Abstract
We report a signal-on, electronic DNA (E-DNA) sensor that is label-free and achieves a subpicomolar detection limit. The sensor, which is based on a target-induced strand displacement mechanism, is composed of a “capture probe” attached by its 5′ terminus to a gold electrode and a 5′ methylene blue-modified “signaling probe” that is complementary at both its 3′ and 5′ termini to the capture probe. In the absence of target, hybridization between the capture and signaling probes minimizes contact between the methylene blue and electrode surface, limiting the observed redox current. Target hybridization displaces the 5′ end of the signaling probe, generating a short, flexible single-stranded DNA element and producing up to a 7-fold increase in redox current. The observed signal gain is sufficient to achieve a demonstrated (not extrapolated) detection limit of 400 fM, which is among the best reported for single-step electronic DNA detection. Moreover, because sensor fabrication is straightforward, the approach appears to provide a ready alternative to the more cumbersome femtomolar electrochemical assays described to date.
Keywords: biosensors, electron transfer, gold electrode, methylene blue, signal-on
The rapid, specific detection of nucleic acid sequences offers the potential for utility in both clinical and research diagnostic applications (1, 2). A wide range of electronic DNA detection schemes have been described to date (3–5), the best of which achieve limits of detection (LOD) ranging from picomolar to femtomolar. Among promising recent examples are approaches based on conductive links produced through the catalytic deposition of silver by nanoparticle-linked secondary probes (LOD 500 fM) (6), the electrocatalytic oxidation of modified bases by Os(bpy)33+ (LOD 400 fM) (7), the electrochemistry of water-soluble, ferrocene-functionalized cationic polythiophenes (LOD 500 pM) (8) or ferrocene-linked triblock copolymer–DNA hybrids (LOD 100 pM) (9), the electron transfer of ferrocenyl-tethered poly(amido-amine)dendrimer in a sandwich-type enzyme-linked DNA sensor (LOD 100 pM) (10), charge transport from electroactive DNA intercalators (with magnetic sample concentration) (LOD 2 pM) (11), the chronopotentiometric detection of micrometer-long indium rod tracer in a DNA sandwich hybridization assay (with magnetic sample concentration) (LOD 2.6 pM) (12), and the anodic stripping voltammetry of silver nanoparticles deposited in a multistep reduction process initiated by a labeled secondary probe (LOD 0.1 fM) (13).
Although the detection limits of the above-described sensor technologies are often impressive, achieving them requires the addition of exogenous, label-containing secondary probes and, typically, complicated, multicomponent deposition/amplification steps. For example, although Hwang et al. (13) report an exceptional 0.1 fM detection limit, achieving it required a five-step assay, including an enzyme-linked secondary probe, enzymatic reduction of p-aminophenyl phosphate, the concomitant reductive deposition of silver, and, finally, anodic stripping voltammetry to quantify the deposited silver. In contrast to these relatively cumbersome assays (6–13), we and others have recently described several reagentless, single-step electrochemical DNA detection methods based on immobilized, redox-tagged single-stranded DNA (14, 15) and DNA stem–loops (16–18). The latter strategy, termed E-DNA (16), is based on a redox-tagged DNA stem–loop structure that self-assembles on a gold electrode by a gold–thiol bond. The hybridization of target with the loop region induces a large conformational change in this surface-confined DNA and thus significantly affects the rate of electron transfer between the redox moiety and the electrode. The associated change in redox current produces a signal indicative of the target without the addition of exogenous reagents.
In addition to being reagentless and single-step, the E-DNA sensor has numerous advantages in terms of its applicability to real-world oligonucleotide detection. It is, for example, readily reusable, sequence-specific, and selective enough to perform even when placed directly in blood serum and in solutions contaminated with soil, foodstuffs, or other complex materials (19). For many applications, however, these advantages are offset by the modest sensitivity of such sensors: the best reported LOD for an E-DNA sensor is, at tens picomolar (16), several orders of magnitude poorer than the best multistep electronic approaches (6, 11, 13). Here, in contrast, we report an E-DNA design that couples the femtomolar detection limits of the best reagent-intensive electrochemical methods with the single-step convenience of the E-DNA-sensing platform.
The LOD of the original E-DNA sensor is, in part, limited by its “signal-off” architecture (target binding reduces the redox current); the gain of signal-off sensors is limited because the target can suppress no more than 100% of the original signal. “Signal-on” sensors for which target binding increases the redox current, in contrast, have the potential for greatly improved sensitivity because, under ideal conditions (as the background signal approaches zero), the gain of such a sensor increases without limit (20). Here we describe an E-DNA sensor that, in contrast to the original E-DNA signal-off architecture, is signal-on, and thus it exhibits enhanced gain and a significantly improved detection limit.
Results
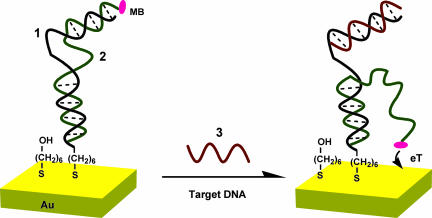
The signal-on E-DNA sensor is based on a target-induced strand-displacement mechanism (Fig. 1). The sensor is composed of two parts. The first part is a single-stranded “capture probe” (1) covalently attached by a 5′ thiol to a gold electrode using standard self-assembled monolayer chemistry (21). As a measure of the surface coverage of this capture probe (1), we have performed quartz crystal microbalance measurements in which the capture probe is immobilized on a gold-coated quartz crystal (resonant frequency, 9 MHz), resulting in a frequency change of 39 Hz that corresponds to a surface coverage of ≈10 pmol·cm−2. The second sensor component is a “signaling probe” (2) with a methylene blue (MB)-redox moiety covalently attached to its 5′ terminus. The signaling probe is complementary to both termini of the capture probe (Fig. 1 Left), hybridizing with 15 bases at the 5′ terminus of the capture probe (1) to link it physically to the electrode and to 7 bases at the 3′ terminus of the capture probe, which, in the absence of target, sequesters the MB from the electrode. Of note, deposition of these probes during fabrication must be carefully controlled to produce optimal signal gain and reproducible sensors. As determined by electrochemical measurements, the surface coverage of the MB-modified signaling probe (2) was maintained within the range of 3.0 ± 0.3 pmol·cm−2 (assuming perfect transfer efficiency), an electrode-loading level at which sensor gain is maximized.
Fig. 1.
Schematic of the signal-on E-DNA sensor, which is based on a conformational change in a MB-modified duplex DNA that occurs after target-induced strand displacement. In the absence of target, the two double-stranded regions formed between the capture and signaling probes sequester the MB from the electrode surface, producing a relatively small MB redox current. When the sensor is challenged with a complementary target, the observed MB redox current increases significantly, presumably because the flexible, single-stranded element liberated in the signaling probe increases the efficiency with which the MB can transfer electrons to the electrode surface. Blue, capture probe (1); green, signaling probe (2); red, target (3).
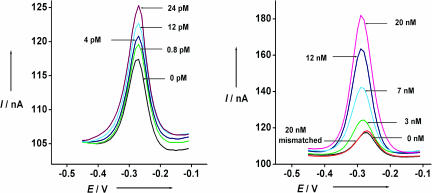
In the absence of target, the two double-stranded regions formed between the capture and signaling probes sequester the MB from the electrode surface, producing a relatively small MB redox current. When the sensor is challenged with a 15-base complementary target (3), the observed MB redox current increases significantly. The proposed mechanism underlying this signaling is as follows. Target hybridization displaces the 7 hybridized bases at the 5′ terminus of the signaling probe. This displacement liberates the MB-modified end of the signaling probe, generating a flexible, single-stranded element that allows the MB to collide with the electrode surface and transfer electrons (Fig. 1 Right). Alternatively, the increased current could reflect improved transfer from the MB when liberation of the single-stranded DNA allows the MB to intercalate within the remaining double-stranded regions of the capture-signaling duplex near the electrode surface (22). It is, however, unclear which of these mechanisms dominates. For example, as expected for a collisional model, the peak potential does not shift when the concentration of complementary target is varied from femtomolar to picomolar (Fig. 2 Left). At the highest target concentrations we have used, however, a shift of up to 25 mV is observed (Fig. 2 Right), which is approximately half the size of the cathodic shift characteristic during MB intercalation within the double-stranded DNA (23).
Fig. 2.
The signal gain of the signal-on E-DNA sensor is relatively large. Shown are the sensor responses using alternating current voltammetry to different concentrations of fully complementary target DNA (3) (Left) and mismatched DNA (4) in 10 mM phosphate buffer including 0.1 M NaCl (pH 7.3). Twenty nanomolar mismatched DNA only shows <5% signal change (Right).
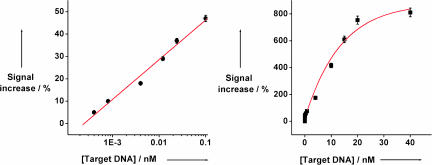
Because of the large signal change induced by the target, the signal-on E-DNA sensor is significantly more sensitive than the corresponding signal-off sensor. As a result of limited electron transfer from the signaling probe we observe only small, ≈10 nA faradaic currents in the absence of target [Fig. 2, 0 nM DNA (3)]. After target (3) binding, the faradaic current increases significantly (Fig. 2), typically reaching ≈7 times that of the original signal at saturating target concentrations (above ≈20 nM; Fig. 2 Right). The signal gain of this new sensor architecture is thus an order of magnitude greater than the ≈35% signal suppression observed at similar target concentrations with a signal-off E-DNA sensor (15–17). This improved signal gain leads in turn to significantly improved sensitivity; the directly measured (not extrapolated) detection limit of the current sensor architecture is 400 fM (Fig. 3 Left), and the dynamic range of the sensor spans four orders of magnitude (Fig. 3 Right). In contrast, control experiments reveal that the addition of a 5-base mismatched target (4) at a concentration of 20 nM (which would be a saturating target concentration for the perfect match) does not produce any significant signal change (Fig. 2 Right).
Fig. 3.
The dynamic range of the signal-on E-DNA sensor covers target concentrations from 400 fM to ≈20 nM. The error bars represent the SD of four AC voltammetric scans conducted with a single electrode at each target DNA concentration. Multiple matched electrodes were used to collect this data set.
The residual reduction peak observed in the absence of target could arise because of limited, long-range transfer from the MB to the electrode surface. Alternatively, a small fraction of the signaling probes might not hybridized with the 3′ terminus of the capture probe, allowing direct, short-range transfer of electrons from the MB to the electrode. Finally, the capture/signaling probe double-stranded complex might be sufficiently flexible to allow the MB to collide occasionally with the electrode surface and perform short-range electron transfer. Using the Laviron equation (24) to calculate the interfacial electron transfer rates between the MB and the electrode we obtain rate constants of 10 s−1 and 58 s−1 before and after reacting with target DNA (3), respectively. The relative similarity in these rate constants (which decay exponentially with increasing distance and which could have differed by orders of magnitude) suggests that the electron transfer mechanism is similar in the both presence and absence of target, supporting the latter two hypothesized mechanisms.
The signal-on E-DNA sensor requires that the 3′ terminus of the signaling probe remain physically associated with the electrode surface after target binding. To ensure that this association occurs we have designed the system to form a 15-base duplex with the 5′ terminus of the immobilized capture probe. In the optimized hybridization we have used, this duplex element is stable for more than 24 h in the absence of target. The described sensor is not, however, regenerated on stringent washing (20 min in distilled, deionized water) without the reintroduction of the signaling probe; the hybridization that retains the signaling probe on the gold surface does not withstand the conditions required to disrupt target (3) binding. The sensor can be regenerated, however, with the reintroduction of a new signaling probe (by hybridization of MB-modified signaling probe for 6 h at room temperature). We speculate that alternative covalent linkages will alleviate this requirement and lead to more readily reusable sensors.
Discussion
The mechanism underlying the new sensor architecture is target-induced strand displacement. The low background currents produced by the initially rigid, double-stranded sensor element allow for significantly improved signal gain over earlier, signal-off E-DNA architectures (16) and provide for a 400 fM detection limit.
A reagentless, signal-on E-DNA sensor architecture was described previously in the elegant approach of Immoos et al. (15). In this work, which utilizes a surface-immobilized, single-stranded oligodeoxynucleotide—poly(ethylene glycol) triblock polymer, signal arises when a large conformational change is induced by the simultaneous hybridization of both the top and bottom oligonucleotide of the immobilized triblock probe with the target. This simultaneous hybridization forces a terminally linked ferrocene redox tag into proximity with the electrode surface, increasing the signaling current. The reported detection limit for the Immoos sensor (15) is, however, three orders of magnitude poorer than that reported here, presumably because the flexibility of the unbound, single-stranded triblock polymer is sufficient to allow the ferrocene to collide with the electrode surface, producing a significant background current. In the approach reported here, in contrast, the sensing DNA forms a relatively rigid double helix in the absence of target, presumably accounting for the orders of magnitude smaller background current we observe. This reduced background current ensures that the signal gain of our sensor is relatively large, thereby lowering our limit of detection to femtomolar levels.
The E-DNA sensor described here works by target-induced strand displacement, with the detection signal arising as a result of a large, binding-induced change in the probe flexibility and thus the electron-transfer distance. The observed detection limit of this simple sensor is among the best reported to date for electronic sensors. Moreover, unlike the few E-DNA detection approaches that approach or exceed this detection limit, the architecture described here is label-free and enables single-step detection. Given the combined sensitivity and simplicity of the signal-on E-DNA architecture, it appears that it may be of utility in a variety of DNA-detection applications.
Materials and Methods
Reagents.
Modified DNA oligonucleotides were synthesized by BioSource, Int. (Foster City, CA), purified by C18 HPLC and PAGE, and confirmed by mass spectroscopy. The sequences of these oligomers used are as follows: (1), 5′-HS-(CH2)6-GCGAGTTAGACCGATCCCCCCCCTTCGTCCAGTCTTTT-3′; (2), 5′-MB-(CH2)6-GACTGGACGCCCCCCCATCGGTCTAACTCGC-3′; (3), 5′-AAAAGACTGGACGAA-3′; (4), 5′-AAAAGACTCCTGAAA-3′.
MB was conjugated to the 5′ end of the probe (2) by succinimide ester coupling (MB-NHS obtained from EMP Biotech, Berlin, Germany) by the fabricator (Biosource) and used as supplied (25). The 6-mercaptohexanol (Sigma–Aldrich, St. Louis, MO) and Tris(2-carboxyethyl)phosphine hydrochloride (Molecular Probes, Eugene, OR) were used as received.
Sensor Preparation and Target Hybridization.
The E-DNA sensor was fabricated by using polycrystalline gold disk electrodes (1.6-mm diameter; BAS, West Lafayette, IN). The electrodes were prepared by polishing them with diamond and alumina (BAS), sonicating them in water, and electrochemically cleaning them (a series of oxidation and reduction cycling in 0.5 M NaOH/0.5 M H2SO4/0.01 M KCl/0.1 M H2SO4/0.05 M H2SO4) before being modified with the thiolated probe DNA. To fabricate our E-DNA sensors, a clean gold surface was reacted with a solution of thiolated DNA (1), 0.5 μM including 5 μM Tris(2-carboxyethyl)phosphine hydrochloride, which is included to reduce disulfide-bonded oligomers (26), in 200 mM Tris·HCl buffer (pH 7.4) for 16 h at room temperature. The resulting surface was washed with the Tris·HCl buffer, and then the (1)-functionalized gold-surface was treated with 1 mM 6-mercaptohexanol in 10 mM Tris·HCl buffer (pH 7.4) for 2 h. The resulting monolayer-functionalized surface was treated with the complementary signaling DNA (2), 2.5 μM, in PerfectHyb Plus hybridization buffer (Sigma, St. Louis, MO) (1×) for 6 h to yield the final capture probe/signaling probe assembly on the surface. The sensor surface was then allowed to hybridize with various concentrations of target DNA (3), in PerfectHyb Plus hybridization buffer (1×), for 5 h at 37°C to obtain the maximum strand displacement on the surface. Time-resolved experiments suggest that this time frame is sufficient to achieve full equilibration at the lowest (femtomolar) concentrations of target we have used; the equilibration time at higher (picomolar) target concentrations is significantly reduced.
Quartz-Crystal Microbalance Measurements.
A quartz-crystal microbalance analyzer (Seiko model QCA 917; EG&G, Oakridge, TN) was used for microgravimetric analyses. Quartz crystals (9 MHz, AT-cut; EG&G) sandwiched between two gold electrodes (area of electrode is 0.19 cm2) were used. The electrode was rinsed with piranha solution (consisting of 70% concentrated sulfuric acid and 30% hydrogen peroxide) for 10 s before being rinsed with ethanol and water. The crystal was dried with nitrogen and its fundamental frequency (fo) was recorded immediately, before modification. The crystal was incubated for 16 h in thiolated DNA (1), 0.5 μM including 5 μM Tris(2-carboxyethyl)phosphine hydrochloride in 200 mM Tris·HCl buffer (pH 7.4) for 16 h at room temperature. The resulting electrode was rinsed with distilled, deionized water. The measurement of frequency was applied immediately after the modified crystal was dried with nitrogen. The mass of the immobilized thiolated DNA was calculated from the change of the crystal's resonant frequency by using the Sauerbery equation (27).
Instrumentation.
All measurements were conducted by using alternating current voltammetry at an alternating current frequency of 10 Hz with a CHI 603 potentiostat (CH Instruments, Austin, TX) in a standard cell with a platinum counter electrode and Ag/AgCl (saturated with 3 M NaCl) reference electrode. Sensor measurements were conducted by monitoring the modified electrode in 0.1 M NaCl in 10 mM phosphate buffer (pH 7.4). Electrodes were incubated in each target DNA sample (200-μl total volume) for 5 h at 37°C before target detection measurements were performed.
Acknowledgments
This work was supported in part by National Institutes of Health Grant EB002046, National Science Foundation Grant DMR 0099843, the Institute for Collaborative Biotechnologies through Grant DAAD19-03-D-0004 from the U.S. Army Research Office, and by the University of California AIDS Research Program Grant ID63-SD-008.
Abbreviations
- E-DNA
electronic DNA
- LOD
limits of detection
- MB
methylene blue
Footnotes
The authors declare no conflict of interest.
References
- 1.Patolsky F, Lichtenstein A, Willner I. J Am Chem Soc. 2001;123:5194–5205. doi: 10.1021/ja0036256. [DOI] [PubMed] [Google Scholar]
- 2.Wang J. Chem Eur J. 1999;5:1681–1685. [Google Scholar]
- 3.Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 4.Thorp HH. Trends Biotechnol. 1998;16:117–121. [Google Scholar]
- 5.Katz E, Willner I, Wang J. J Electroanal. 2004;16:19–44. [Google Scholar]
- 6.Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 7.Gore MR, Szalai VA, Ropp PA, Yang IV, Silverman JS, Thorp HH. Anal Chem. 2003;75:6586–6592. doi: 10.1021/ac034918v. [DOI] [PubMed] [Google Scholar]
- 8.Floch LF, Ho HA, Harding-Lepage P, Bedard M, Neagu-Plesu R, Leclerc M. Adv Mater. 2005;17:1251–1254. [Google Scholar]
- 9.Gibbs JM, Park SJ, Anderson DR, Watson KJ, Mirkin CA, Nguyen ST. J Am Chem Soc. 2005;127:1170–1178. doi: 10.1021/ja046931i. [DOI] [PubMed] [Google Scholar]
- 10.Kim E, Kim K, Yang H, Kim YT, Kwak J. Anal Chem. 2003;75:5665–5672. doi: 10.1021/ac034253x. [DOI] [PubMed] [Google Scholar]
- 11.Kerman K, Matsubara Y, Morita Y, Takamura Y, Tamiya E. Sci Technol Adv Mater. 2004;5:351–357. [Google Scholar]
- 12.Wang J, Liu GD, Zhu QY. Anal Chem. 2003;75:6218–6222. doi: 10.1021/ac034730b. [DOI] [PubMed] [Google Scholar]
- 13.Hwang S, Kim E, Kwak J. Anal Chem. 2005;77:579–584. doi: 10.1021/ac048778g. [DOI] [PubMed] [Google Scholar]
- 14.Anne A, Bouchardon A, Moiroux J. J Am Chem Soc. 2003;125:1112–1113. doi: 10.1021/ja028640k. [DOI] [PubMed] [Google Scholar]
- 15.Immoos CE, Lee SJ, Grinstaff MW. J Am Chem Soc. 2004;126:10814–10815. doi: 10.1021/ja046634d. [DOI] [PubMed] [Google Scholar]
- 16.Fan CH, Plaxco KW, Heeger AJ. Proc Natl Aad Sci USA. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Immoos CE, Lee SJ, Grinstaff MW. ChemBioChem. 2004;5:1100–1103. doi: 10.1002/cbic.200400045. [DOI] [PubMed] [Google Scholar]
- 18.Mao YD, Luo CX, Ouyang Q. Nucleic Acids Res. 2003;31:e108. doi: 10.1093/nar/gng108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubin AA, Lai RY, Baker BR, Heeger AJ, Plaxco KW. Anal Chem. 2006;78:5671–5677. doi: 10.1021/ac0601819. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 21.Levicky R, Herne TM, Tarlov MJ, Satija SK. J Am Chem Soc. 1998;120:9787–9792. [Google Scholar]
- 22.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 23.Kelley SO, Barton JK. Bioconjugate Chem. 1997;8:31–37. doi: 10.1021/bc960070o. [DOI] [PubMed] [Google Scholar]
- 24.Laviron E. J Electroanal Chem. 1979;101:19–28. [Google Scholar]
- 25.Hermanson GT. Bioconjugate Techniques. San Diego: Academic; 1996. [Google Scholar]
- 26.Ruegg UT, Rudinger J. Methods Enzymol. 1977;47:111–116. doi: 10.1016/0076-6879(77)47012-5. [DOI] [PubMed] [Google Scholar]
- 27.Cohen Y, Levi S, Rubin S, Willner I. J Electroanal Chem. 1996;417:65–75. [Google Scholar]