Abstract
Understanding the genetic basis of HIV-1 drug resistance is essential to developing new antiretroviral drugs and optimizing the use of existing drugs. This understanding, however, is hampered by the large numbers of mutation patterns associated with cross-resistance within each antiretroviral drug class. We used five statistical learning methods (decision trees, neural networks, support vector regression, least-squares regression, and least angle regression) to relate HIV-1 protease and reverse transcriptase mutations to in vitro susceptibility to 16 antiretroviral drugs. Learning methods were trained and tested on a public data set of genotype–phenotype correlations by 5-fold cross-validation. For each learning method, four mutation sets were used as input features: a complete set of all mutations in ≥2 sequences in the data set, the 30 most common data set mutations, an expert panel mutation set, and a set of nonpolymorphic treatment-selected mutations from a public database linking protease and reverse transcriptase sequences to antiretroviral drug exposure. The nonpolymorphic treatment-selected mutations led to the best predictions: 80.1% accuracy at classifying sequences as susceptible, low/intermediate resistant, or highly resistant. Least angle regression predicted susceptibility significantly better than other methods when using the complete set of mutations. The three regression methods provided consistent estimates of the quantitative effect of mutations on drug susceptibility, identifying nearly all previously reported genotype–phenotype associations and providing strong statistical support for many new associations. Mutation regression coefficients showed that, within a drug class, cross-resistance patterns differ for different mutation subsets and that cross-resistance has been underestimated.
Keywords: antiviral therapy, HIV, linear regression, machine learning
Twenty antiretroviral drugs are approved for treating HIV-1 infection: eight protease inhibitors (PIs), seven nucleoside and one nucleotide reverse transcriptase (RT) inhibitors (NRTIs), three nonnucleoside RT inhibitors (NNRTIs), and one fusion inhibitor. Resistance to these drugs is caused by mutations in their molecular targets. Understanding the genetic basis of cross-resistance is essential for designing new antiviral drugs and for using genotypic drug resistance testing to select optimal therapy. Despite the large number of PIs and RT inhibitors, therapy is challenging because drug resistance arises from complex patterns of mutations and because of the high degree of cross-resistance within each drug class.
Approaches for using HIV-1 drug resistance mutations to predict changes in drug susceptibility have included decision trees (1), linear regression (2), linear discriminant analysis (3), neural networks (4), and support vector regression (SVR) (5). Here, we compare five statistical learning methods each using four different sets of input mutations to develop quantitative models associating HIV-1 protease and RT mutations with changes in susceptibility to 16 antiretroviral drugs. The analyses are performed on a curated publicly available data set (6) generated with a highly reproducible drug susceptibility assay (7, 8). The results provide insight into the performance of different statistical learning methods at predicting phenotypic characteristics of highly polymorphic proteins and into the genetic mechanisms of HIV-1 antiretroviral cross-resistance.
Results
Drug Susceptibility Results, Input Mutations, and Learning Methods.
For each of the three drug classes, we created four mutation sets that included (i) a complete set of all mutations present in ≥2 sequences, (ii) an expert panel mutation set (9), and (iii) a set of nonpolymorphic treatment-selected mutations (TSMs) derived from a database linking protease and RT sequences to the treatment histories of persons from whom the sequenced viruses were obtained (10) (Table 1). A control set of the 30 most common mutations in the data set was also created (see Supporting Text, which is published as supporting information on the PNAS web site). Predictions using these 30 mutations were consistently inferior to those using the other three mutation sets (data not shown).
Table 1.
Sets of protease and RT mutations used as input features for predicting drug susceptibility
| Mutation set | Description | Mutations |
|---|---|---|
| Complete | Mutations occurring ≥ 2 times in the data set |
|
| Expert panel | IAS-USA mutation panel (9). The complete list of mutations for each drug class is shown here. The subset of mutations associated with each drug in the primary publication was used for prediction. |
|
| Nonpolymorphic TSMs | Nonpolymorphic mutations significantly more common in treated compared with untreated persons (10) |
|
IAS-USA, International AIDS Society–USA.
Table 2 shows the number of isolates for which sequences and susceptibility results were available. Thirty-seven to 60% of isolates had reduced drug susceptibility to one or more PIs. Thirty-one to 70% had reduced drug susceptibility to one or more NRTIs. Thirty-three to 41% had reduced drug susceptibility to one or more NNRTIs. The distribution in the number of nonpolymorphic TSMs and expert panel mutations per isolate is shown in Fig. 2, which is published as supporting information on the PNAS web site.
Table 2.
Summary of HIV-1 isolates with genotype and phenotype correlations according to drug tested and level of resistance
| Drug | Isolate phenotypes* |
Total | ||
|---|---|---|---|---|
| Susc, % | Low/int, % | High, % | ||
| Protease inhibitors | ||||
| Amprenavir (APV) | 61 | 29 | 10 | 768 |
| Atazanavir (ATV) | 48 | 30 | 22 | 329 |
| Indinavir (IDV) | 51 | 28 | 21 | 827 |
| Lopinavir (LPV) | 45 | 23 | 32 | 517 |
| Nelfinavir (NFV) | 40 | 25 | 35 | 844 |
| Ritonavir (RTV) | 48 | 20 | 32 | 795 |
| Saquinavir (SQV) | 60 | 18 | 22 | 826 |
| Nucleoside RT inhibitors | ||||
| Lamivudine (3TC) | 32 | 15 | 53 | 633 |
| Abacavir (ABC) | 30 | 45 | 25 | 628 |
| Zidovudine (AZT) | 52 | 23 | 25 | 630 |
| Stavudine (D4T) | 57 | 28 | 15 | 630 |
| Didanosine (DDI) | 56 | 37 | 7 | 632 |
| Tenofovir (TDF) | 70 | 18 | 12 | 353 |
| Nonnucleoside RT inhibitors | ||||
| Delavirdine (DLV) | 64 | 14 | 21 | 732 |
| Efavirenz (EFV) | 64 | 16 | 22 | 734 |
| Nevirapine (NVP) | 56 | 7 | 37 | 746 |
*Susc, susceptible; Low/int, low/intermediate; High, high-level. The cut-offs (levels of fold-decreased susceptibility) for distinguishing susceptible from low/intermediate resistance and low/intermediate from high-level resistance are described in Methods.
We applied five statistical learning methods [decision trees, neural networks, least-squares regression (LSR), SVR, and least angle regression (LARS)] to classify isolates as susceptible, low/intermediate, or highly resistant to the drugs used for testing. The regression methods were used to predict the level of reduced drug susceptibility for each isolate.
Classification.
The mean prediction accuracy (5 methods × 3 mutation sets) was highest for the NNRTIs (83.0%) compared with the PIs (78.2%; P < 0.001) and NRTIs (75.9%; P < 0.001) (Table 3). Among the PIs, the highest accuracy was for ritonavir (85.4%) and the lowest was for atazanavir (70.6%), the PI with the fewest test results. Among the NRTIs, the highest accuracies were for lamivudine (88.6%) and the lowest were for tenofovir (TDF) (67.7%), the NRTI with the fewest test results. There was minimal variation in prediction accuracy among the NNRTIs.
Table 3.
Predictive accuracy of LSR, SVR, LARS, decision trees, and neural networks using the nonpolymorphic TSMs, complete set (Comp), and expert panel mutations (Expert)
| Drug* | LSR |
SVR |
LARS |
Decision trees |
Neural networks |
Drug mean† | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSM | Comp | Expert | TSM | Comp | Expert | TSM | Comp | Expert | TSM | Comp | Expert | TSM | Comp | Expert | ||
| Protease inhibitors | ||||||||||||||||
| APV | 0.84 | 0.81 | 0.8 | 0.84 | 0.82 | 0.8 | 0.84 | 0.81 | 0.8 | 0.78 | 0.77 | 0.77 | 0.76 | 0.74 | 0.76 | 0.80 |
| ATV | 0.77 | 0.68 | 0.73 | 0.76 | 0.69 | 0.73 | 0.77 | 0.76 | 0.7 | 0.65 | 0.71 | 0.65 | 0.7 | 0.64 | 0.65 | 0.71 |
| IDV | 0.79 | 0.78 | 0.75 | 0.79 | 0.77 | 0.76 | 0.79 | 0.77 | 0.75 | 0.75 | 0.75 | 0.75 | 0.76 | 0.73 | 0.71 | 0.76 |
| LPV | 0.81 | 0.79 | 0.82 | 0.81 | 0.8 | 0.83 | 0.81 | 0.83 | 0.81 | 0.74 | 0.77 | 0.73 | 0.76 | 0.76 | 0.73 | 0.79 |
| NFV | 0.82 | 0.79 | 0.74 | 0.82 | 0.79 | 0.76 | 0.81 | 0.8 | 0.73 | 0.8 | 0.76 | 0.8 | 0.77 | 0.73 | 0.73 | 0.78 |
| RTV | 0.89 | 0.86 | 0.85 | 0.89 | 0.86 | 0.86 | 0.88 | 0.88 | 0.86 | 0.85 | 0.84 | 0.84 | 0.82 | 0.81 | 0.82 | 0.85 |
| SQV | 0.84 | 0.81 | 0.76 | 0.85 | 0.81 | 0.77 | 0.82 | 0.82 | 0.74 | 0.8 | 0.75 | 0.77 | 0.8 | 0.76 | 0.75 | 0.79 |
| Avg‡ | 0.82 | 0.79 | 0.78 | 0.82 | 0.79 | 0.79 | 0.82 | 0.81 | 0.77 | 0.77 | 0.76 | 0.76 | 0.77 | 0.74 | 0.74 | 0.78 |
| Nucleoside RT inhibitors | ||||||||||||||||
| 3TC | 0.89 | 0.83 | 0.87 | 0.88 | 0.84 | 0.87 | 0.87 | 0.88 | 0.88 | 0.92 | 0.9 | 0.92 | 0.92 | 0.9 | 0.92 | 0.89 |
| ABC | 0.77 | 0.63 | 0.77 | 0.77 | 0.65 | 0.77 | 0.78 | 0.77 | 0.78 | 0.74 | 0.69 | 0.75 | 0.71 | 0.66 | 0.71 | 0.73 |
| AZT | 0.78 | 0.64 | 0.74 | 0.78 | 0.7 | 0.75 | 0.77 | 0.76 | 0.72 | 0.71 | 0.7 | 0.72 | 0.73 | 0.71 | 0.71 | 0.73 |
| D4T | 0.8 | 0.66 | 0.78 | 0.8 | 0.68 | 0.78 | 0.79 | 0.78 | 0.79 | 0.76 | 0.75 | 0.77 | 0.75 | 0.72 | 0.76 | 0.76 |
| DDI | 0.77 | 0.61 | 0.73 | 0.77 | 0.67 | 0.73 | 0.77 | 0.75 | 0.74 | 0.75 | 0.74 | 0.75 | 0.72 | 0.71 | 0.71 | 0.73 |
| TDF | 0.76 | 0.46 | 0.66 | 0.76 | 0.69 | 0.66 | 0.72 | 0.7 | 0.65 | 0.69 | 0.68 | 0.67 | 0.69 | 0.73 | 0.63 | 0.68 |
| Avg‡ | 0.80 | 0.64 | 0.76 | 0.79 | 0.71 | 0.76 | 0.78 | 0.77 | 0.76 | 0.76 | 0.74 | 0.76 | 0.75 | 0.74 | 0.74 | 0.75 |
| Nonnucleoside RT inhibitors | ||||||||||||||||
| DLV | 0.82 | 0.73 | 0.82 | 0.81 | 0.78 | 0.78 | 0.8 | 0.84 | 0.82 | 0.84 | 0.84 | 0.82 | 0.84 | 0.78 | 0.82 | 0.81 |
| EFV | 0.86 | 0.78 | 0.82 | 0.86 | 0.82 | 0.82 | 0.85 | 0.87 | 0.8 | 0.85 | 0.84 | 0.8 | 0.84 | 0.77 | 0.8 | 0.83 |
| NVP | 0.87 | 0.74 | 0.87 | 0.86 | 0.78 | 0.85 | 0.88 | 0.87 | 0.86 | 0.91 | 0.91 | 0.89 | 0.91 | 0.81 | 0.89 | 0.86 |
| Avg‡ | 0.85 | 0.75 | 0.84 | 0.84 | 0.79 | 0.82 | 0.84 | 0.86 | 0.83 | 0.87 | 0.86 | 0.84 | 0.86 | 0.79 | 0.84 | 0.83 |
Shown is the proportion of phenotypes for which the predicted result matched the actual result: susceptible vs. low/intermediate vs. high-level resistance.
*The list of drug abbreviations is shown in Table 2.
†Mean accuracy for each drug averaged over mutation set and learning method.
‡Mean accuracy for drug class according to mutation set and learning method.
When the prediction accuracies of the possible combinations of drug and learning method were averaged over the different mutation sets, the mean accuracies of the learning methods ranged from 76.1% (neural networks) to 79.7% (LARS). The superiority of LARS was due to its accuracy in using the complete mutation set (80.3%), which was significantly higher than decision trees (77.2%; P = 0.02), SVR (76.1%; P < 0.001), neural networks (74.6%; P < 0.001), and LSR (72.3%; P < 0.001). Regression methods (79.9%) had higher accuracy for the PIs than the decision tree and neural network methods (75.5%; P < 0.001).
Averaged over the different learning methods and drugs, the nonpolymorphic TSM set had the highest prediction accuracy (80.1%) followed by the expert panel (77.5%; P < 0.001) and complete mutation set (76.1%; P < 0.001). However, the use of all of the expert panel mutations for a drug class (pooling the mutations associated with all of the drugs of a class) increased the accuracy from 77.5% to 79.3%.
Table 4, which is published as supporting information on the PNAS web site, shows the number of highly discordant results according to the mutation data set and learning method. For 10,624 predictions obtained by applying LARS to the nonpolymorphic TSM data set, only 33 (0.31%) were highly discordant with the measured phenotype (Table 5, which is published as supporting information on the PNAS web site).
Regression.
The TSM set had the highest correlation coefficients (r2) between actual and predicted susceptibility compared with the complete and expert panel mutation set (Table 6, which is published as supporting information on the PNAS web site). The r2 values for the TSM set determined by SVR, LSR, and LARS ranged from 0.83 to 0.84 averaged over the PIs, 0.76 to 0.77 averaged over the NRTIs, and 0.77 to 0.79 averaged over the NNRTIs.
Averaged over each of the 16 drugs and the three regression methods, the mean-squared errors (MSEs) were 0.22 for the TSMs and 0.32 for the expert panel mutations (Table 7, which is published as supporting information on the PNAS web site). However, the use of the complete set of expert panel mutations for a drug class reduced the overall MSE from 0.32 to 0.26. The MSEs of the regression methods using the TSMs were inversely proportional to the number of samples used for testing and training (Fig. 3, which is published as supporting information on the PNAS web site).
Regression coefficients for each of the PI, NRTI, and NNRTI TSMs determined by the different regression methods were highly correlated (Tables 8–10 and Fig. 4, which are published as supporting information on the PNAS web site). For the PIs, the mean r2 between the LSR and SVR coefficients was 0.98 and between LSR and LARS was 0.96. For the NRTIs, the mean r2 between LSR and SVR was 0.94 and between LSR and LARS was 0.91.
PI-Resistance Mutations.
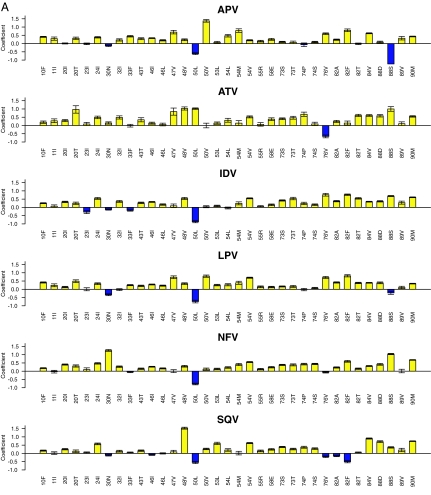
Fig. 1A shows the LSR coefficients for 35 PI TSMs occurring in ≥10 sequences and significantly associated with decreased susceptibility to one or more PIs (regression coefficient >3.0 standard deviations above or below 0). The substrate cleft mutations G48V and I84V; the flap mutations I54V and Q58E; and the mutations L24I, G73S, and L90M were associated with decreased susceptibility to all seven PIs. The substrate cleft mutations V32I, I50V, and V82A/T/F; the flap mutations K43T, M46I/L, I47V, F53L, and I54M/L; and the mutations L10F, K20I/T, G73T, T74S, and N88D/S were associated with decreased susceptibility to four or more PIs.
Fig. 1.
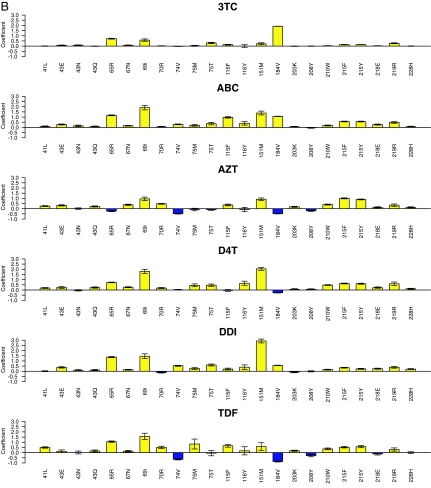
LSR coefficients for PI (A) and NRTI (B) TSMs. Shown are regression coefficients of the LSR models for PI susceptibility using nonpolymorphic PI TSMs (A) and NRTI using nonpolymorphic NRTI TSMs (B). The y axis indicates the magnitude of the coefficient. Positive coefficients (yellow histograms) indicate mutations that decrease drug susceptibility; negative coefficients (blue histograms) indicate mutations that increase drug susceptibility. The y axis has no units because the log-fold susceptibility changes were normalized before regression analysis. The error bars indicate the standard deviation of the mean generalized error determined 50 times (10 repetitions of 5-fold cross-validation). For the PIs (n = 35) and NRTIs (n = 23), the mutations shown are those that occurred ≥10 times in the data set and for which the absolute value of the coefficient was ≥3.0 times the standard deviation for one or more drugs. The regression coefficients for the PI ritonavir and for the NNRTIs are shown in Tables 8 and 10, respectively.
The TSM coefficients provided quantitative confirmation of each nonpolymorphic expert panel mutation. In addition, six non-expert-panel TSMs (V11I, K43T, Q58E, T74S, L76V, and L89V) were significantly associated with decreased susceptibility to one or more PIs.
NRTI- and NNRTI-Resistance Mutations.
Fig. 1B shows the LSR regression coefficients for 23 NRTI TSMs occurring in ≥10 sequences and significantly associated with decreased susceptibility to one or more NRTIs. The TSM coefficients provided quantitative confirmation of each nonpolymorphic expert panel mutation. In addition, 8 non-expert-panel TSMs (K43E/Q, V75M/T, E203K, D218E, K219R, and L228H) decreased susceptibility to one or more NRTIs. As previously reported, M184V increased susceptibility to zidovudine (AZT), stavudine (d4T), and TDF; L74V increased susceptibility to AZT and TDF; and K65R increased susceptibility to AZT (11–13).
The LSR coefficients for 24 NNRTI TSMs occurring in ≥2 sequences and associated with decreased susceptibility to one or more NNRTIs are shown in Table 10. Most mutations decreased susceptibility to each of the NNRTIs. Seven non-expert-panel TSMs (K101E/P, K103S, Y181V, G190E/Q, and K238T) decreased susceptibility to one or more NNRTIs.
To explore the effect of NNRTI-resistance mutations on NRTI susceptibility and NRTI-resistance mutations on NNRTI susceptibility, we used LSR and the combined set of NRTI and NNRTI TSMs to predict both NRTI and NNRTI susceptibility. Two NNRTI-resistance mutations, L100I and Y181C, had significantly negative regression coefficients for AZT (−0.45 and −0.45) and TDF (−0.43 and −0.33) consistent with previous reports (12, 14). Five NRTI-resistance mutations had significantly negative regression coefficients for efavirenz (EFV) including M41L, D67N, M184V, L210W, and K219Q (range of −0.08 to −0.17), consistent with previous reports (15).
Discussion
Association studies with HIV-1 genotype are complicated by the high dimensionality of the data caused by HIV-1's high mutation rate. We used several types of external feature selection to limit the dimensionality of HIV-1 genotypic data and five statistical learning methods to create models of how genotype influences susceptibility. LARS, the only regression method that performed its own feature selection, was superior to the other regression methods at predicting decreased susceptibility in the absence of external feature selection (i.e., using the complete set of mutations present in ≥2 sequences in the data set). However, LARS was not superior to the other regression methods when externally derived features were used.
The most successful approach to external feature selection was to use a set of mutations previously identified as absent in viruses from treatment-naïve individuals and occurring at significantly increased frequencies in viruses from treated individuals (10). This set of nonpolymorphic TSMs had the highest accuracy (80.1%) averaged over the five learning methods when classifying isolates as susceptible, low/intermediate, and high-level resistant, and had the lowest MSEs compared with the other mutation data sets when used for regression. The success of TSMs at predicting susceptibility follows from Darwinian principles: the TSMs emerge during therapy presumably because they facilitate virus escape from drug inhibition. Exploiting the results of an analysis on a separate data set containing genotype-treatment correlations thus proved superior to relying completely on the genotype–phenotype data set to predict phenotype.
The fact that prediction accuracy was lowest for the drugs (atazanavir and TDF) with the fewest samples suggests that accuracy depends to a large extent on sample size. The learning curves show that for most drugs, ≈400 genotype–phenotype examples were required for optimal predictive accuracy and that the MSE then saturates at 0.15–0.20, suggesting that factors other than TSMs influence susceptibility. For example, polymorphic sites influence susceptibility either directly or indirectly by affecting virus replication capacity (16). In addition, protease cleavage site mutations, primarily those in the gag gene, also influence viral replication capacity and drug susceptibility (17).
Although it is likely that the effect of individual mutations on drug susceptibility depends on the presence of other mutations, we did not demonstrate an improvement in predictive accuracy using a LARS regression model that included all possible two-way interactions or using a polynomial kernel for SVR (data not shown). This may be because the process of exploring interactions without model selection to choose sets of interacting mutations often leads to decreased performance due to over-fitting. Alternatively, the main effects may have already included the effects of interactions because, in clinical virus isolates, only those combinations of mutations that reduce susceptibility and allow the virus to replicate are likely to emerge.
The regression coefficients provide evidence for the high level of cross-resistance within the PI class and identified new mutations that were associated with decreased susceptibility to one or more drugs. Because the coefficients were derived from a model in which susceptibility results were standardized (i.e., corrected for the different ranges in susceptibility for different drugs), their magnitude indicates the mutation's contribution to resistance relative to other mutations affecting the same drug and relative to the mutation's effect on other drugs. The high correlation among the coefficients derived from three different regression methods suggests that the mutation regression coefficients represent a reproducible effect that is a real property of the data.
Although drugs of the same class shared high levels of cross-resistance, the patterns of cross-resistance were often different for different mutations. This may reflect the physical interactions inhibitors make with more than one part of the target molecule. For example, PIs bind to four or more binding pockets in the protease substrate cleft, whereas NRTIs interact with several parts of RT both before and after being added to a growing DNA chain. The genetic mechanisms of cross-resistance and of mutational antagonism should be exploited in selecting antiretroviral drug regimens by combining drugs with the least cross-resistance and in designing new compounds containing moieties that select for antagonistic mutations.
Methods
HIV-1 Isolates and Genotypes.
HIV-1 sequences used for this study were from publicly available isolates in the Stanford HIV Drug Resistance Database for which both sequences and in vitro susceptibility results were available (6). Isolates included viruses from the plasma of HIV-1-infected persons and laboratory viruses with drug-resistance mutations resulting from site-directed mutagenesis or in vitro passage. Up to two isolates from a small number of individuals were included provided their isolates differed at two or more drug-resistance positions. Isolates with electrophoretic evidence of more than one amino acid at a nonpolymorphic drug-resistance position were excluded from analysis.
Genotypes were derived from the amino acid sequences of positions 1–99 in protease and 1–240 in RT. Mutations were defined as amino acid differences from the subtype B consensus wild-type sequence (http://hivdb.stanford.edu/pages/asi/releaseNotes/). Mutations were classified as nonpolymorphic if they occurred with a frequency of ≤0.5% in untreated persons.
Drug Susceptibility Results.
For consistency, only susceptibility results generated by the PhenoSense method (Monogram Biosciences, South San Francisco, CA) were analyzed (7, 8). Drug susceptibility results were expressed as fold change in susceptibility defined as the ratio of the IC50 of an isolate and a standard wild-type control isolate. Results were classified into three categories: susceptible, low/intermediate resistance, and high-level resistance. The cut-off between susceptible and low/intermediate resistance was based on the distribution of results in HIV-1 isolates from untreated persons lacking drug-resistance mutations (18). The cut-off between low/intermediate and high-level resistance was based partly on the drug's dynamic susceptibility range (fold difference of the most highly resistance isolates) and partly based on levels of resistance associated with markedly reduced clinical activity.
For the PIs, <3.0-fold resistance was considered susceptible, 3.0- to 20-fold was considered low/intermediate resistant, and >20-fold was considered highly resistant. For the NNRTIs and the NRTIs AZT and lamivudine, <3.0-fold resistance was considered susceptible, 3.0- to 25-fold was considered low/intermediate, and >25-fold was considered highly resistant. For the NRTIs ddI, d4T, and TDF, a fold resistance of <1.5 was considered susceptible, 1.5–3.0 was considered low/intermediate, and >3.0 was considered highly resistant. For the NRTI ABC, <2.0-fold resistance was considered susceptible, 2.0- to 6.0-fold was considered low/intermediate, and >6.0-fold was considered highly resistant. Susceptibility results were log-transformed and standardized before analysis.
Prediction Algorithms.
Decision trees.
The C4.5 algorithm (19) was used to construct decision trees for the 16 drugs in the study. Mutations were the attributes, and phenotypic classifications were the decision variables.
Neural network.
Feed-forward neural network models with a single hidden layer were trained to predict susceptibility by using the R package AMORE (http://cran.r-project.org). The input layer contained a node for each of the mutation attributes. The hidden layer was assigned 12 nodes for all drugs. The output layer consisted of three nodes, one for each of the susceptibility classes. The weights were learned by using the error back propagation algorithm.
Support vector regression.
SVR was used to learn a regression function of the form: ƒ(x) = ΣαiK(x,xi) + b, where ƒ(x) is the logarithm of the fold value for the training sample and x is the binary vector of mutations (20). K is the kernel function, the nonzero αi correspond to the support vectors, and b is the bias term. Both linear and polynomial kernels were used. The latter was used to model interactions between mutations. SVR was performed with the PyML package (available at http://pyml.sourceforge.net).
Linear regression.
LSR and LARS (20, 21) were used to predict the logarithm of the fold decrease in susceptibility. For each of the regression methods, the coefficients were learned after scaling the log-fold values to zero mean and unit variance to analyze the relative impact of a mutation on different drugs. LARS is a constrained model building procedure similar to the LASSO (23) that constructs a model by first finding the mutation most correlated with susceptibility and then incrementally builds the model by following the “equi-angular vector” until another variable is equally correlated with the residual. The process would eventually terminate at a least-squares solution. However, a validation set (20% of the data) was used to decide when to stop adding variables to the model. With LARS, second-order polynomials were also used to model interactions among each of the input mutations.
Cross-Validation.
Five-fold cross-validation was used to determine the mean generalization accuracy of each learning method on test data. Five-fold cross-validation was run 10 times on different subdivisions of the data set to estimate variability of the mean generalization accuracy. For decision trees, LSR, and SVR, 80% of the data were used for training and 20% for testing. For neural networks and LARS, 60% of the data were used for training, 20% for validating the selected model, and 20% for testing.
Performance Criteria.
Accuracy was the proportion of correctly predicted samples. The regression methods were also evaluated by using the MSE and r2 between actual and predicted standardized log-fold values. Both measurements indicate how much of the variability in the response variable (the standardized log-transformed reduction susceptibility) was explained by the regression model.
All comparisons between drug classes, drugs, mutation sets, or learning methods were done in a pairwise fashion, using paired differences of the averaged accuracy or MSE for each repeated run of cross-validation. For instance, the accuracy for mutations sets was averaged over all drugs and learning methods, resulting in a single observation per run of cross-validation. A one-sample permutation test on the 10 paired differences was used to ensure the correct null distribution was used for computing P values.
To assess the effect of the number of training examples (genotype–phenotype correlations) on prediction accuracy, we created sets of randomly selected training examples that were multiples of 50 and ranging in size from 50 to 600 for testing and training using 5-fold cross-validation.
Supplementary Material
Acknowledgments
S.-Y.R., G.W., J.T., and R.W.S. were supported in part by National Institute of Allergy and Infectious Diseases Grant AI46148-01 and National Institute of General Medical Sciences Grant 5P01GM066524-03. A.B.-H. and D.L.B. were supported by a Stanford Bio-X Interdisciplinary Grant.
Abbreviations
- AZT
zidovudine
- LARS
least angle regression
- LSR
least-squares regression
- MSE
mean-squared error
- PI
protease inhibitor
- RT
reverse transcriptase
- NRTI
nucleoside RT inhibitor
- NNRTI
nonnucleoside RT inhibitor
- SVR
support vector regression
- TDF
tenofovir
- TSM
treatment-selected mutation.
Footnotes
The authors declare no conflict of interest.
References
- 1.Beerenwinkel N, Schmidt B, Walter H, Kaiser R, Lengauer T, Hoffmann D, Korn K, Selbig J. Proc Natl Acad Sci USA. 2002;99:8271–8276. doi: 10.1073/pnas.112177799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K, Jenwitheesuk E, Samudrala R, Mittler JE. Antivir Ther. 2004;9:343–352. [PubMed] [Google Scholar]
- 3.Sevin AD, DeGruttola V, Nijhuis M, Schapiro JM, Foulkes AS, Para MF, Boucher CA. J Infect Dis. 2000;182:59–67. doi: 10.1086/315673. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Larder B. J Infect Dis. 2003;188:653–660. doi: 10.1086/377453. [DOI] [PubMed] [Google Scholar]
- 5.Beerenwinkel N, Daumer M, Oette M, Korn K, Hoffmann D, Kaiser R, Lengauer T, Selbig J, Walter H. Nucleic Acids Res. 2003;31:3850–3855. doi: 10.1093/nar/gkg575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Rhee SY, Taylor J, Shafer RW. J Acquired Immune Defic Syndr. 2005;38:439–444. doi: 10.1097/01.qai.0000147526.64863.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson VA, Brun-Vezinet F, Clotet B, Conway B, Kuritzkes DR, Pillay D, Schapiro J, Telenti A, Richman D. Top HIV Med. 2005;13:51–57. [PubMed] [Google Scholar]
- 10.Rhee SY, Fessel WJ, Zolopa AR, Hurley L, Liu T, Taylor J, Nguyen DP, Slome S, Klein D, Horberg M, et al. J Infect Dis. 2005;192:456–465. doi: 10.1086/431601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St Clair M, Martin JL, Tudor-Williams G, Bach MC, Vavro CL, King DM, Kellam P, Kemp SD, Larder BA. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 12.Parkin N, Chappey C, Petropoulos C, Hellmann N. Antivir Ther. 2003;8:S34. [Google Scholar]
- 13.Parikh UM, Koontz DL, Chu CK, Schinazi RF, Mellors JW. Antimicrob Agents Chemother. 2005;49:1139–1144. doi: 10.1128/AAC.49.3.1139-1144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larder BA. J Gen Virol. 1994;75:951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- 15.Shulman NS, Bosch RJ, Mellors JW, Albrecht MA, Katzenstein DA. AIDS. 2004;18:1781–1785. doi: 10.1097/00002030-200409030-00006. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Picado J, Wrin T, Frost SD, Clotet B, Ruiz L, Brown AJ, Petropoulos CJ, Parkin NT. J Virol. 2005;79:5907–5913. doi: 10.1128/JVI.79.10.5907-5913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguire MF, Guinea R, Griffin P, Macmanus S, Elston RC, Wolfram J, Richards N, Hanlon MH, Porter DJ, Wrin T, et al. J Virol. 2002;76:7398–7406. doi: 10.1128/JVI.76.15.7398-7406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkin NT, Hellmann NS, Whitcomb JM, Kiss L, Chappey C, Petropoulos CJ. Antimicrob Agents Chemother. 2004;48:437–443. doi: 10.1128/AAC.48.2.437-443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan J. Sydney: Morgan Kaufmann; 1993. C4.5: Programs for Machine Learning. [Google Scholar]
- 20.Schoelkopf B, Smola A. Cambridge, MA: MIT Press; 2002. Learning with Kernels. [Google Scholar]
- 21.Efron B, Hastie T, Johnstone IF, Tibshirani R. Ann Stat. 2004;32:407–499. [Google Scholar]
- 22.Tibshirani R. J R Stat Soc. 1996;58:267–288. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.