Abstract
Hyper-IgM syndrome (HIGM) is a heterogeneous condition characterized by impaired Ig class-switch recombination (CSR). The molecular defects that have so far been associated with this syndrome — which affect the CD40 ligand in HIGM type 1 (HIGM1), CD40 in HIGM3, and activation-induced cytidine deaminase (AID) in HIGM2 — do not account for all cases. We investigated the clinical and immunological characteristics of 15 patients with an unidentified form of HIGM. Although the clinical manifestations were similar to those observed in HIGM2, these patients exhibited a slightly milder HIGM syndrome with residual IgG production. We found that B cell CSR was intrinsically impaired. However, the generation of somatic hypermutations was observed in the variable region of the Ig heavy chain gene, as in control B lymphocytes. In vitro studies showed that the molecular defect responsible for this new HIGM entity (HIGM4) occurs downstream of the AID activity, as the AID gene was induced normally and AID-induced DNA double-strand breaks in the switch μ region of the Ig heavy chain locus were detected during CSR as normal. Thus, HIGM4 is probably the consequence of a selective defect either in a CSR-specific factor of the DNA repair machinery or in survival signals delivered to switched B cells.
Introduction
Hyper-IgM syndrome (HIGM) is a primary immunodeficiency disease, characterized by normal or elevated serum IgM levels associated with low or absent IgG, IgA, and IgE serum levels, indicating a defect in the class-switch recombination (CSR) process (1). It is a heterogeneous condition, as several molecular defects result in HIGM. The X-linked form, HIGM type 1 (HIGM1), is caused by mutations in the gene that encodes the CD40 ligand (CD40L, CD154) (2–5). A deficiency in the CD40L receptor (CD40) results in HIGM3 (6). CD40L is expressed on activated Th cells, and its interaction with CD40, which is constitutively expressed on B cells, is required for B cell terminal differentiation within the germinal centers of secondary lymphoid organs. CD40 is also expressed by monocytes and dendritic cells, and the CD40/CD40L interaction is involved in T cell responses (7). Patients with CD40L or CD40 deficiency are thus susceptible to bacterial infections, as are patients affected with other severe B cell deficiencies, and they are also vulnerable to some opportunistic infections (8). Another form of X-linked HIGM, associated with anhydrotic ectodermal dysplasia, is caused by missense mutations in the gene encoding the NF-κB essential modulator (NEMO, or IKKγ), which is required, as in several other transduction pathways, for the CD40-induced activation of the transcription factor NF-κB (9, 10).
A B cell–specific defect that causes susceptibility only to bacterial pathogens has been found in other HIGM conditions (11). One subset, characterized by autosomal recessive transmission (HIGM2), is related to mutations in the gene that encodes the activation-induced cytidine deaminase (AID), which is selectively expressed in germinal center B cells (12). The AID defect leads not only to profoundly impaired CSR but also to the defective generation of somatic hypermutations (SHMs) in the Ig variable-region genes, resulting in overall defective antibody-affinity maturation. This finding, together with the HIGM phenotype of AID–/– mice, helps to elucidate the molecular mechanism involved in the secondary genetic rearrangements that occur in B cells within germinal centers (13).
Nonetheless, a fraction of patients affected with a CD40L+CD40+ form of HIGM do not carry mutations in the AID gene. This new entity, HIGM4, is described herein. HIGM4 patients exhibit a slightly milder HIGM syndrome than HIGM2 patients. One difference between HIGM4 and HIGM2 is that SHMs are generated as normal in the former but not in the latter. The molecular defect, although yet undefined, must occur downstream of the AID-dependent CSR-induced DNA breaks, which occur as normal in HIGM4 B cells in the switch region of the IgM heavy chain locus. Thus, HIGM4 may be caused by either a CSR-specific DNA repair abnormality or a defect in survival signals for switched B cells.
Methods
Patient selection and data collection.
Between December 1999 and September 2002, we recruited 103 HIGM patients with normal expression and sequence of CD40L and normal expression of CD40 and without any abnormality suggesting anhidrotic ectodermal dysplasia. Thirty-nine patients (34%) had HIGM2 with AID gene mutation. Peripheral blood samples were available for only 38 out of 64 patients. Patients with low peripheral blood B cell counts (<1%) (n = 8), patients with CD40- and B cell receptor–activation defect (n = 8) or with associated T cell–activation defect (n = 7) were excluded from this study. Thus we report herein data from 15 patients from 14 different families. Informed consent was obtained from patients or their parents. Clinical course and laboratory data were retrospectively reviewed by a questionnaire sent to all referring physicians. Information was collected on family history, age at disease onset, diagnosis of HIGM, intravenous Ig (IVIG) treatment initiation, clinical features before and after diagnosis, serum Ig levels at diagnosis, therapeutic features, and clinical status at last follow-up.
B cell activation and in vitro CSR.
B cell activation and in vitro CSR were performed as described previously (12, 14). We used a combination of soluble CD40L (sCD40L) and recombinant interleukin-4 (rIL-4), which strongly induces B cell proliferation and CSR toward IgE. Peripheral blood B cells from healthy adult volunteers or age-matched children (after parents’ consent) were studied as controls. PBMCs were separated by the Ficoll-Hypaque density centrifugation method (Lymphoprep; Axis-Shield PoC AS, Oslo, Norway) and then activated for 5 days with sCD40L (500 ng/ml; a kind gift from Immunex, Seattle, Washington, USA) and rIL-4 (100 U/ml; R&D Systems Inc., Minneapolis, Minnesota, USA). Proliferation was assessed by measurement of the uptake of [3H]thymidine. CSR toward IgE and IgG was assessed after 12 days of culture in the presence of sCD40L+rIL-4 by an ELISA technique (12).
Detection of RNA transcripts in activated B cells.
RNA was extracted from B cells that had been incubated in the presence of sCD40L+rIL-4 for 5 days by use of the Trizol reagent (Invitrogen BV, Groningen, The Netherlands). cDNA was produced using reverse transcriptase (SuperScript II; Invitrogen BV) with an oligo-dT primer (Invitrogen BV) according to the manufacturer’s instructions. RNA transcripts of CD19, AID genes and IgE gene germ-line (εGLTs), circle (εCTs), and functional (εFTs) transcripts, and IgG gene functional transcripts (γFTs) were detected by RT-PCR as previously described (11, 13). Serial dilutions of cDNA (1:1 up to 1:1,000) were performed for titration of the GLTε transcript amounts in HIGM4 patients and controls.
Detection of DNA double-strand breaks in the switch region of the μ heavy chain.
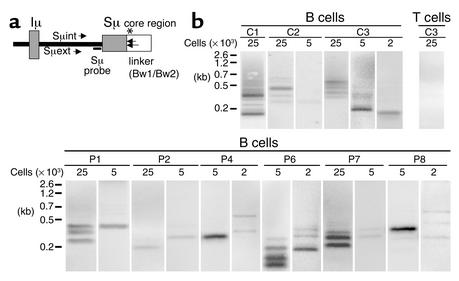
A ligation-mediated PCR (LM-PCR) method was used to identify double-strand breaks (DSBs) in genomic DNA extracted in agarose plugs from activated B cells after FACS sorting, as previously described (14). Briefly, B cells that had been activated by incubation in sCD40L+rIL-4 for 5 days were stained with FITC-conjugated anti-CD19 mAb’s (Immunotech, Marseilles, France). The cells were then sorted in the presence of propidium iodide (PI), which stains dead cells (Sigma-Aldrich, Taufkirchen, Germany), using a FACStarPLUS cell sorter (Becton Dickinson and Co., Franklin Lakes New Jersey, USA). Sorted, viable, activated B cells (CD19+PI–) were more than 94% pure. T cells activated with anti-CD3 mAb and recombinant interleukin-2 (rIL-2) for 7 days were used as a negative control. DSBs were then detected by LM-PCR (14). Briefly, two oligonucleotides (Bw1: 5′-GCGGTGACCCGGGAGATCTGAATTC-3′; and Bw2: 5′-GAATTCAGATC-3′) were allowed to form duplexes and thus to generate a double-stranded blunt-end linker (15). T4-DNA ligase (Promega Corp., Madison, Wisconsin, USA) was used to ligate DNA to the linker. Specific amplification was achieved for genomic DNA isolated from sorted viable cells (corresponding to 2 × 103, 5 × 103, and 25 × 103 cells per lane) using a semi-nested PCR, with the Bw1 primer and two primers in the switch region of the μ heavy chain (Sμ region) (Sμext: 5′-ATGGAAGCCAGCCTGGCTGT-3′; and Sμint: 5′-AGCCTGGCTGTGCAGGAACC-3′) (see Figure 4a). Twenty-five cycles of first PCR (94°C for 1 minute, 64°C for 1 minute, and 72°C for 2 minutes) were performed. One-tenth of the first PCR product was added to the reaction of second PCR, and 30 cycles of PCR (94°C for 1 minute, 67°C for 1 minute, and 72°C for 2 minutes) were performed (15). LM-PCR products were separated on 2% agarose gels, vacuum-transferred with 0.4N NaOH onto GeneScreen Plus membranes (Perkin-Elmer Life Sciences Inc., Boston, Massachusetts, USA), and hybridized to a gene-specific oligonucleotide probe labeled with 32Pγ-ATP (Amersham Biosciences AB, Uppsala, Sweden) and recognizing the Sμ region (Sμ probe: 5′-TCAGAAATGGACTCAGATGG-3′) (see Figure 4a).
Figure 4.
Detection of double-strand DNA breaks (DSBs) during CSR in HIGM4. (a) Positions of primers and probe used in DSB assay. Directions and positions of the primers and radioactive Sμ-specific probe used in the DSB assay are shown. The asterisk shows the putative position of DSBs in the Sμ region. (b) DSBs were detected by LM-PCR in activated B cells of HIGM4 patients. Genomic DNA of sCD40L+rIL-4–activated B cells (2 × 103, 5 × 103, or 25 × 103 cells per lane) from controls (C1, C2, C3) or HIGM4 patients (n = 6, P1, P2, P4, P6–P8) was ligated with a double-stranded linker and amplified by a semi-nested PCR using the linker (Bw1) as a primer as well as two specific primers in the Sμ region (Sμext and Sμint). The PCR products were subjected to electrophoresis and transferred onto a membrane before being incubated with a radiolabeled Sμ-specific probe. Radioactivity was detected by use of a PhosphorImager. The negative control consisted of anti-CD3+rIL-2–activated T cells (25 × 103 cells per lane, C3).
In another set of experiments, an aliquot of the purified LM-PCR products was ligated into pCRII vector and was transformed into chemically competent TOP10 cells by use of the TA Cloning Kit (Invitrogen BV). The DNA sequences of individual clones were determined by use of the BigDye DNA sequencing kit (Perkin-Elmer Life Sciences) with M13 forward and reverse primers and an automated ABI PRISM 377 genetic analyzer (Applied Biosystems, Foster City, California, USA).
Detection of SHMs in the variable region of Ig heavy chain.
We studied the frequency and characteristics of SHMs in the variable region of the IgM heavy chain (VH region) in purified CD19+CD27+ B cells as previously described (16). PBMCs were labeled with anti-CD19 mAb and anti-CD27 mAb (Immunotech), and then purified by FACS sorting. The purity of sorted CD19+CD27+ B cells was more than 99%. SHMs were identified in the VH3-23 (GenBank accession no. AB019439) region after PCR amplification using VH3-23 and Cμ primers and cloning as previously described (16).
Sequence analysis of the human uracil-DNA glycosylase gene.
Genomic DNA from the patients was amplified by PCR using primers that bound to intronic and noncoding sequences. PCR products were sequenced using the BigDye DNA sequencing kit (Perkin-Elmer Life Sciences), internal primers and an automated ABI PRISM 377 genetic analyzer (Applied Biosystems) as previously described (17).
Results
Phenotypic characteristics of patients with HIGM4.
Patients originated from six countries (five from Italy, four from France, two from Hungary, two from England, one from Turkey, and one from Japan). Most of the cases were sporadic, with no family history of immunodeficiency. One of the 15 patients was born to a consanguineous family (P1) and two patients (P2 and P3) were brothers born to a non-consanguineous family. The sex ratio of 11 males to 4 females likely resulted from a bias in the recruitment. Mean age at diagnosis of immunodeficiency was 8.8 years (range: 0.3–23 years). Mean age at the time of the study was 18.7 years (range: 2–32 years). All patients were alive and receiving IVIG treatment.
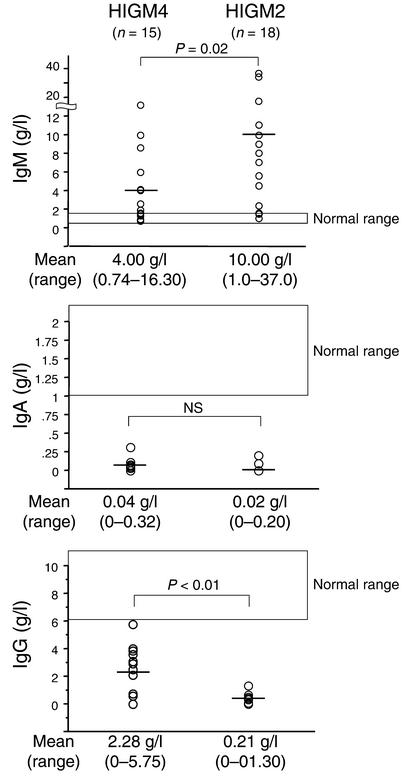
The clinical manifestations of the patients were very similar to those of HIGM2 patients (Table 1). Before the initiation of IVIG, all patients had recurrent upper respiratory tract infections and 14 patients had recurrent lower respiratory tract infections. Other severe infections were occasionally reported (gastrointestinal tract infections, sepsis, lymphadenitis, and osteomyelitis). Lymphoid hyperplasia was frequently observed (seven patients, 47%). A biopsy of a mesenteric lymph node from one patient (P4) had follicular hyperplasia, but no giant germinal centers characteristic of HIGM2 were observed (13) (data not shown). Autoimmune manifestations, especially autoimmune cytopenia, were found in four patients (P4, P7, P10, P13) (Table 1). Serum Ig levels were typical of a HIGM. IgM was elevated in ten patients and within the normal range in five patients (P5, P6, P11, P13, P14). IgM levels were significantly lower (P = 0.02), and IgG levels were significantly higher (P < 0.01), than in HIGM2 patients. Serum IgG level was very low (<1.0 g/l) in five patients (P1, P7, P8, P12, P15), while residual IgG levels (>2.0 g/l) were found in eight patients. In four of them (P4, P5, P11, P14), levels of IgG isotypes were the following: IgG1, 1.8–2.8 g/l; IgG2, undetectable to 0.6 g/l; IgG3, undetectable to 1.2 g/l; and IgG4; undetectable to 0.1 g/l. IgG levels before IVIG were not available in two patients (P6, P10). IgE was not detectable in the six tested patients (P5–P8, P11, P14). IgA levels were also undetectable (<0.10 g/l) in all but one patient (P8) (Figure 1). All patients exhibited normal numbers of circulating lymphocytes and B cells. The percentage of CD27+ memory B cells was normal (>10% of CD19+ B cells, range: 10.0–41.9%) in five patients (P5, P9–P12) and low (<10% of CD19+ B cells, range: 2.3–9.5%) in eight patients. The number and tested functions of T cells and NK cells were normal (data not shown).
Table 1.
Clinical manifestations of patients with HIGM4
Figure 1.
Serum Ig levels in HIGM4 and HIGM2 patients. Horizontal bars indicate the mean levels for each group of patients. Levels from age-matched controls are indicated.
sCD40L+rIL-4–induced proliferation and CSR of B cells from HIGM4 patients.
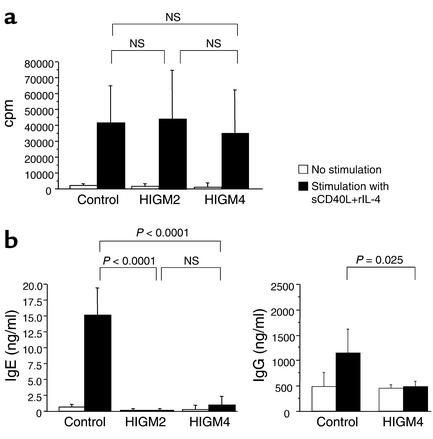
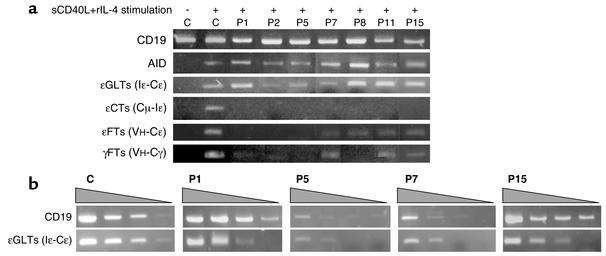
sCD40L+rIL-4–induced B cell proliferation was normal in all tested patients (n = 15), contrasting with a complete lack of CSR toward IgE (n = 15) and IgG (n = 4) (Figure 2), a defect present even in patients exhibiting a residual in vivo CSR. Since HIGM2 (AID-deficient) B cells exhibit the same phenotype (14), and although the AID gene sequence was normal in HIGM4 patients, we investigated AID mRNA transcript levels in activated B cells from seven patients. AID transcripts were found in all tested patients (Figure 3a), indicating that the CSR defect is located downstream of the induction of AID expression. Induction of Ig gene germ-line transcripts (GLTs) is required to enable CSR. To identify the stage at which the CSR defect occurs, we analyzed εGLTs (Iε-Cε) from HIGM4 B cells. In all seven tested cases, εGLTs were induced following sCD40L+rIL-4 activation, showing that this early step of CSR occurs normally. Titration of εGLTs as compared with CD19 transcripts performed in four cases (P1, P5, P7, P15) did not reveal any obvious difference with controls (Figure 3b). In contrast, circle transcripts of IgE (εCTs, the transcribed RNA from excised circular DNA during CSR) was not found in the B cells of any of the patients tested (Figure 3a). IgE gene functional transcripts (εFTs, VH-Cε) were also not found in three patients (P1, P2, P5) and were very faintly positive in four patients (P7, P8, P11, P15). IgG gene functional transcripts (γFTs, VH-Cγ) were also not detected in four cases (P1, P2, P5, P8) and were faintly positive in three (P7, P11, P15). However, unlike in controls, no induced production of IgE and IgG was detected in culture supernatants (Figure 2b). This reflects that γFTs detection is a more sensitive technique than IgG detection by ELISA.
Figure 2.
B cell responses to sCD40L+rIL-4 activation. (a) Normal B cell proliferative response to sCD40L+rIL-4 stimulation in HIGM4 patients. PBMCs from age-matched controls (n = 24), HIGM2 patients (n = 16), and HIGM4 patients (n = 13) were stimulated with sCD40L and rIL-4 for 5 days. White bars, no stimulation; black bars, 12-day stimulation with sCD40L+IL-4. Mean uptakes of [3H]thymidine (cpm) are shown. Error bars show the SD. (b) Defective IgE and IgG CSR following stimulation by sCD40L+rIL-4 in HIGM4 patients. PBMCs from normal controls (n = 20 for IgE, n = 5 for IgG), HIGM2 patients (n = 20), and HIGM4 patients (n = 13 for IgE, n = 4 for IgG) were stimulated with sCD40L+rIL-4 for 12 days. White bars, no stimulation; black bars, 12-day stimulation with sCD40L+IL-4. Concentrations of IgE and IgG in the culture supernatants were quantified by ELISA. Mean values are shown. Error bars show the SD.
Figure 3.
Detection of RNA transcripts of CSR-related genes. (a) RNA transcripts of the CD19 (as control), AID, germ-line (εGLTs), circular (εCTs), and functional (εFTs) transcripts for IgE, and of the functional transcripts for IgG (γFTs) were detected by RT-PCR after stimulation with sCD40L+rIL-4 for 5 days in control subjects (C) and in HIGM4 patients (P1, P2, P5, P7, P8, P11, P15). (b) CD19 transcripts and εGLTs levels were analyzed using different quantities of cDNA (1:1, 1:10, 1:100, and 1:1,000 dilution) in RT-PCR for selected cases (P1, P5, P7, P15).
Detection of CSR-induced DNA DSBs in the switch region of the μ heavy chain of B cells from HIGM4 patients.
As DNA DSBs specifically occur in Sμ regions during CSR (15) in normal conditions but are defective in activated AID-deficient B cells (N. Catalan et al., manuscript submitted for publication) (18), we sought DSBs in the Sμ region of purified B cells from HIGM4 patients after a 5-day culture in the presence of sCD40L+rIL-4. Activated, viable CD19+PI– B cells were purified by FACS sorting, and LM-PCR was used to detect DSBs in Sμ regions (14). After hybridizing the LM-PCR products to a 32P-labeled Sμ-specific DNA probe, several bands were detected in the B cells (2 × 103 to 25 × 103 cells per lane) from the six tested HIGM4 patients (P1, P2, P4, P6–P8) and in B cells from the controls but not in activated T cells (Figure 4b). Cloning and sequencing of the LM-PCR products of the sCD40L+rIL-4–activated B cells from patients and controls revealed that the linker was ligated to the Sμ region at different sites in 58 of 103 clones (56.3%) and 36 of 64 clones (56.2%), respectively. In other clones as well as in all clones from activated T cells, nonspecific linker-linker ligation products were detected. These data demonstrate that, in contrast to HIGM2 (18), Sμ DSBs occur in CSR-induced B cells from patients with HIGM4.
In summary, the selective CSR defect of HIGM4 B cells is neither the consequence of faulty AID gene induction nor caused by the inability to transcribe the Ig locus; the defect must be located downstream of the AID-induced DNA cleavage of switch regions.
Analysis of SHMs in CD27+ B cells from HIGM4 patients.
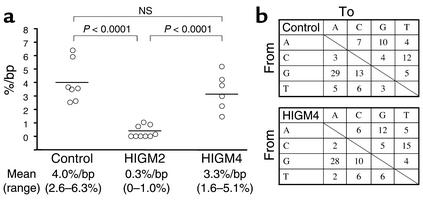
We next studied the generation of SHMs in the VH region of IgM in purified CD19+CD27+ B cells from the patients and controls. The generation of SHMs was normal in CD27+ B cells from all tested patients (n = 6, P2, P5–P9), in terms of both percentage of mutations per bp (mean 3.4%, range: 1.6–5.1%) in the VH3-23 region and percentage of mutated clones (mean 85.1%, range: 71.4–100%) (Figure 5a). This finding sharply contrasts with the strongly defective generation of SHMs observed in AID-deficient patients (Figure 5a) (13). The mean frequency of mutations at dG or dC residues was 64.8% (range: 54–80%) in HIGM4 and 65.0% (range: 62–68%) in controls. The mean frequency of transition mutations at dG or dC (dG > dA, dC > dT) in patients was 65.1% (range: 59–76%) similar to control values(63.1%, range: 58–72%). The mean frequency of transition mutations at dA or dT (dA > dG, dT > dC) in patients was 48.8% (range: 40–75%) was also similar to control values (45.7%, range: 33–57%) (Figure 5b). Thus, HIGM4 is characterized by the normal generation of SHMs that exhibit normal patterns.
Figure 5.
Generation of SHMs in the VH region of IgM in HIGM4. (a) The frequency of SHMs in the VH region of IgM is normal in HIGM4 patients. SHMs in the VH3-23 region of the IgM were analyzed using cDNA from CD19+CD27+ B cells from controls (n = 7), HIGM2 patients (n = 9), and HIGM4 patients (n = 6, P2, P5–P9). Horizontal bars indicate the mean values. (b) The pattern of SHMs in the VH region of IgM is normal in HIGM4 patients. Mean percentages of nucleotide changes and targets of SHMs in the VH3-23 region of IgM in controls (n = 7) and HIGM4 patients (n = 6, P2, P5–P9) are shown.
Sequence of the uracil-DNA-glycosylase gene.
As uracil-DNA-glycosylase (UNG) gene–deficient mice exhibit a CSR defect with quantitatively normal SHMs in vivo (19), we sequenced the UNG gene from HIGM4 patients. The sequence of the UNG gene was normal for all 15 patients (data not shown).
Discussion
We herein describe a new HIGM entity, HIGM4, with a clinical phenotype that is similar to that of AID deficiency (HIGM2) but without any mutations in the AID gene. Like HIGM2 patients, HIGM4 patients suffer from recurrent bacterial infections from childhood, and they do not suffer from opportunistic infections, indicating that T cell immunity is unaffected. Lymphoid organ hyperplasia appears to be less severe in HIGM4 than in HIGM2, although it affects half of the patients. Like in HIGM2, autoimmune features, especially cytopenias, are found in a quarter of patients. The HIGM is slightly less pronounced in HIGM4 than in HIGM2, as serum IgG levels are less markedly diminished.
B cells proliferated normally following sCD40L+rIL-4 activation. However, in vitro CSR toward IgE and IgG was profoundly defective, as in HIGM2. Like in HIGM2, the defect must occur downstream of the induction of GLTs, since GLTs were normally induced after sCD40L+rIL-4 activation. However the defect must occur upstream of the recombination process, as no excision loops (CTs) were found. Thus, although the AID gene sequence was normal, we considered that the defective expression of AID might account for HIGM4. This hypothesis was ruled out by the finding of normal amounts of AID RNA transcripts in activated B cells from HIGM4 patients.
The normal (or elevated) IgM levels exclude the diagnosis of common variable immunodeficiency (CVID), which is characterized by a decrease in the serum concentration of all Ig isotypes, including IgM (20). In addition, in vitro CSR was reported to be normal when B cells from CVID patients were analyzed (21). However, as CVID is a heterogeneous and poorly defined syndrome, we cannot exclude the possibility that there is some overlap between CVID and HIGM4.
As most identified cases of HIGM4 are sporadic, we cannot be totally sure of how or whether this syndrome is inherited. The early onset is suggestive of an inherited disease. An X-linked inheritance can be ruled out by the observed sex ratio. An autosomal dominant mode of inheritance is unlikely, given the family history. An autosomal recessive mode of inheritance is suggested by the facts that one patient was from a consanguineous family and that two brothers from a non-consanguineous family were affected. However, we cannot exclude the possibility that HIGM4 is a multifactorial disease that involves predisposition genes and/or environmental factors.
The immunological investigations strongly suggest that HIGM4 is a syndrome, characterized in all tested patients by a CSR defect but by the normal generation of SHMs of the Ig genes. These data suggest, in sharp contrast to AID deficiency, the existence of a defect in a CSR-specific cofactor for AID. This hypothesis seems, however, unlikely. The CSR defect observed in HIGM4 occurs downstream of the CSR-associated double-strand DNA breaks, a step that occurs only in the presence of a functional AID gene (N. Catalan et al., manuscript submitted for publication) (18, 22, 23). Thus, the HIGM4-associated block in CSR must occur downstream of AID function.
How AID induces DNA breaks is controversial. As AID has a homologous sequence to the RNA-editing enzyme APOBEC-1, it has been proposed that AID edits an RNA that encodes an endonuclease (12). However, recent data strongly suggest that AID acts directly on DNA, deaminating dC into dU residues. dU is subsequently removed from DNA by the uracil-DNA glycosylase (UNG), leading to DNA breaks (24–26). The nicks that occur on dG or dC residues located nearby on two opposite DNA strands can lead to the DSBs that we observe during CSR after end processing (27).
A UNG deficiency can be suggested in HIGM4 patients. However, UNG-deficient mice present with a mild CSR defect and a normal generation of SHMs characterized by a skewed pattern to transitions at the dG and dC residues. This is clearly not the case in HIGM4 patients, who present with a more pronounced CSR defect and a normal generation of SHMs with a normal pattern. UNG deficiency has been excluded in all patients by gene sequencing.
A stronger argument is given by the observation that CSR-induced HIGM4 B cells undergo normal DNA DSBs, suggesting a defect located downstream of this step, likely in DNA repair. AID-induced DNA breaks are necessary for both CSR and SHMs, but the DNA repair processes involved differ (28). Nonhomologous end joining (NHEJ) machinery, which is required for CSR DNA repair but not for SHM DNA repair (29–31), is also required for V(D)J recombination of T and B cell receptors (32). Therefore, it seems unlikely that NHEJ factors are involved, as T and B cells from HIGM4 patients exhibit functional antigen-specific receptors. As molecules from the mismatch repair pathway, especially Msh2, Mlh1, and Pms2, participate in CSR (33–36), it is possible that HIGM4 patients carry a defect in one or several of these molecules. However, the drastic defect in CSR together with the normal frequency and pattern of SHMs in HIGM4 does not favor the involvement of these molecules in the pathogenesis of the disease. It can thus be proposed that one or several other undefined factor(s) involved in the CSR process are defective in HIGM4. Alternatively, HIGM4 may result from a deficiency in a factor required for the survival of switched B cells.
In summary, this new HIGM entity is characterized by a selective CSR defect, located downstream of AID activity, with normal SHM generation. Further analysis of HIGM4 should provide new insights into the final steps of CSR.
Acknowledgments
We are grateful to Immunex (Seattle, Washington, USA) for kindly providing the recombinant soluble CD40L. We thank I. Tezcan and F. Ersoy (Ankara, Turkey) for taking care of patients. This work was supported by grants from INSERM, l’Association de la Recherche Contre le Cancer (ARC), la Ligue Contre le Cancer, the European Economic Community (contract no. QLG1-CT-2001-01536- IMPAD), the Louis Jeantet Foundation, and the Hungarian Scientific Research Fund (grant no. OTKA 38095). P. Revy is a scientist from Centre National de Recherche Scientifique (Paris, France). N. Catalan is supported by l’ARC.
Footnotes
See the related Commentary beginning on page 19.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: hyper-IgM syndrome (HIGM); class-switch recombination (CSR); CD40 ligand (CD40L); soluble CD40L (sCD40L); Ig gene germ-line transcript (GLT); activation-induced cytidine deaminase (AID); somatic hypermutation (SHM); intravenous Ig (IVIG); recombinant interleukin-4 (rIL-4); Ig gene circle transcript (CT); Ig gene functional transcripts (FT); ligation-mediated PCR (LM-PCR); propidium iodide (PI); double-strand break (DSB); switch region of the μ heavy chain (Sμ region); variable region of the Ig heavy chain (VH region); uracil-DNA glycosylase (UNG); common variable immunodeficiency (CVID); nonhomologous end joining (NHEJ).
References
- 1.Durandy A, Honjo T. Human genetic defects in class-switch recombination (hyper-IgM syndromes) Curr. Opin. Immunol. 2001;13:543–548. doi: 10.1016/s0952-7915(00)00256-9. [DOI] [PubMed] [Google Scholar]
- 2.Korthauer U, et al. Defective expression of T-cell CD40 ligand causes X-linked immunodeficiency with hyper-IgM. Nature. 1993;361:539–541. doi: 10.1038/361539a0. [DOI] [PubMed] [Google Scholar]
- 3.DiSanto JP, Bonnefoy JY, Gauchat JF, Fischer A, de Saint Basile G. CD40 ligand mutations in x-linked immunodeficiency with hyper-IgM. Nature. 1993;361:541–543. doi: 10.1038/361541a0. [DOI] [PubMed] [Google Scholar]
- 4.Aruffo A, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 5.Allen RC, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259:990–993. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari S, et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12614–12619. doi: 10.1073/pnas.221456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banchereau J, et al. The CD40 antigen and its ligand. Annu. Rev. Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 8.Seyama K, et al. Mutations of the CD40 ligand gene and its effect on CD40 ligand expression in patients with X-linked hyper IgM syndrome. Blood. 1998;92:2421–2434. [PubMed] [Google Scholar]
- 9.Doffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat. Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 10.Jain A, et al. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat. Immunol. 2001;2:223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 11.Durandy A, et al. Abnormal CD40-mediated activation pathway in B lymphocytes from patients with hyper-IgM syndrome and normal CD40 ligand expression. J. Immunol. 1997;158:2576–2584. [PubMed] [Google Scholar]
- 12.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 13.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 14.Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 15.Wuerffel RA, Du J, Thompson RJ, Kenter AL. Ig Sgamma3 DNA-specifc double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J. Immunol. 1997;159:4139–4144. [PubMed] [Google Scholar]
- 16.Levy Y, et al. Defect in IgV gene somatic hypermutation in common variable immuno-deficiency syndrome. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13135–13140. doi: 10.1073/pnas.95.22.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kvaloy K, et al. Sequence variation in the human uracil-DNA glycosylase (UNG) gene. Mutat. Res. 2001;461:325–338. doi: 10.1016/s0921-8777(00)00063-x. [DOI] [PubMed] [Google Scholar]
- 18.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rada C, et al. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin. Immunol. 1999;92:34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 21.Nonoyama S, Farrington M, Ishida H, Howard M, Ochs HD. Activated B cells from patients with common variable immunodeficiency proliferate and synthesize immunoglobulin. J. Clin. Invest. 1993;92:1282–1287. doi: 10.1172/JCI116701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bross L, Muramatsu M, Kinoshita K, Honjo T, Jacobs H. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J. Exp. Med. 2002;195:1187–1192. doi: 10.1084/jem.20011749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 25.Faili A, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 26.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Kinoshita K, Honjo T. Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13860–13865. doi: 10.1073/pnas.241524898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin A, Scharff MD. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2002;2:605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 29.Rolink A, Melchers F, Andersson J. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- 30.Casellas R, et al. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manis JP, et al. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blunt T, et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrenstein MR, Neuberger MS. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrader CE, Vardo J, Stavnezer J. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]