaw and sonication, insoluble material was removed by ultracentrifugation (100,000 × g for 1 h at 4°C).
Biochemical Analysis of FGF-2 Binding to Heparin.
The concentration of FGF-2-GFP fusions in cell-free supernatants prepared from CHO cell lines was determined based on GFP fluorescence, which was measured with a fluorescence plate reader (Molecular Devices, Sunnyvale, CA). Supernatants normalized for fusion-protein content were incubated with heparin Sepharose 6 Fast Flow beads (Amersham Biosciences, Piscataway, NJ) for 1 h at 4°C. After washing with buffer (50 mM Tris, pH 7.5/150 mM NaCl/1 mM EDTA), bound material was eluted with SDS sample buffer and analyzed along with samples from the input and the flow-through fractions (unbound material) by Western blotting with affinity-purified anti-GFP antibodies.
Quantitative Analysis.
For quantitative analysis of FGF-2 export from CHO cells by flow cytometry, FGF-2 binding to cell surfaces by flow cytometry, and FGF-2 export from CHO cells by cell-surface biotinylation, see Supporting Experimental Methods.
Biochemical Analysis of FGF-2 Release from CHO Cells Using FGF-2 Affinity Purification from Cell Culture Supernatants.
CHO cells that express truncated or full-length FGF-2-GFP were induced by doxycycline for 16 h at 37°C. Cell culture medium was collected, centrifuged at 500 × g (20 min at 4°C) to remove detached cells, and incubated with Ni-NTA Agarose (truncated FGF-2; Qiagen, Valencia, CA) or heparin Sepharose (full-length FGF-2; Amersham Biosciences) for 1 h at 4°C. Alternatively, immunoprecipitations were performed by using affinity-purified rabbit anti-GFP antibodies bound to protein A Sepharose (Amersham Biosciences). As a standard, defined amounts of total lysates were also subjected to affinity purification. After washing with PBS (Ni-NTA-Agarose) or buffer (50 mM Tris, pH 7.5/150 mM NaCl/1 mM EDTA (heparin Sepharose and immunoprecipitations), bound material was analyzed by Western blotting using affinity-purified anti-FGF-2 or monoclonal anti-GFP antibodies.
Supplementary Material
Acknowledgments
We thank Julia Lenz and Blanche Schwappach (University of Heidelberg) for support with FACS analyses, Koen Temmerman (University of Heidelberg) for providing recombinant FGF-2-GFP, Jeffrey D. Esko (University of California, San Diego, CA) for providing CHO-745 mutant cells and for critical and helpful comments on the manuscript, and Tracy LaGrassa for critical reading of the manuscript. This study was supported by a grant from the German Research Foundation and by the Dietmar Hopp Foundation.
Abbreviations
- APC
allophycocyanin
- HSPGs
heparan sulfate proteoglycans.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Walter P, Gilmore R, Blobel G. Cell. 1984;38:5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport TA, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 3.Palade G. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 4.Rothman JE, Wieland FT. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 5.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 6.Mellman I, Warren G. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- 7.Nickel W, Brügger B, Wieland FT. J Cell Sci. 2002;115:3235–3240. doi: 10.1242/jcs.115.16.3235. [DOI] [PubMed] [Google Scholar]
- 8.Cleves AE. Curr Biol. 1997;7:R318–R320. doi: 10.1016/s0960-9822(06)00148-5. [DOI] [PubMed] [Google Scholar]
- 9.Hughes RC. Biochim Biophys Acta. 1999;1473:172–185. doi: 10.1016/s0304-4165(99)00177-4. [DOI] [PubMed] [Google Scholar]
- 10.Nickel W. Eur J Biochem. 2003;270:2109–2119. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 11.Prudovsky I, Mandinova A, Soldi R, Bagala C, Graziani I, Landriscina M, Tarantini F, Duarte M, Bellum S, Doherty H, Maciag T. J Cell Sci. 2003;116:4871–4881. doi: 10.1242/jcs.00872. [DOI] [PubMed] [Google Scholar]
- 12.Nickel W. Traffic. 2005;6:607–614. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 13.Rogelj S, Klagsbrun M, Atzmon R, Kurokawa M, Haimovitz A, Fuks Z, Vlodavsky I. J Cell Biol. 1989;109:823–831. doi: 10.1083/jcb.109.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mignatti P, Rifkin DB. J Cell Biochem. 1991;47:201–207. doi: 10.1002/jcb.240470303. [DOI] [PubMed] [Google Scholar]
- 15.Jackson A, Friedman S, Zhan X, Engleka KA, Forough R, Maciag T. Proc Natl Acad Sci USA. 1992;89:10691–10695. doi: 10.1073/pnas.89.22.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mignatti P, Morimoto T, Rifkin DB. J Cell Physiol. 1992;151:81–93. doi: 10.1002/jcp.1041510113. [DOI] [PubMed] [Google Scholar]
- 17.Florkiewicz RZ, Majack RA, Buechler RD, Florkiewicz E. J Cell Physiol. 1995;162:388–399. doi: 10.1002/jcp.1041620311. [DOI] [PubMed] [Google Scholar]
- 18.Jackson A, Tarantini F, Gamble S, Friedman S, Maciag T. J Biol Chem. 1995;270:33–36. doi: 10.1074/jbc.270.1.33. [DOI] [PubMed] [Google Scholar]
- 19.Shin JT, Opalenik SR, Wehby JN, Mahesh VK, Jackson A, Tarantini F, Maciag T, Thompson JA. Biochim Biophys Acta. 1996;1312:27–38. doi: 10.1016/0167-4889(96)00013-4. [DOI] [PubMed] [Google Scholar]
- 20.Trudel C, Faure-Desire V, Florkiewicz RZ, Baird A. J Cell Physiol. 2000;185:260–268. doi: 10.1002/1097-4652(200011)185:2<260::AID-JCP11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 21.Engling A, Backhaus R, Stegmayer C, Zehe C, Seelenmeyer C, Kehlenbach A, Schwappach B, Wegehingel S, Nickel W. J Cell Sci. 2002;115:3619–3631. doi: 10.1242/jcs.00036. [DOI] [PubMed] [Google Scholar]
- 22.Rubartelli A, Cozzolino F, Talio M, Sitia R. EMBO J. 1990;9:1503–1510. doi: 10.1002/j.1460-2075.1990.tb08268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, Rubartelli A. Mol Biol Cell. 1999;10:1463–1475. doi: 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, Rubartelli A. Proc Natl Acad Sci USA. 2004;101:9745–9750. doi: 10.1073/pnas.0308558101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flieger O, Engling A, Bucala R, Lue H, Nickel W, Bernhagen J. FEBS Lett. 2003;551:78–86. doi: 10.1016/s0014-5793(03)00900-1. [DOI] [PubMed] [Google Scholar]
- 26.Cooper DN, Barondes SH. J Cell Biol. 1990;110:1681–1691. doi: 10.1083/jcb.110.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindstedt R, Apodaca G, Barondes SH, Mostov KE, Leffler H. J Biol Chem. 1993;268:11750–11757. [PubMed] [Google Scholar]
- 28.Sato S, Burdett I, Hughes RC. Exp Cell Res. 1993;207:8–18. doi: 10.1006/excr.1993.1157. [DOI] [PubMed] [Google Scholar]
- 29.Cho M, Cummings RD. J Biol Chem. 1995;270:5207–5212. doi: 10.1074/jbc.270.10.5207. [DOI] [PubMed] [Google Scholar]
- 30.Cho M, Cummings RD. J Biol Chem. 1995;270:5198–5206. doi: 10.1074/jbc.270.10.5198. [DOI] [PubMed] [Google Scholar]
- 31.Cleves AE, Cooper DN, Barondes SH, Kelly RB. J Cell Biol. 1996;133:1017–1026. doi: 10.1083/jcb.133.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehul B, Hughes RC. J Cell Sci. 1997;110(Pt 10):1169–1178. doi: 10.1242/jcs.110.10.1169. [DOI] [PubMed] [Google Scholar]
- 33.Seelenmeyer C, Wegehingel S, Lechner J, Nickel W. J Cell Sci. 2003;116:1305–1318. doi: 10.1242/jcs.00312. [DOI] [PubMed] [Google Scholar]
- 34.Seelenmeyer C, Wegehingel S, Tews I, Kunzler M, Aebi M, Nickel W. J Cell Biol. 2005;171:373–381. doi: 10.1083/jcb.200506026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calandra T, Bernhagen J, Mitchell RA, Bucala R. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutomski D, Fouillit M, Bourin P, Mellottee D, Denize N, Pontet M, Bladier D, Caron M, Joubert-Caron R. Glycobiology. 1997;7:1193–1199. doi: 10.1093/glycob/7.8.1193. [DOI] [PubMed] [Google Scholar]
- 37.Gloe T, Sohn HY, Meininger GA, Pohl U. J Biol Chem. 2002;277:23453–23458. doi: 10.1074/jbc.M203889200. [DOI] [PubMed] [Google Scholar]
- 38.Baird A, Klagsbrun M. Cancer Cells. 1991;3:239–243. [PubMed] [Google Scholar]
- 39.Klagsbrun M, Baird A. Cell. 1991;67:229–231. doi: 10.1016/0092-8674(91)90173-v. [DOI] [PubMed] [Google Scholar]
- 40.Pellegrini L. Curr Opin Struct Biol. 2001;11:629–634. doi: 10.1016/s0959-440x(00)00258-x. [DOI] [PubMed] [Google Scholar]
- 41.Yayon A, Klagsbrun M. Cancer Metastasis Rev. 1990;9:191–202. doi: 10.1007/BF00046360. [DOI] [PubMed] [Google Scholar]
- 42.Rusnati M, Urbinati C, Tanghetti E, Dell'Era P, Lortat-Jacob H, Presta M. Proc Natl Acad Sci USA. 2002;99:4367–4372. doi: 10.1073/pnas.072651899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backhaus R, Zehe C, Wegehingel S, Kehlenbach A, Schwappach B, Nickel W. J Cell Sci. 2004;117:1727–1736. doi: 10.1242/jcs.01027. [DOI] [PubMed] [Google Scholar]
- 44.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 45.Faham S, Linhardt RJ, Rees DC. Curr Opin Struct Biol. 1998;8:578–586. doi: 10.1016/s0959-440x(98)80147-4. [DOI] [PubMed] [Google Scholar]
- 46.Raman R, Venkataraman G, Ernst S, Sasisekharan V, Sasisekharan R. Proc Natl Acad Sci USA. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommer A, Rifkin DB. J Cell Physiol. 1989;138:215–220. doi: 10.1002/jcp.1041380129. [DOI] [PubMed] [Google Scholar]
- 48.Coltrini D, Rusnati M, Zoppetti G, Oreste P, Isacchi A, Caccia P, Bergonzoni L, Presta M. Eur J Biochem. 1993;214:51–58. doi: 10.1111/j.1432-1033.1993.tb17895.x. [DOI] [PubMed] [Google Scholar]
- 49.Baeuerle PA, Huttner WB. Biochem Biophys Res Commun. 1986;141:870–877. doi: 10.1016/s0006-291x(86)80253-4. [DOI] [PubMed] [Google Scholar]
- 50.Safaiyan F, Kolset SO, Prydz K, Gottfridsson E, Lindahl U, Salmivirta M. J Biol Chem. 1999;274:36267–36273. doi: 10.1074/jbc.274.51.36267. [DOI] [PubMed] [Google Scholar]
- 51.Conrad HE. Methods Mol Biol. 2001;171:325–328. doi: 10.1385/1-59259-209-0:325. [DOI] [PubMed] [Google Scholar]
- 52.Esko JD, Stewart TE, Taylor WH. Proc Natl Acad Sci USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esko JD. Curr Opin Cell Biol. 1991;3:805–816. doi: 10.1016/0955-0674(91)90054-3. [DOI] [PubMed] [Google Scholar]
- 54.Chiang CF, Okou DT, Griffin TB, Verret CR, Williams MN. Arch Biochem Biophys. 2001;394:229–235. doi: 10.1006/abbi.2001.2537. [DOI] [PubMed] [Google Scholar]
- 55.Schäfer T, Zentgraf H, Zehe C, Brügger B, Bernhagen J, Nickel W. J Biol Chem. 2004;279:6244–6251. doi: 10.1074/jbc.M310500200. [DOI] [PubMed] [Google Scholar]
- 56.Robinson C, Bolhuis A. Biochim Biophys Acta. 2004;1694:135–147. doi: 10.1016/j.bbamcr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Osborne AR, Rapoport TA, van den Berg B. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 58.Matlack KE, Misselwitz B, Plath K, Rapoport TA. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.