Abstract
Environmental arsenic is a world-wide health issue, making it imperative for us to understand mechanisms of metalloid uptake and detoxification. The predominant intracellular form is the highly mephitic arsenite, which is detoxified by removal from cytosol. What prevents arsenite toxicity as it diffuses through cytosol to efflux systems? Although intracellular copper is regulated by metallochaperones, no chaperones involved in conferring resistance to other metals have been identified. In this article, we report identification of an arsenic chaperone, ArsD, encoded by the arsRDABC operon of Escherichia coli. ArsD transfers trivalent metalloids to ArsA, the catalytic subunit of an As(III)/Sb(III) efflux pump. Interaction with ArsD increases the affinity of ArsA for arsenite, thus increasing its ATPase activity at lower concentrations of arsenite and enhancing the rate of arsenite extrusion. Cells are consequently resistant to environmental concentrations of arsenic. This report of an arsenic chaperone suggests that cells regulate the intracellular concentration of arsenite to prevent toxicity.
Keywords: antimony, resistance pump, metalloid, ArsD, ArsA
All cells possess regulatory mechanisms that control the intracellular concentration of metals, including essential metals, which become toxic in excess (copper and zinc), toxic metals (cadmium and lead), and toxic metalloids (arsenite and antimonite) (1–3). For example, metallothioneins are small proteins found in both eukaryotes and prokaryotes that bind transition metal ions such as copper and zinc for storage and heavy metals such as cadmium for detoxification (4). In plants and some fungi phytochelatins are cysteine-rich peptides that bind heavy metal ions for detoxification (5). In contrast to these types of metal-binding peptides and polypeptides are metallochaperones, which sequester metals in the cytoplasm and deliver them to protein targets (1, 6, 7). In some cases the targets are metalloenzymes such as superoxide dismutase, where the Saccharomyces cerevisiae metallochaperone Lys-7 (CCS in humans) is required for insertion of copper ions (8). Although the best characterized metallochaperones are for copper, UreE is a nickel chaperone that delivers Ni2+ to urease (9), and frataxin is a chaperone that delivers iron to proteins for assembly of iron–sulfur clusters and heme (10). Other targets are membrane transporters, predominantly transport ATPases, that participate in metal ion homeostasis and detoxification such as S. cerevisiae Atx1 (Hah1 in humans), the metallochaperone for the yeast Ccc2p (ATP7A and ATP7B in humans) Cu(I)-translocating P-type ATPase (11). To date, no metallochaperones involved in detoxification of metals other than copper have been identified.
Arsenic is the most prevalent environmental toxic metal and is first on the Superfund list of hazardous substances (www.atsdr.cdc.gov/cercla/05list.html). Health effects associated with exposure to low levels of arsenite include cardiovascular and peripheral vascular disease, neurological disorders, diabetes mellitus, and various forms of cancer (12, 13). Chronic exposure to the low levels of arsenic that exist naturally in both water and soil (14) provides competitive pressure to maintain arsenic tolerance or detoxification systems in most, if not all, living organisms, including humans (15). To date, nearly every prokaryote with a sequenced genome has an ars operon, as do a number of archaea, and some budding and filamentous fungi have clusters of arsenic detoxification genes. ars operons are diverse but most often contain three genes for an arsenic-responsive transcriptional repressor (arsR), an arsenite efflux membrane protein (arsB), and an arsenate reductase (arsC) (16). Some have two additional genes, arsD and arsA, first identified in the Escherichia coli plasmid R773 ars operon (17, 18). The 26-kDa homodimeric ArsD (120 aa residues per monomer) is a cytosolic protein with three vicinal cysteine pairs per monomer (Cys-12-Cys-13, Cys-112-Cys-113, and Cys-119-Cys-120) (see Fig. 5, which is published as supporting information on the PNAS web site). ArsD has weak As(III)-responsive transcriptional regulatory activity (18). ArsA is an ATPase that associates with ArsB to form an As(III)-translocating ATPase that extrudes arsenite from cells, conferring an additional level of resistance over cells with only ArsB (19). It is striking that the arsD and arsA genes always are found together in bacterial and archaeal arsenic resistance operons or gene clusters (see Table 1, which is published as supporting information on the PNAS web site), which led us to propose that five-gene arsRDABC operons evolved from three-gene arsRBC operons by insertion of the arsD and arsA genes as a unit (16). The linkage of ArsD and ArsA further suggests that they have associated functions in arsenic detoxification. In this report, we demonstrate that the ArsD serves as a chaperone for intracellular arsenite, transferring metalloid to the pump.
Results
ArsD Confers a Competitive Advantage to Cells Growing in Subtoxic Concentrations of Arsenite.
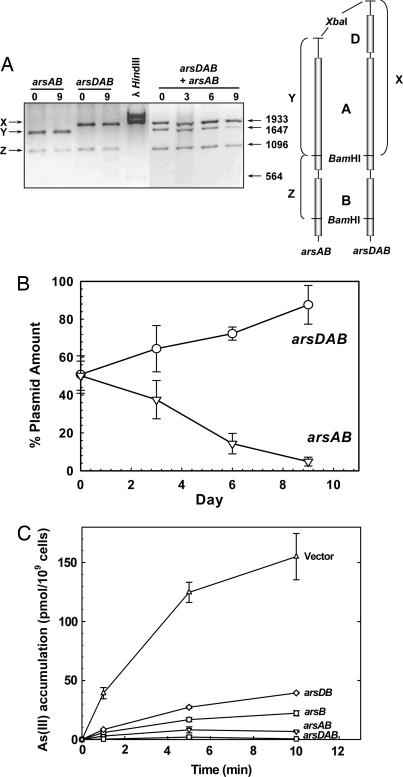
When arsenite resistance was compared between cells expressing the arsAB genes or the arsDAB genes, the latter showed a modest increase in resistance, with the greatest differences observed at toxic (millimolar) concentrations of arsenite (see Fig. 6, which is published as supporting information on the PNAS web site). Does ArsD confer a competitive advantage for growth in subtoxic concentrations of arsenite? A molecular competition experiment was devised to examine this question. We used cells of E. coli strain AW3110, in which the chromosomal ars operon had been deleted, bearing either a plasmid with either arsAB or arsDAB in the same vector. Note that the E. coli chromosome has neither arsD nor arsA genes. Equal amounts of the two types of cells were mixed together and grown in the presence of subtoxic arsenite (10 μM), which is in the range of what is found in the environment (14). Although expression was under the control of the tac promoter in this experiment, when under the control of the ars promoter, the ars operon is induced fully by 1 μM arsenate or arsenite (20). Each day the mixed culture was diluted 1,000-fold, and the relative amounts of the arsDAB and arsAB plasmids were determined by quantitative densitometry of the bands produced by restriction enzyme digestion with XbaI and BamHI (Fig. 1A). Digestion of both plasmids produced a 1.1-kbp BamHI fragment. The size of the XbaI–BamHI fragment in the arsAB plasmid was 1.6 kbp. The presence of arsD in arsDAB plasmid increased the size of this fragment to 1.9 kbp. After 9 days, cells with arsDAB had largely replaced those with only arsAB (Fig. 1B), indicating that ArsD provides a competitive advantage for growth in the low concentrations of arsenite that are ubiquitous in soil or surface waters.
Fig. 1.
ArsD enhances the activity of the ArsAB pump in vivo. (A) Molecular competition between cells with arsDAB and arsAB. Cells of E. coli strain AW3110 bearing either pSE-AB or pSE-DAB were grown separately or in mixed culture of each for 9 days with daily dilutions into fresh LB medium containing 10 μM sodium arsenite, as described in Materials and Methods. The plasmids from equal amounts of cells were extracted and digested with restriction enzymes XbaI and BamHI, and equal amounts of DNA containing the restriction fragments were separated by electrophoresis on a 1% agarose gel. Both plasmids produced a large fragment (data not shown) of vector DNA, and both produced a small arsB-containing BamHI fragment of 1.1 kb (fragment Z). Digestion of pSE-AB also produced a 1.6-kb fragment containing most of arsA (fragment Y), and digestion of pSE-DAB yielded the 1.9-kb fragment X containing arsD and most of arsA. The restriction fragments were from cells with only pSE-AB before growth (lane 1) or after 9 days (lane 2), only pSE-DAB before growth (lane 3) or after 9 days (lane 4), or from a mixed culture of cells with either pSE-AB or pSE-DAB before growth (lane 6) or after 3 days (lane 7), 6 days (lane 8), or 9 days (lane 9). Lane 5 contains a standard of λ HindIII-digested DNA. (B) Cells with only arsAB are lost from the population. The fraction of each plasmid was calculated by quantifying bands X, Y, and Z by densitometry. The percentage of the cells with each plasmid was calculated as follows. arsDAB: X/((vector + Z)/2); arsAB: Y/((vector + Z)/2). The data are the mean of values from five separate gels representing three independent experiments. The error bars represent the standard deviation of the mean calculated by using SigmaPlot 9.0. (C) Cells expressing arsDAB maintain lower intracellular arsenite than do cells expressing arsAB. Transport of As(III) was assayed with AW3110 bearing vector plasmids pSE380 and pACBAD (▵) or plasmids with arsB (○), arsAB (▿), arsD and arsAB (□), or arsD and arsB (◇).
The effect of the ars genes on arsenite accumulation was examined. Higher rates of extrusion result in lower accumulation (19). Cells of the arsenite-hypersensitive strain AW3110 with no ars genes (vector plasmids pSE380 and pACBAD) accumulated ≈150 pmol As(III) per 109 cells per 10 min (Fig. 1C). Cells expressing only arsB accumulated ≈22 pmol As(III) per 109 cells per 10 min. Expression of arsAB resulted in decreased accumulation to ≈7 pmol As(III) per 109 cells per 10 min, reflecting more efficient arsenite extrusion by the ArsAB pump than by ArsB alone (19). Cells coexpressing arsD and arsAB accumulated substantially less arsenite (≈1 pmol As(III) per 109 cells per 10 min) than did those with arsAB. Cells expressing arsD with only arsB accumulated arsenite to approximately the same level as cells expressing only arsB, indicating that ArsD does not affect expression or activity of ArsB. Immunoblotting showed that arsD does not affect the levels of ArsA produced (see Fig. 7, which is published as supporting information on the PNAS web site). These results clearly show that ArsD enhances the ability of the ArsAB pump to reduce intracellular arsenite.
Interaction of ArsD with ArsA.
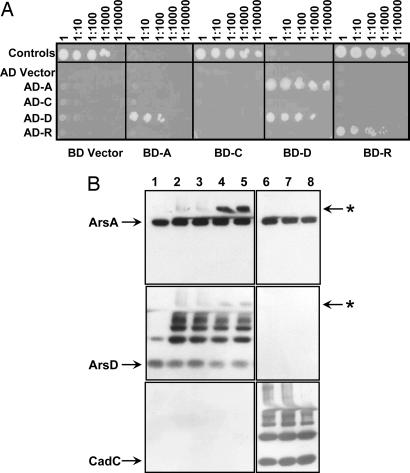
Yeast two-hybrid analyses were used to show that ArsD and ArsA physically interact (Fig. 2A). ArsA interacted with ArsD but not with ArsR or ArsC. ArsD interacted with ArsA and with itself, as expected because ArsD is a homodimer (21), but not with ArsR or ArsC. Homodimeric ArsR also interacts with itself but not with ArsD or ArsA. These results indicate specific interaction of ArsD and ArsA.
Fig. 2.
ArsD and ArsA interact in vivo and in vitro. (A) Yeast two-hybrid assays with wild-type ars genes. Yeast strain AH109 bearing both GAL4 AD and BD fusion plasmids was grown in SD medium overnight and inoculated on agar plates with SD lacking histidine with 10-fold serial dilutions. The plates were incubated at 30°C for 2–3 days. As a positive control, pVA3 (BD-p53) was expressed with pTD1 (AD-T antigen); as a negative control, vector plasmid pGBT9 was expressed with pACT2 (alternating in the top row). (B) bBBr cross-linking. The indicated proteins (16 μM concentrations each) were incubated with 0.5 mM bBBr and/or 1 mM concentrations each of potassium antimonyl tartrate, MgCl2, and ATP. Samples were analyzed by SDS/PAGE and immunoblotting with anti-ArsA (Top), anti-ArsD (Middle), or anti-CadC (Bottom). Lane 1, ArsA + ArsD; lane 2, ArsA + ArsD + bBBr; lane 3, ArsA + ArsD + Sb(III) + bBBr; lane 4, ArsA + ArsD + MgATP + bBBr; lane 5, ArsA + ArsD + Sb(III) + MgATP + bBBr; lane 6, ArsA + CadC + bBBr; lane 7, ArsA + CadC + MgATP + bBBr; lane 8, ArsA + CadC + Sb(III) + MgATP + bBBr. The position of individual proteins is indicated by arrows. The location of the band that reacts with both antibodies (the putative ArsD–ArsA adduct) is indicated with an asterisk.
Because both ArsD and ArsA have metalloid binding sites composed of cysteine residues (19), direct physical interaction at those sites was examined by cross-linking with dibromobimane (bBBr), a thiol reagent that becomes fluorescent when it cross-links cysteine residues within 3–6 Å of each other (22). When ArsD was treated with bBBr, dimers and higher-order species were observed by SDS/PAGE (Fig. 2B Middle). All ArsD bands became fluorescent, showing formation of both intra- and intermolecular cross-links (data not shown). When equimolar ArsD and ArsA were reacted with bBBr, a cross-linked species was detected that reacted with both anti-ArsA and anti-ArsD antibodies (indicated by an asterisk in Fig. 2B Top and Middle, lane 2). This species migrated as a band with an apparent mass of ≈90 kDa. Although we cannot definitively rule out other combinations of ArsD and ArsA, the mass of this band corresponds to the predicted mass of an ArsD dimer cross-linked to an ArsA monomer. Note that the intensity of the bands from one blot to the next cannot directly be compared because different polyclonal antibodies react with their antigens differently and because the stripping process removes variable amounts of the antigenic species. As a control, ArsA was reacted with CadC, a Cd(II)-responsive regulatory protein of similar size to ArsD with a metal-binding site formed by cysteines that react intramolecularly with bBBr (23). No ArsA–CadC adducts were observed by using anti-ArsA (Fig. 2B Top, lanes 6–8) or anti-CadC (Fig. 2B Bottom, lanes 6–8) antibodies. The amount of cross-linked ArsD–ArsA increased with MgATP (Fig. 2B Top and Middle, lane 4), but Sb(III) did not increase cross-linking because the thiol groups that coordinate the metalloid had already reacted with bBBr (Fig. 2B Top and Middle, lane 3). Sb(III) was used rather than As(III) because both ArsD and ArsA have higher affinity for trivalent antimony, a softer and more thiophilic metal than arsenic.
Transfer of Metalloids from ArsD to ArsA.
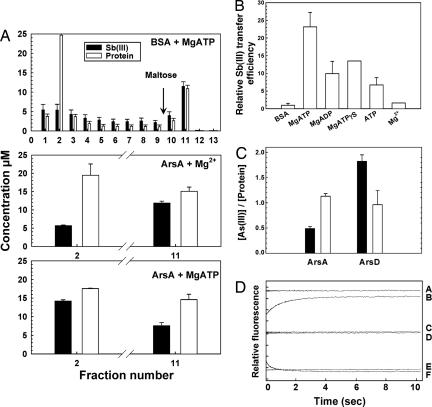
Cytosol from cells expressing a maltose-binding protein (MBP)–ArsD fusion was incubated with Sb(III). The MBP–ArsD–Sb(III) complex was bound to an amylose column. When the column was eluted with BSA and MgATP, little Sb(III) came off in the BSA-containing fractions (Fig. 3A Top). Subsequent application of buffer with maltose eluted nearly homogeneous MBP–ArsD protein in fraction 11 in a 1:1 molar complex with Sb(III). Similarly, little Sb(III) eluted with ArsA and Mg2+ without ATP in fraction 2, and most of the metalloid eluted with ArsD in fraction 11 (Fig. 3A Middle). In contrast, when the column was eluted with ArsA and MgATP, more Sb(III) came off with ArsA in fraction 2 and less with ArsD in fraction 11, consistent with transfer of metalloid from ArsD to ArsA (Fig. 3A Bottom). The effect of nucleotides on Sb(III) transfer was MgATP > MgATPγS > MgADP, indicating that nucleotide enhances transfer, but hydrolysis is not required (Fig. 3B). Mg2+ enhanced transfer with ATP but was not effective alone. Furthermore, when ArsA was incubated with MgATPγS, ArsA bound more As(III) in the presence of excess ArsD than in its absence (Fig. 3C). In contrast, ArsD bound less As(III) with excess ArsA than in its absence, consistent with transfer of metalloid from ArsD to ArsA (Fig. 3C).
Fig. 3.
ArsD transfers metalloid to and activates the ArsA ATPase. (A) ArsA releases Sb(III) from ArsD. Sb(III)–MBP–ArsD was bound to a 2-ml amylose column. One milliliter of either 20 μM ArsA or BSA preincubated with 1 mM MgCl2 or MgATP was applied to the column. The column was washed with 8 ml of column buffer, and MBP–ArsD was eluted with 4 ml of 10 mM maltose. The protein in each fraction was identified by SDS/PAGE. BSA or ArsA eluted primarily in fraction 2, and most of the MBP–ArsD eluted in fraction 11. The molar concentration of each protein in the fractions was estimated from the absorption at 280 nm (white bars), and amount of Sb(III) was determined by inductively coupled plasma mass spectroscopy (black bars). (B) A nucleotide enhances the ArsA-induced release of Sb(III) from ArsD. Sb(III) release from ArsD was assayed in the presence of the indicated nucleotides and is expressed relative to the values with BSA. The values are the average of two independent assays. (C) ArsD transfers As(III) to ArsA. The molar ratio of As(III) to either ArsA or ArsD monomer was measured with protein either alone (black bars) or in the presence of the partner protein (white bars). The values are the mean of three independent assays. (D) The transfer of Sb(III) from ArsD to ArsA time-resolved by stopped-flow fluorescence spectroscopy. Equal volumes of the following reagents were mixed in a stopped-flow device, and the changes in protein fluorescence (excitation = 285 nm; emission >340 nm) monitored. Curve A, 2 μM ArsD + 10 μM ArsA; curve B, 2 μM ArsD/10 μM Sb(III) + 10 μM ArsA; curve C, 10 μM ArsA + buffer; curve D, 10 μM ArsA + 10 μM Sb(III); curve E, 2 μM ArsD + 10 μM Sb(III); curve F, 2 μM ArsD/10 μM Sb(III) + buffer. Each division on the y axis represents a fluorescence change of 5% relative to that of 2 μM ArsD/10 μM ArsA.
Release of Sb(III) from ArsD is slow (24). Dissociation produced by dilution of the ArsD–Sb(III) complex leads to slow enhancement of ArsD fluorescence as Sb(III) is released, requiring more than 8 h to reach completion. If metalloid transfer from ArsD to ArsA were the result of dissociation and reassociation, there should be little transfer on the time scale of these experiments, indicating that interaction facilitates metalloid transfer. Indicative of the change in affinity of ArsA for Sb(III) induced by nucleotide binding, Sb(III) has little effect on ArsA fluorescence in the absence of nucleotide but induces a substantial quench when MgATP is bound (25). This difference in the Sb(III)-induced quench in ArsA fluorescence was used to detect transfer of metalloids from ArsD in the absence of nucleotides. In contrast to the slow release of Sb(III) upon dilution of the ArsD–Sb(III) complex with buffer, when the ArsD–Sb(III) complex was diluted with ArsA, reversal of the Sb(III)-induced quench in the fluorescence of ArsD occurred within seconds (Fig. 3D, trace B). This fluorescence change did not occur in the absence of Sb(III) when ArsD was mixed with ArsA (Fig. 3D, trace A), consistent with ArsA-induced release of Sb(III) from ArsD during transfer to ArsA. Although binding of nucleotide to ArsA would be expected to facilitate transfer from ArsD (compare Fig. 3 A and B), it was not feasible in this fluorescence assay to monitor metalloid transfer from ArsD to ArsA in the presence of MgATP because binding of Sb(III) to the ArsA–MgATP complex induces substantial quenching of ArsA fluorescence that offsets the increase in fluorescence of ArsD as Sb(III) is released.
ArsD Enhances the Catalytic Activity of ArsA.
The ATPase activity of ArsA is stimulated by As(III) (19). When ArsD was added to the ATPase assay, the apparent affinity of ArsA for arsenite increased 60-fold, from ≈1.2 mM to 20 μM (Fig. 4A). A similar increase in affinity for Sb(III) was observed (data not shown). This finding was not the result of increased thiol buffering of arsenite because dithiothreitol could not replace ArsD (data not shown). ArsD did not greatly affect the Km of ArsA for ATP at a concentration of arsenite (0.5 mM) below saturation in the absence of ArsD but was sufficient to saturate the enzyme in the presence of ArsD (Fig. 4B). Significantly, at this concentration of arsenite, ArsD increased the Vmax with ATP by ≈3-fold. Thus, the functional consequence of the ArsD–ArsA interaction is to increase the efficiency of the catalytic subunit of the ArsAB pump at low concentrations of the pump substrate, arsenite.
Fig. 4.
ArsD increases the affinity of the ArsA ATPase for As(III). (A) ArsD lowers the K1/2 of ArsA for As(III). ATPase activities were measured in the absence and presence of MBP–ArsD at varying concentrations of sodium arsenite. (B) ArsD does not affect the affinity of ArsA for ATP. ArsA ATPase activities were measured at varying concentrations of ATP in the presence of 0.5 mM sodium arsenite and the presence or absence of ArsD. The values in each plot are the mean of three independent assays. The error bars represent the standard deviation of the mean, which were calculated by using SigmaPlot 9.0, as were the Vmax, K1/2, and Km values.
Discussion
Metallochaperones protect both prokaryotic and eukaryotic cells from excess of copper by sequestering the metal for delivery to efflux ATPases (7). Resistance to other toxic metals is conferred by efflux pumps, including transport ATPases and resistance–nodulation–cell division (RND) transporters (26, 27). However, to date, the only other metallochaperones identified appear to be involved in delivery to metal-containing proteins. For example, a nickel chaperone delivers Ni2+ to the enzyme urease (9), and the chaperone frataxin delivers iron to partners for assembly of iron–sulfur clusters and heme (10). In this article, we report the existence of a chaperone for the toxic metalloid arsenic that enhances resistance by delivering the metalloid to the ArsAB efflux pump. Although there are superficial similarities with copper chaperones, ArsD is unrelated to any other chaperones in sequence or mechanism. Our yeast two-hybrid data suggest that ArsD interacts with ArsA in the absence of metalloid. We propose that ArsD interacts with ArsA with low affinity in the absence of metalloid and high affinity when metalloid is bound. ArsD has been shown to bind metalloid with higher affinity than ArsA does. For transfer from a high-affinity binding site on one protein to a relatively lower-affinity site on another protein, interaction of the two proteins must produce conformational changes in ArsD and/or ArsA. Indeed, binding of Sb(III) to ArsD is so tight that dissociation takes hours in the absence of ArsA, but the release rate is stimulated by more than four orders of magnitude by ArsA (Fig. 3D). We further propose that cysteine residues on ArsD are involved in the interaction with ArsA and that they are brought into close proximity to the metalloid binding sites of ArsA, allowing transfer of metalloid directly from the binding site on ArsD to the binding site on ArsA. It is plausible that this interaction destabilizes the metalloid binding sites on ArsD, reducing its affinity, while stabilizing binding to ArsA by occluding metalloid within the complex, thus facilitating transfer to ArsA. In this manner, the thermodynamically unfavorable process of transferring metalloid from a high- to a low-affinity site is overcome.
Our stopped-flow studies of the transfer of Sb(III) from ArsD to ArsA clearly are consistent with a mechanism in which this metallochaperone accelerates the rate of transfer to its partner. Because ArsD has higher affinity for metalloid than ArsA, it can scavenge the cytosol for free metalloid for delivery to ArsA, allowing the ArsAB pump to confer resistance at significantly lower concentrations of As(III). The Km of ArsB as a secondary carrier is 140 μM, but natural waters range in concentration of total inorganic arsenic from 7 nM to 70 μM, and concentrations of arsenic in drinking water in the worst arsenic-contaminated wells in West Bengal and Bangladesh are ≈40 μM (14). The ars operon is induced fully by 1 μM arsenate or arsenite (20), so resistance is limited by the activity, not the expression, of the pump at low concentrations. By lowering the concentration of substrate at which the pump functions efficiently, ArsD and ArsA provide cells with a mechanism to respond to environmental concentrations of metalloid.
Materials and Methods
Strains, Plasmids, and Media.
E. coli strain JM109 [recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ (traD36 proAB+ lacIq lacZΔM15)] and JM110 [rps (Strr) thr leu thi-1 lacY galK galT ara tonA tsx dam dcm supE44 Δ(lac-proAB) F′ traD36 proAB lacIq ZΔM15] were used for molecular cloning. E. coli strain BL21(DE3) [hsdS gal(ΔcIts857 ind1 Sam7 nin5 lacUV5-T7 gene1)] was used for protein expression and purification. E. coli strain AW3110 [K12 F− IN(rrnD-rrnE) ΔarsRBC] (28) was used for resistance and transport assays. S. cerevisiae strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ) was used for two-hybrid analyses (Clontech, Mountain View, CA). Plasmids pET28a (Kmr) (Novagen, Madison, WI) and pSE380 (Apr) (Invitrogen, Carlsbad, CA) were used as cloning vectors, and plasmid pGBT9 and pACT2 (Apr) were used as S. cerevisiae/E. coli shuttle vectors (Clontech). E. coli cells were grown in Luria–Bertani (LB) medium (29) at 37°C. Ampicillin (100 μg/ml), tetracycline (10 μg/ml), chloramphenicol (50 μg/ml), kanamycin (40 μg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1–0.3 mM) were added as required. Yeast was grown in yeast extract/peptone/dextrose (YPD) or minimal SD media (30) with appropriate supplements at 30°C. Growth in liquid culture was estimated from the absorbance at 600 nm.
DNA Manipulations.
Molecular methods were performed as described (29). Transformation of yeast cells was carried out by using a Geno FAST-Yeast transformation kit (Geno Technologies, St. Louis, MO).
Plasmid Construction and Yeast Two-Hybrid Assays.
The details of the construction of plasmids with the arsDAB, arsAB, arsB, and arsD genes are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, as are the details of the yeast two-hybrid assays.
Molecular Competition Assays.
Molecular competition growth assays were performed as described (31). Cells of E. coli strain AW3110 bearing either pSE-AB or pSE-DAB were grown overnight and mixed at a 1:1 ratio. The mixture was diluted 1:1,000 in LB medium containing 10 μM sodium arsenite at 37°C daily for 9 days. The plasmids were extracted from the mixed culture, and equal amounts of DNA were analyzed by digestion with XbaI and BamHI. The digested DNA fragments were run on a 1% agarose gel containing 0.5 μg/ml ethidium bromide. The bands were visualized at 302 nm and digitized by using UN-SCAN-IT software (Silk Scientific, Orem, UT). The percentage of the cells with each plasmid was calculated from five separate gels representing three independent experiments, and the mean values calculated by using SigmaPlot 9.0.
Transport Assays.
For transport assays, E. coli strain AW3110 was cotransformed with pSE380 series and pACBAD series plasmids. Cultures were grown overnight in LB medium and diluted 50-fold into LB medium at 37°C. After 1 h, 0.05% arabinose was added to induce ArsD expression, and the cells were harvested at an A600 of 1. The cells were washed and suspended in 1/10 of the original volume in a buffer consisting of 75 mM Hepes-KOH, 0.15 M KCl, and 1 mM MgSO4 (pH 7.5) at 22°C. Transport assays were performed with 10 μM sodium arsenite. Protein expression levels were determined by immunoblotting using anti-ArsA and anti-ArsD antibodies. Metalloids were quantified by inductively coupled plasma mass spectroscopy with a PerkinElmer (Wellesley, MA) ELAN 9000.
Protein Purification.
Proteins were purified as described in Supporting Materials and Methods.
Crosslinking Assays.
Crosslinking studies with bBBr (4,6-bis(bromomethyl)-3,7-di-methyl-1,5-diazabicyclo[3.3.0]octa-3,6-diene-2,8-dione) (Invitrogen) were described previously (23, 32). Purified wild-type ArsD and ArsA were buffer-exchanged into 50 mM Mops/0.2 M NaCl (pH 7.5) by using a microspin gel-filtration column (Bio-Rad, Hercules, CA) and are expressed as molar concentration of ArsA or ArsD monomer. Proteins (16 μM concentrations each) were incubated with 0.5 mM bBBr and/or 1 mM concentrations each of potassium antimonyl tartrate, MgCl2, and ATP for 30 min at room temperature. Samples were analyzed by SDS/PAGE using a step-gradient gel with 8% (to resolve ArsA) and 16% (to resolve ArsD) acrylamide. Formation of fluorescent cross-linked products was visualized at 365 nm, and by immunoblotting with antiserum directed against ArsA, ArsD, or CadC. The membranes were stripped for reaction with the next antibody by incubation in a buffer containing 62.5 mM Tris·HCl (pH 6.8), 0.1 M 2-mercaptoethanol, and 2% SDS at 50°C for 30 min.
Metalloid Transfer Assays.
Cells of E. coli BL21(DE) expressing MBP–ArsD were lysed in the presence of 1 mM potassium antimonyl tartrate in buffer A lacking DTT. The cytosol containing the MBP–ArsD–Sb(III) complex was applied to a 2-ml amylose column, which was washed with 20 ml of the same buffer at room temperature. BSA or purified ArsA (1 ml of 20 μM) was applied to the columns with 1 mM ATP, ADP, ATPγS, and/or MgCl2, as indicated. The column was washed with 8 ml of buffer, and MBP–ArsD was eluted with 4 ml of 10 mM maltose. As shown from SDS/PAGE, fraction 2 contained nearly all of the BSA or ArsA with little MBP–ArsD, and fraction 11 contained nearly all of the MBP–ArsD with little or no BSA or ArsA. Antimony was quantified by inductively coupled plasma mass spectroscopy. Transfer activity was calculated as ([Sb(III)ArsA]/[ArsA])/([Sb(III)ArsD]/[ArsD]).
To further demonstrate that ArsA accepts As(III) from ArsD, the amount of arsenic on each protein was determined after interaction. To determine the amount of As(III) bound to ArsA, a mixture of 3 μM his-tagged ArsA, 25 μM sodium arsenite, 2.5 mM MgCl2, and 2 mM ATPγS with or without 9 μM MBP–ArsD was incubated at 37°C for 10 min. To isolate the As(III)–ArsA complex, MBP–ArsD and free As(III) were removed with a microspin gel-filtration column with a 0.3-ml layer of amylose resin applied at the top of the spin column. ArsA and bound arsenic concentrations were quantified. The amount of As(III) bound to ArsD was determined similarly by adding excess ArsA (9 μM) to ArsD (3 μM). His-tagged ArsA and free As(III) were removed by using a gel-filtration spin column with a 0.3-ml layer of Ni-nitrilotriacetic acid resin applied at the top of the spin column, and the amounts of ArsD and As(III) were determined as described above. The details of fluorescence measurements and ATPase assays with these proteins are described in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by U.S. Public Health Service Grant AI45428 (to B.P.R.) and a grant from the Wellcome Trust (U.K.) (to A.R.W.).
Abbreviations
- bBBr
dibromobimane
- MBP
maltose-binding protein
- LB
Luria–Bertani.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Finney LA, O’Halloran TV. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 2.Gatti D, Mitra B, Rosen BP. J Biol Chem. 2000;275:34009–34012. doi: 10.1074/jbc.R000012200. [DOI] [PubMed] [Google Scholar]
- 3.Solioz M, Stoyanov JV. FEMS Microbiol Rev. 2003;27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 4.Romero-Isart N, Vasak M. J Inorg Biochem. 2002;88:388–396. doi: 10.1016/s0162-0134(01)00347-6. [DOI] [PubMed] [Google Scholar]
- 5.Cobbett C, Goldsbrough P. Annu Rev Plant Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 6.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 7.Rosenzweig AC. Chem Biol. 2002;9:673–677. doi: 10.1016/s1074-5521(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 8.Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 9.Mulrooney SB, Hausinger RP. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansy SS, Cowan JA. Acc Chem Res. 2004;37:719–725. doi: 10.1021/ar0301781. [DOI] [PubMed] [Google Scholar]
- 11.Lin SJ, Pufahl RA, Dancis A, O’Halloran TV, Culotta VC. J Biol Chem. 1997;272:9215–9220. [PubMed] [Google Scholar]
- 12.Abernathy CO, Thomas DJ, Calderon RL. J Nutr. 2003;133:1536S–1538S. doi: 10.1093/jn/133.5.1536S. [DOI] [PubMed] [Google Scholar]
- 13.Beane Freeman LE, Dennis LK, Lynch CF, Thorne PS, Just CL. Am J Epidemiol. 2004;160:679–687. doi: 10.1093/aje/kwh267. [DOI] [PubMed] [Google Scholar]
- 14.Smedley PL, Kinniburgh DG. Appl Geochem. 2002;17:517–568. [Google Scholar]
- 15.Rosen BP. Comp Biochem Physiol A Mol Integr Physiol. 2002;133:689–693. doi: 10.1016/s1095-6433(02)00201-5. [DOI] [PubMed] [Google Scholar]
- 16.Rosen BP. Trends Microbiol. 1999;7:207–212. doi: 10.1016/s0966-842x(99)01494-8. [DOI] [PubMed] [Google Scholar]
- 17.Chen CM, Misra TK, Silver S, Rosen BP. J Biol Chem. 1986;261:15030–15038. [PubMed] [Google Scholar]
- 18.Wu J, Rosen BP. Mol Microbiol. 1993;8:615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 19.Rosen BP. FEBS Lett. 2002;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Rosen BP. J Biol Chem. 1993;268:52–58. [PubMed] [Google Scholar]
- 21.Chen Y, Rosen BP. J Biol Chem. 1997;272:14257–14262. doi: 10.1074/jbc.272.22.14257. [DOI] [PubMed] [Google Scholar]
- 22.Kosower NS, Newton GL, Kosower EM, Ranney HM. Biochim Biophys Acta. 1980;622:201–209. [PubMed] [Google Scholar]
- 23.Wong MD, Lin YF, Rosen BP. J Biol Chem. 2002;277:40930–40936. doi: 10.1074/jbc.M206536200. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Rosen BP, Borges-Walmsley MI, Walmsley AR. J Biol Chem. 2002;277:25992–26002. doi: 10.1074/jbc.M201619200. [DOI] [PubMed] [Google Scholar]
- 25.Walmsley AR, Zhou T, Borges-Walmsley MI, Rosen BP. J Biol Chem. 1999;274:16153–16161. doi: 10.1074/jbc.274.23.16153. [DOI] [PubMed] [Google Scholar]
- 26.Legatzki A, Franke S, Lucke S, Hoffmann T, Anton A, Neumann D, Nies DH. Biodegradation. 2003;14:153–168. doi: 10.1023/a:1024043306888. [DOI] [PubMed] [Google Scholar]
- 27.Silver S, Phung le T. J Ind Microbiol Biotechnol. 2005;32:587–605. doi: 10.1007/s10295-005-0019-6. [DOI] [PubMed] [Google Scholar]
- 28.Carlin A, Shi W, Dey S, Rosen BP. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 30.Adams A, Gottschling DE, Kaiser C, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1998. [Google Scholar]
- 31.Ruan X, Bhattacharjee H, Rosen BP. J Biol Chem. 2006;281:9925–9934. doi: 10.1074/jbc.M600125200. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharjee H, Rosen BP. J Biol Chem. 1996;271:24465–24470. doi: 10.1074/jbc.271.40.24465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.