Abstract
Quantitative analyses of biological sequences generally proceed under the assumption that individual DNA or protein sequence elements vary independently. However, this assumption is not biologically realistic because sequence elements often vary in a concerted manner resulting from common ancestry and structural or functional constraints. We calculated intersite associations among aligned protein sequences by using mutual information. To discriminate associations resulting from common ancestry from those resulting from structural or functional constraints, we used a parametric bootstrap algorithm to construct replicate data sets. These data are expected to have intersite associations resulting solely from phylogeny. By comparing the distribution of our association statistic for the replicate data against that calculated for empirical data, we were able to assign a probability that two sites covaried resulting from structural or functional constraint rather than phylogeny. We tested our method by using an alignment of 237 basic helix–loop–helix (bHLH) protein domains. Comparison of our results against a solved three-dimensional structure confirmed the identification of several sites important to function and structure of the bHLH domain. This analytical procedure has broad utility as a first step in the identification of sites that are important to biological macromolecular structure and function when a solved structure is unavailable.
Quantitative analyses of biological sequences are the cornerstone for studies in bioinformatics and molecular evolution. Such analyses generally proceed assuming that the sites in individual DNA or protein sequences vary independently, i.e., amino acid replacements at site X occur independently of those at site Y (1). Biochemical and biophysical studies show this assumption is not biologically realistic because sequence elements often change in a concerted manner (2–6). Nonrandom associations among sites within sequences arise from at least three sources: (i) chance, (ii) common ancestry (= phylogeny), and (iii) structural or functional constraints. (For simplicity, associations resulting from structure and function are considered to be equivalent.) Effectively discriminating among these underlying causes facilitates understanding the origin and magnitude of associations observed among sites in biological sequences and clarifying the role of such associations in evolution.
The first step in resolving questions about the origins of associations among sequence elements is to generate replicate data sets that vary according to specific underlying evolutionary models. For biological sequences, the typical model components are a reconstructed phylogeny and a nucleotide or amino acid substitution matrix. These components are relevant because sequence diversity has been generated by a process of descent with modification from a common ancestor.
Historical associations between sequences are represented by the reconstructed phylogeny. The topology of the evolutionary tree specifies the cladistic relationships among sequences, whereas the branch lengths reflect the amount of change that has occurred among sequences. The specific changes that occur in the various sequences are summarized by the substitution matrix. This matrix can consist of uniform substitution probabilities [e.g., Jukes–Cantor model for DNA substitution (7)], be partially parameterized [Kimura two-parameter model for DNA (8)], or completely parameterized [Jones–Taylor–Thornton (9) substitution matrix for proteins]. In combination, the phylogeny and substitution matrix provide the parameters necessary to generate stochastic data having historical relationships and substitution classes reflecting specific conditions. The parametric bootstrap procedure (10–12) uses this data-generation algorithm to create replicate data sets that can be used to investigate the underlying properties of aligned biological sequences.
Herein, we describe a general analytical method based on parametric bootstrap simulations for the discrimination of intersite associations resulting from stochastic and phylogenetic sources from those resulting from structural and functional associations. When a general substitution matrix (i.e., one derived from a broad survey of protein sequences rather than the specific data set being analyzed) is used, data generated with the parametric bootstrap procedure will have intersite associations arising only from shared evolutionary history. Therefore, an intersite association statistic calculated for data sets generated by using the parametric bootstrap will reflect only associations among aligned sequence sites resulting from phylogeny or chance. From the distribution of this statistic, one can calculate a threshold value above which the statistic will have a specific probability of resulting from causes other than phylogeny. Comparison of association statistic values calculated for the empirical data alignment against this parametric bootstrap threshold allows identification of pairs of sites having a specific probability of interaction resulting from structure or function.
To demonstrate the utility of this approach, we analyzed a set of 237 sequences containing the basic helix–loop–helix (bHLH) DNA binding and dimerization domain. The bHLH proteins have a well-described structure and are represented by a large number of diverse sequences (13, 14). Having a well-defined three-dimensional structure permits direct comparison of the physical structure of the molecule with numeric data of intersite associations. Thus, sites of known functional and structural importance can be compared against the association statistics involving these sites. The availability of a large number of bHLH sequences increases confidence in the results by reducing the effect of spurious associations.
Methods
Sequence Alignment and Phylogeny Construction.
An alignment of 237 bHLH domains was generated by using clustal w (15) and improved by eye. A phylogenetic tree was then derived by using the neighbor-joining algorithm (16) with mean pairwise distances. We used a substitution matrix generated from a broad collection of protein sequences using the Jones–Taylor–Thornton (JTT) algorithm (9). As a consequence, our model for amino acid substitution is not unduly influenced by the idiosyncrasies of a particular protein family. Further, the resulting model has broad generality because the JTT algorithm accounts for the underlying phylogeny of the sequences when calculating the probability of change between amino acids. Thus, data generated from a random ancestral sequence using this general substitution matrix and a specified phylogeny should have either chance or phylogeny as their only sources of observed association.
Alternatively, one could use a substitution matrix derived from the specific data set being analyzed. To demonstrate the effect a substitution matrix of this type would have on the parametric bootstrap analysis, we used the rind program (17) to calculate a maximum-likelihood substitution matrix based on the bHLH protein sequences. It is expected that a matrix of this type would reflect the biases resulting from phylogeny, structure, and function that are inherent in the empirical data being analyzed.
Calculation of Intersite Associations.
The next step is to accurately estimate the magnitude of association between pairs of amino acid sites. Because sequence elements are symbol variables with no underlying metric, conventional statistical procedures for estimating correlation among sites cannot be used (14). Thus, intersite associations were estimated by using the mutual information statistic (MI) from information theory (18, 19). Mutual information measures the extent of association between two positions in a sequence beyond that expected resulting from chance. The mutual information MIXY between sites X and Y is calculated as:
 |
1 |
where P(Xi) is the probability of i at site X, P(Yj) is the probability of j at site Y, and P(Xi, Yj) is the joint probability of i at site X and j at site Y (X ≠ Y). The double summation runs over all possible symbols at those sites. This formula has the property that when symbols vary independently [i.e., P(Xi)P(Yj) = P(Xi, Yj)], so that knowledge of j at site Y does not reduce the uncertainty of i at site X, the mutual information is zero (0).
The minimum MI value of 0 also occurs for invariant sites. Generally, the less variable a site is, the smaller its associated MI values will be. The maximum MI value will occur when the variation at two sites is perfectly correlated. Using a base-20 logarithm (n = 20 in Eq. 1, corresponding to the 20 peptide-forming amino acids) scales the maximum possible MI value to unity, which will occur when the residues at these sites are uniformly distributed. The maximum MI value will decline as the distribution of residues at each site departs from uniformity.
Results and Discussion
MI Distributions.
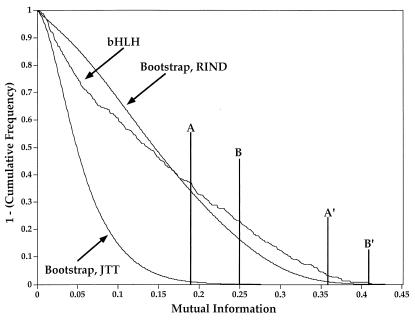
Fig. 1 provides inverted cumulative frequency distributions of MI values calculated for the alignment of 237 bHLH domains and 1,000 parametric bootstrap replicates calculated using two different types of substitution models. Inverted cumulative distributions are calculated by subtracting from unity the cumulative frequency within a particular range of MI values. In this way, one achieves a distribution that declines in value as the independent variable increases.
Figure 1.
Inverse cumulative frequency distribution of MI values for the alignment of 237 bHLH protein sequences and 1,000 parametric bootstrap replicates using either the JTT substitution matrix or the rind substitution matrix. MI values were calculated by using Eq. 1 with n = 20 so that the maximum possible value is unity. Line A is the P < 0.01 threshold for the JTT replicates at MI = 0.188. Line B is the P < 0.001 threshold for the JTT replicates at MI = 0.250. Line A′ is the P < 0.01 threshold for the rind replicates at MI = 0.359. Line B′ is the P < 0.001 threshold for the JTT replicates at MI = 0.408. These were the MI values that were >99% (for P < 0.01) or 99.9% (for P < 0.001) of the MI values calculated in the parametric bootstrap replicates. Because MI is a pairwise measure, x(x − 1)/2 values were calculated in each replicate, where x is the number of nongapped sites in the alignment. For the alignment of 237 bHLH sequences, there were 32 sites without gaps, resulting in 496 MI values per replicate.
The inverted cumulative frequency distribution of MI values for the parametric bootstrap replicates is then used to calculate a threshold for acceptability of a false-positive result, as described in the Fig. 1 legend. Setting a statistical acceptability threshold permits the identification, within a quantifiable error, of those intersite associations most probably arising from structural/functional causes. For example, any pair of amino acid sites within the bHLH domain alignment having an MI value >0.188 has a probability of <0.01 of resulting from phylogeny or chance and, consequently, a >0.99 probability of reflecting an association resulting from structural/functional constraints. These probabilities are reduced and increased by an order of magnitude (0.001 and 0.999, respectively) for any pair of sites having MI >0.250. Because these MI values have been calculated using a base-20 logarithm, the maximum possible MI value is unity, although the largest MI value calculated for any pair of sites in the bHLH domain was 0.413. The sites having MI values >0.188 are presented in Table 1.
Table 1.
MI values calculated for 237 bHLH domains and arranged by site number
| Site no. | MI | Site no. | MI | Site no. | MI | |||
|---|---|---|---|---|---|---|---|---|
| 30 | 37 | 0.3235 | 39 | 44 | 0.2365 | 47 | 65 | 0.2139 |
| 30 | 38 | 0.3269 | 39 | 49 | 0.1896 | 47 | 68 | 0.2087 |
| 30 | 39 | 0.1912 | 39 | 62 | 0.2378 | 47 | 72 | 0.2068 |
| 30 | 41 | 0.2856 | 39 | 65 | 0.2053 | 48 | 49 | 0.3129 |
| 30 | 42 | 0.2559 | 39 | 72 | 0.2123 | 48 | 50 | 0.2229 |
| 30 | 44 | 0.3599 | 41 | 42 | 0.2652 | 48 | 59 | 0.2747 |
| 30 | 45 | 0.2751 | 41 | 44 | 0.3496 | 48 | 61 | 0.2695 |
| 30 | 47 | 0.2328 | 41 | 45 | 0.2120 | 48 | 62 | 0.3184 |
| 30 | 48 | 0.2759 | 41 | 48 | 0.2674 | 48 | 65 | 0.2533 |
| 30 | 49 | 0.3022 | 41 | 49 | 0.2569 | 48 | 68 | 0.2148 |
| 30 | 50 | 0.1954 | 41 | 61 | 0.1978 | 48 | 69 | 0.2511 |
| 30 | 57 | 0.1935 | 41 | 62 | 0.2666 | 48 | 72 | 0.3086 |
| 30 | 59 | 0.2671 | 41 | 65 | 0.2070 | 49 | 50 | 0.2471 |
| 30 | 61 | 0.2415 | 41 | 68 | 0.2395 | 49 | 57 | 0.2059 |
| 30 | 62 | 0.3151 | 41 | 69 | 0.2637 | 49 | 59 | 0.2775 |
| 30 | 65 | 0.2802 | 41 | 72 | 0.2969 | 49 | 61 | 0.3044 |
| 30 | 68 | 0.2676 | 42 | 44 | 0.3545 | 49 | 62 | 0.3388 |
| 30 | 69 | 0.2365 | 42 | 45 | 0.2092 | 49 | 65 | 0.2965 |
| 30 | 72 | 0.3481 | 42 | 47 | 0.1918 | 49 | 68 | 0.2590 |
| 37 | 38 | 0.3764 | 42 | 48 | 0.3140 | 49 | 69 | 0.2734 |
| 37 | 39 | 0.2360 | 42 | 49 | 0.2693 | 49 | 72 | 0.3608 |
| 37 | 41 | 0.3063 | 42 | 59 | 0.2582 | 50 | 57 | 0.2109 |
| 37 | 42 | 0.3476 | 42 | 61 | 0.2440 | 50 | 59 | 0.2229 |
| 37 | 43 | 0.1935 | 42 | 62 | 0.3418 | 50 | 61 | 0.1962 |
| 37 | 44 | 0.4094 | 42 | 65 | 0.3091 | 50 | 62 | 0.2073 |
| 37 | 45 | 0.2957 | 42 | 68 | 0.2128 | 50 | 65 | 0.1910 |
| 37 | 47 | 0.2347 | 42 | 69 | 0.2616 | 50 | 72 | 0.2462 |
| 37 | 48 | 0.3469 | 42 | 72 | 0.2802 | 57 | 62 | 0.2023 |
| 37 | 49 | 0.3716 | 44 | 45 | 0.3231 | 57 | 65 | 0.2046 |
| 37 | 50 | 0.2314 | 44 | 47 | 0.3008 | 57 | 72 | 0.1962 |
| 37 | 57 | 0.1992 | 44 | 48 | 0.3627 | 59 | 61 | 0.3254 |
| 37 | 59 | 0.3462 | 44 | 49 | 0.3565 | 59 | 62 | 0.3526 |
| 37 | 61 | 0.3638 | 44 | 50 | 0.2659 | 59 | 65 | 0.2890 |
| 37 | 62 | 0.3865 | 44 | 57 | 0.2744 | 59 | 68 | 0.1936 |
| 37 | 65 | 0.3569 | 44 | 59 | 0.3535 | 59 | 70 | 0.1953 |
| 37 | 67 | 0.1940 | 44 | 61 | 0.3206 | 59 | 71 | 0.1920 |
| 37 | 68 | 0.3018 | 44 | 62 | 0.4099 | 59 | 72 | 0.3097 |
| 37 | 69 | 0.2834 | 44 | 65 | 0.3866 | 61 | 62 | 0.3435 |
| 37 | 70 | 0.1915 | 44 | 68 | 0.3132 | 61 | 65 | 0.2963 |
| 37 | 72 | 0.3769 | 44 | 69 | 0.2869 | 61 | 68 | 0.2501 |
| 38 | 41 | 0.2871 | 44 | 72 | 0.4130 | 61 | 69 | 0.2422 |
| 38 | 42 | 0.2640 | 45 | 49 | 0.2949 | 61 | 72 | 0.3415 |
| 38 | 44 | 0.3756 | 45 | 59 | 0.2154 | 62 | 65 | 0.4131 |
| 38 | 45 | 0.2419 | 45 | 61 | 0.2426 | 62 | 68 | 0.3092 |
| 38 | 47 | 0.2150 | 45 | 62 | 0.2503 | 62 | 69 | 0.2448 |
| 38 | 48 | 0.2812 | 45 | 65 | 0.2537 | 62 | 70 | 0.1886 |
| 38 | 49 | 0.3269 | 45 | 68 | 0.1919 | 62 | 71 | 0.1925 |
| 38 | 50 | 0.2317 | 45 | 69 | 0.1973 | 62 | 72 | 0.3757 |
| 38 | 59 | 0.2906 | 45 | 72 | 0.2514 | 65 | 68 | 0.2528 |
| 38 | 61 | 0.3135 | 47 | 48 | 0.1966 | 65 | 69 | 0.2338 |
| 38 | 62 | 0.3477 | 47 | 49 | 0.2128 | 65 | 70 | 0.1967 |
| 38 | 65 | 0.2997 | 47 | 57 | 0.2227 | 65 | 72 | 0.3412 |
| 38 | 68 | 0.2936 | 47 | 59 | 0.2227 | 68 | 69 | 0.2018 |
| 38 | 69 | 0.2384 | 47 | 61 | 0.2325 | 68 | 72 | 0.3048 |
| 38 | 72 | 0.3569 | 47 | 62 | 0.2671 | 69 | 72 | 0.3032 |
Only pairs of sites having MI > 0.188 (P < 0.01) are included.
Comparison Against Three-Dimensional Structure.
To gauge the efficacy of this algorithm, we compared the sites presented in Table 1 with the solved three-dimensional structure of a representative bHLH domain. Crystal structure studies have been carried out on the bHLH domains of six proteins: Max (20), E47 (21), MyoD (22), USF (23), PHO4 (24), and SREBP (25). As the bHLH domains in these molecules all have the same general organization of a DNA-binding, predominantly basic α-helix (b), an amphipathic α-helix contiguous with the basic region (H1), a variable length loop, and a second α-helix (H2), we used the bHLH domain of the Max protein as our representative bHLH structure. All site numbers refer to the Max structure as presented by Ferre-D'Amare et al. (20).
Each turn in an α-helix requires approximately 3.6 residues. Therefore, residues that are seven sites apart will lie on the same face of the helix. Also, residues that are three or four sites apart will lie approximately above or below each other. In the initial α-helix (b/H1), site pairs (30, 37), (30, 44), (38, 45), (41, 48), and (42, 49) would be on the same face of the helix and have significant MI values. In this same region, site pairs (37, 41), (38, 41), (38, 42), (41, 44), (41, 45), (42, 45), (44, 47), (44, 48), and (45, 49) would be spatially adjacent in the helix and have significant MI values. In H2, the site pairs (61, 65), (62, 65), (65, 68), (65, 69), (68, 72), and (69, 72) are spatially adjacent and have significant MI values. Site pairs (61, 68), (62, 69), and (65, 72) are on the same face of the helix and have significant MI values. In both helical regions, many of the same sites are involved in these interactions separated by three, four, and seven residues, prompting speculation that these sites are important to helical integrity.
Ferre-D'Amare et al. (20) identified several sites having important interactions within the molecule, with the dimerization partner, or with the DNA recognition sequence. Sites 47 and 57, which have a significant association at P ≤ 0.003, were identified as being important to the stability of the loop conformation. Sites 70 and 71 were shown to be involved in several packing interactions. Many associations involving these two sites [(37, 70), (59, 70), (59, 71), (62, 71), and (65, 70)] were significant at 0.009 ≤ P ≤ 0.007. However, many of the sites involved in the specific packing interactions identified in ref. 20 did not have significant MI values because of the lack of variability at one or both of the sites.
Effect of an Alternative Substitution Model.
In any numerical simulation of a physical process, the validity of the results depends on the assumptions of the underlying models. For phylogenetic analyses, the results are dependent on the confidence one has that the tree is a realistic description of the history of the data being analyzed. The parametric bootstrap also depends on the tree as the source of information about the level and distribution of sequence variation. The residue substitution matrix specifies the probabilities of specific amino acid changes that occur between sequences in the simulation. Biases in this matrix can affect the potential associations measured in the resulting simulated sequences. However, a matrix having no biases (i.e., a matrix of identical substitution probabilities) would ignore the biology of the substitution process.
As seen in Fig. 1, the distribution of MI values generated using the parametric bootstrap with the rind substitution matrix is much more similar to the distribution of the empirical MI values than the distribution generated using the JTT substitution matrix. The MI values for the two statistical thresholds (P < 0.01 and P < 0.001) are increased to 0.359 and 0.408, respectively, for the rind matrix distribution. Although there are empirical MI values greater than these thresholds, several of the significant associations identified above have MI values that fall below the rind thresholds. This reduction in sensitivity is the result of the specificity of the rind substitution matrix to the bHLH sequence data, which guarantees that any biases because of structural and functional constraints on substitution will be incorporated into the substitution matrix. For this reason, any analyses incorporating constraints on the evolution of sites in biological sequences should use a substitution matrix derived from a broad sample of sequences.
The way in which structural and functional constraints act on the evolutionary process will influence the variation seen in existing molecular sequences. These influences will be incorporated into the reconstructed phylogeny by the algorithm used to derive it. This leads to a certain level of circularity in the use of the parametric bootstrap to partition sources of association. However, the existence of empirical values greater than reasonable statistical thresholds for acceptance of false-positive results, and the divergence of the empirical and bootstrapped JTT MI distributions, lead us to believe that the problem of circularity is not insurmountable.
Statistical Identification of Structurally and Functionally Important Sites.
We used the parametric bootstrap algorithm to construct a statistical distribution that reflects the associations between sites in a biological sequence exclusively resulting from a specific phylogeny (and chance). This distribution was then used to calculate a threshold, above which the calculated statistic should (with a specific probability) reflect structural and functional associations. Several sites identified from the solved three-dimensional structure as being important to bHLH domain structure and function were found to correlate with predictions based on MI values. It is possible that pairs of sites with values less than the threshold could be exhibiting associations resulting from structure or function. However, based on the distribution from the parametric bootstrap replicates, the level of confidence one would have in making this assertion would be reduced.
Using this parametric bootstrap-based algorithm to differentiate phylogenetic and chance associations from those resulting from structure and function will be quite useful for any sequence analysis that requires knowledge of higher-level structure. For example, in phylogenetic studies, this approach allows one to construct a character weighting scheme so that the resulting analysis more closely reflects the primary assumption of intersite independence. For molecular function analyses, the statistical threshold permits identification of sites of possible importance in site-directed mutagenesis analyses. For protein structural analyses, the statistical threshold allows the identification of sites important to structure without having a solved structure. Comparison against a solved structure could identify sites important to secondary or tertiary structure that may not be obvious by inspection of the solved structure.
Acknowledgments
We thank Jim Rosinski, Brian Rhees, Jason Lowry, and two anonymous reviewers for comments that have strengthened this work. We also thank Walter Fitch for his advice and assistance. This work was supported by National Institutes of Health Grant GM-5-46472 (to W.R.A.).
Abbreviations
- bHLH
basic helix–loop–helix
- MI
mutual information
- JTT
Jones–Taylor–Thornton
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070154797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070154797
References
- 1.Swofford D L, Olsen G J, Waddell P J, Hillis D M. In: Molecular Systematics. 2nd Ed. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 407–514. [Google Scholar]
- 2.Chelvanayagam G, Eggenschwiler A, Knecht L, Gonnet G H, Benner S A. Protein Eng. 1997;10:307–316. doi: 10.1093/protein/10.4.307. [DOI] [PubMed] [Google Scholar]
- 3.Pollock D D, Taylor W R. Protein Eng. 1997;10:647–657. doi: 10.1093/protein/10.6.647. [DOI] [PubMed] [Google Scholar]
- 4.Thompson M J, Goldstein R A. Proteins. 1996;25:28–37. doi: 10.1002/(SICI)1097-0134(199605)25:1<28::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 5.Gobel U, Sander C, Schneider R, Valencia A. Proteins Struct Funct Genet. 1994;18:309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 6.Taylor W R, Hatrick K. Protein Eng. 1994;7:341–348. doi: 10.1093/protein/7.3.341. [DOI] [PubMed] [Google Scholar]
- 7.Jukes T H, Cantor C. In: Mammalian Protein Metabolism. Munro H N, editor. Vol. 3. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 8.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 9.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 10.Efron B, Tibshirani R J. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 11.Goldman N. J Mol Evol. 1993;36:182–198. doi: 10.1007/BF00166252. [DOI] [PubMed] [Google Scholar]
- 12.Huelsenbeck J P, Hillis D M, Jones R. In: Molecular Zoology: Advances, Strategies, and Protocols. Ferraris J D, Palumbi S R, editors. New York: Wiley–Liss; 1996. pp. 19–45. [Google Scholar]
- 13.Atchley W R, Fitch W M. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atchley W R, Terhalle W, Dress A W. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 17.Bruno W. Mol Biol Evol. 1996;13:1368–1374. doi: 10.1093/oxfordjournals.molbev.a025583. [DOI] [PubMed] [Google Scholar]
- 18.Shannon C, Weaver W. The Mathematical Theory of Information. Urbana, IL: Univ. of Illinois Press; 1949. [Google Scholar]
- 19.Applebaum D. Probability and Information: an Integrated Approach. New York: Cambridge Univ. Press; 1996. [Google Scholar]
- 20.Ferre-D'Amare A R, Prendergast G C, Ziff E B, Burley S K. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 21.Ellenberger T, Fass D, Arnaud M, Harrison S C. Genes Dev. 1994;15:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 22.Ma P C, Rould M A, Weintraub H, Pabo C O. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 23.Ferre-D'Amare A R, Pognonec P, Roeder R G, Burley S K. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parraga A, Bellsolell L, Ferre-D'Amare A R, Burley S K. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]