Abstract
The human β-globin locus is home to five genes that are regulated in a tissue-specific and developmental stage-specific manner. While the exact mode of expression remains somewhat enigmatic, a significant effort has been focused at the locus control region (LCR). The LCR is marked by five DNase I-hypersensitive sites (HS) ∼15 kb upstream of the ε-globin gene. Nuclear matrix-associated regions (MARs) organize chromatin into functional domains and at least one of the HS appears bound to the nuclear matrix. We have employed an in vivo based PCR MAR assay to investigate the role of MAR-mediated regulation of the β-globin locus. This was facilitated with a novel reaction efficiency based quantitative real-time PCR analysis software tool, Target Analysis Quantification. Using a log-linear regression strategy, discordances were eliminated. This allowed us to reliably estimate the relative amount of initial template associated with the nuclear matrix at 15 unique regions spanning the β-globin locus in both non-expressing and expressing cell lines. A dynamic association dependent on expression status was revealed both at the LCR/5′HS region and within the second intron of the β-globin gene. These results provide the first evidence that nuclear matrix association dynamically mediates the looping of the β-globin locus to achieve transcriptional control.
INTRODUCTION
With complete sequences available for the human genome (1,2), an enormous amount of attention has been directed towards understanding the relationships that exist between chromatin structure and function (3–7). An elaborate chromatin structure is not only thought to package and protect the genome but also provide a mechanism to regulate gene expression.
Individual chromosomes occupy distinct territories within the cell nucleus (4,8,9). Until recently, the nuclear matrix was regarded as a static structure. It was thought to arrange chromatin into transcriptional domains, by providing a place for the transcriptional machinery and active genes to intermingle (7,10). In some cases these anchors can also function to shield specific domains from neighboring enhancers and the silencing effects of heterochromatin (11,12). While these and other roles of nuclear matrix–DNA interaction have been scrutinized (3), the exact nature of their association and the role provided by the nuclear matrix remains to be fully characterized.
The β-globin gene cluster has long been a prototype for studying the regulation of multigenic loci (13–16). This locus spans a region of ∼70 kb and contains five developmentally regulated and tissue-specific genes, i.e. ε→Gγ→Aγ→δ→β. Hematopoeiesis originates in the embryonic yolk sac expressing the ε-globin gene. As development proceeds, the γ-globin genes are expressed in the fetal liver, the δ-globin gene is expressed in the neonate and the β-globin gene in the adult bone marrow (15). The classic ‘looping’ model of regulation purports that the five most proximal DNase I-hypersensitive sites (5′HS5–5′HS1) of the LCR work in concert to interact with various regions of the domain at definitive times during development through a looping mechanism (17–21). Each 5′HS performs a different function in promoting the proper spatial and temporal regulation of the domain, as summarized in Table 1. A computational search and sequence comparison has provided additional evidence to support the looping model and offered a mechanism mediated by alterations in gene-specific nuclear matrix attachment (22). This model predicted that the LCR was consistently located at the nuclear matrix and that specific genes within the locus are either recruited to or away from the nuclear matrix-bound LCR in a developmental and tissue-specific manner. It was proposed that this movement either enhanced or attenuated transcription by altering the three-dimensional proximity of the genes to the LCR.
Table 1. Summary of regulatory roles of the five most proximal 5′ hypersensitive sites of the β-globin LCR.
| Mode of regulation | Gene/region | cis-element | Time/tissue | Tissue | Factors/mode of action | References |
|---|---|---|---|---|---|---|
| Insulating | LCR/5′HS5 | Ub | NM binding insulates chromatin domains | (13,45,46) | ||
| Silencing | LCR/5′HS5 | (GATA)7 | All | Er | Binds large complexes of GATA-1, silencing enhancer activity of LCR/5′HS2 | (48) |
| Recruitment, enhancer, chromatin remodeling | γ, β | LCR/5′HS4 | A | Er | Binds NF-E2, GATA-1, Sp1; helps form holocomplex, can be functionally replaced by 5′HS3, but not v/v | (21,49,50) |
| Chromatin remodeling | domain | LCR/5′HS3 | All | Er | Binds EKLF; data is conflicting as to chromatin remodeling activity of this and neighboring HSs. | (16,45,51,52) |
| Enhancing | ε, γ, β | LCR/5′HS2 core | All | Er | Sp1, NF-E2, GATA-1, USF, Tal-1 (SCF) | (21,47) |
| Recruitment | ε, γ, β | LCR/5′HS2 flanking | All | Er | Flanking regions recruit other HSs in looping model to form LCR holocomplex | (47) |
| Architectural? activation? enhancer? | ε, γ, β | LCR/5′HS1 | Unknown | ? | Binds GATA-1, Sp1; no transcriptional activation activity, but deletions have variegated expression | (14,49,53,54) |
LCR, locus control region; HS, DNase I-hypersensitive site; A, adult; Er, erythroid; Ub, ubiquitous; NM, nuclear matrix.
Nuclear matrix association has been assessed using Southern hybridization (23), reassociation (5,24) and standard PCR assays (25). These techniques are not amenable to the rapid genome-wide analysis afforded by real-time PCR. However, to reliably measure differences between matrix-associated and loop enriched DNA samples with quantitative real-time PCR, another dimension inherent to all PCR assays must be addressed. Under ideal conditions, a doubling of PCR products should be observed with each amplification cycle (26,27). This efficiency is seldom achieved (28–30). It usually differs within and among reactions and can vary by as much as 50%. When the initial amount of template in several samples is equal, deviations as small as 5% in reaction efficiency can lead to discordant estimates in the relative amount of starting material (31). However, as we show, the relative amount of initial template can be accurately and reliably determined using a log-linear regression strategy that considers amplification efficiency. This has been implemented in the Target Analysis Quantification (TAQ) software (http://vortex.cs.wayne.edu/Projects.html). The utility of this software is demonstrated by its ability to detect differences in the association of specific regions of the human β-globin locus with the nuclear matrix in both non-expressing and expressing cell lines. The results of this study provide the first evidence that nuclear matrix association mediates the looping transcriptional control of the β-globin locus.
MATERIALS AND METHODS
In silico prediction of matrix attachment
Three different matrix-associated region prediction algorithms were used to detect candidate MARs within 81.1 kb surrounding the β-globin locus (NCBI accession no. NG_ 000007.2). These were MARFINDER, now known as MARWIZ (http://www.futuresoft.org/MarFinder/) (32), SMARTest (http://www.genomatix.de/cgi-bin/smartest_pd/smartest.pl) (33) and MARSCAN (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/) (34).
In vivo determination of matrix attachment
Prior to the in vivo determination of matrix attachment, an endonuclease restriction site map was developed using DS Gene ver. 1.1 (Accelrys, Burlington, MA). Appropriate PCR primer pairs were designed to interrogate the predicted MAR regions using OLIGO ver. 6 (Molecular Biology Insights, Cascade, CO).
An optimal time for nuclear protein extraction was resolved. Human fetal fibroblast (GM00468) and human erythromyeloid (K562) cells stored at –80°C in frozen storage buffer (25) were gently thawed and washed in ∼2 ml phosphate-buffered saline (PBS) supplemented with 1 mg/ml bovine serum albumin (BSA) and Roche Biochemicals Complete Proteinase Inhibitor (CPI) (protease inhibitor cocktail inhibiting serine proteases, cysteine proteases, metalloproteases and calpains, 1 mini tablet per 10 ml solution; Roche Biochemicals, Indianapolis, IN). The cells were resuspended in ∼2 ml of PBS supplemented only with CPI. The cells were lysed on ice for 15 min using 2 ml of nuclei buffer (10 mM Tris pH 7.7, 100 mM NaCl, 0.3 M sucrose, 3 mM MgCl2, 0.5% Triton® X-100, supplemented with CPI). The concentration of nuclei was adjusted to 1.5 × 105/ml with PBS supplemented with CPI and 1.25 × 104 nuclei were attached to slides using a Cytospin 2 cytofuge (Thermo Shandon, Pittsburgh, PA). To determine the optimal extraction time, 50 µl of Halo Buffer (10 mM Tris pH 7.7, 10 mM EDTA, 2 M NaCl, 1 mM dithiothreitol, supplemented with CPI) was placed on the nuclei and incubated for varying times ranging from 0 to 10 min. The reaction was stopped by washing each slide for 1 min in each of a graded series of ice-cold 10×, 5×, 2× and 1× PBS washing solutions then dehydrated using a series of 1 min washes in 50, 70, 95 and 100% graded ice-cold ethanol baths. The nucleic acids were fixed to the slide by baking for 2 h at 70°C. Each halo preparation was stained with DAPI (Vysis, Downers Grove, IL). Nuclei were visualized using a Leica DMRXA2 under a 40× Fluotar objective with a numerical aperture of 0.75 (Leica Microsystems, Wetzlar, Germany). A Retiga EX CCD camera was used to obtain images (Qimaging, Burnaby, BC, Canada) that were subsequently analyzed with Image-Pro Express (Media Cybernetics, Inc., Silver Spring, MD). The time required for nuclear protein extraction was determined as a function of the distance from the edge of the nuclear matrix to the perimeter of the extracted DNA. The minimum time required to reach maximum halo diameter was considered optimal. In this fashion, it was determined that a 3 min extraction was optimal for both the GM00468 and K562 cells.
Preparation of matrix-associated and loop enriched DNA samples
Fractionation of cellular DNA into matrix-associated (MA) and loop enriched (LE) fractions was carried out essentially as described (25). In brief, thawed GM00468 and K562 cells were washed in PBS supplemented with 1 mg/ml BSA and CPI. The cells were pelleted at 200 g for 7 min, at 4°C, washed in PBS supplemented with CPI then repelleted, this time leaving a small volume (∼50 µl) of PBS behind. The cells were resuspended in that small volume then lysed on ice for 15 min following the addition of 2 ml of nuclei buffer supplemented with CPI. Nuclei (1.5 × 107) were pelleted at 200 g for 10 min, at 4°C, and the nuclei buffer was aspirated leaving a small volume to resuspend the nuclei prior to nuclear protein extraction. It is crucial that nuclei are not excessively clumped together during the extraction. Two milliliters of halo buffer supplemented with CPI was then added and the nuclear proteins extracted on ice for 3 min. Extraction was stopped with the addition of 50 mM Tris–HCl pH 8.0, 10 mM MgCl2 to achieve a final concentration of NaCl suitable for restriction endonuclease digestion. The halo preparations were centrifuged at 200 g for 5 min, at 4°C. This mild centrifugation prevented the collapse of the loop onto the matrix structure. The supernatant was then removed leaving ∼2 ml of buffer containing a very loose and gelatinous matrix pellet. The restriction endonucleases were added (∼100 U each, EcoRI/HindIII or EcoRI/BamHI) and the DNA digested at 37°C for 4 h. These preparations were centrifuged at 16 000 g for 3 min, at 4°C, to ensure the collection of all matrix-associated DNA. The supernatant containing the LE and the pellet containing the MA fractions were placed in separate tubes. The MA fraction was washed three times with 500 µl of restriction buffer then centrifuged at 16 000 g for 3 min, at 4°C. After each wash, the supernatant was removed and discarded. Both samples were digested with 120 µg of proteinase K in a proteinase K buffer (50 mM Tris–HCl pH 8.0 buffer containing 50 mM NaCl, 25 mM EDTA, 0.5% SDS) overnight at 55°C, under slow oscillation. The DNA was purified from both fractions using an affinity matrix (25) and concentration determined by fluorescence analysis.
PCR amplification and analysis
Three hundred picograms of MA and LE DNA were used in quadruplicate 20 µl parallel real-time PCR assays using the MJ Research Opticon Monitor (MJ Research, Waltham, MA). The fluorescence of 0.25× SYBR-Green (Molecular Probes, Eugene, OR) was measured at each amplification step. Sixty cycles of PCR were carried out using the HotStar Taq polymerase system (Qiagen, Valencia, CA) with the primer pairs shown in Table 2 at the specified annealing temperatures.
Table 2. Real-time PCR primer information.
| Amplicon | Primer sequence | Product length | Ta | Optical read temperature |
|---|---|---|---|---|
| HS5 f | GTC ATA TAT TTA TTC AAA AGG GTA A | 393 | 56.2 | 77.3 |
| HS5 r | CAA GCA TTT ATT CTA TCT ATC ATC T | |||
| HS3 f | GAC ACG AGG AAA TAG TGT AGA T | 596 | 61 | 79.7 |
| HS3 r | TCT GAG TAT TGG TGT GAG TAA A | |||
| HS2 f | ACC TAG AAG CGG CAG AAT C | 514 | 61.5 | 81.2 |
| HS2 r | GAA ACA TCT TTG CCA CTG TG | |||
| HS1 f | AAT GAA TGA GCA GTC AAA C | 445 | 55.6 | 78.7 |
| HS1 r | CAT GAA GAT GGA TGA ATA AG | |||
| C f | ACA GGC AGG AAT TAA TCT TCA TCG | 443 | 61 | 76.9 |
| C r | AGG GAG GAG CTT CAA GAA AGG TTA T | |||
| 30rep f | GTT GGG TAG AAT GTC CTG TA | 434 | 61 | 78.1 |
| 30rep r | GAA TTC ACC AAC CAG TTA TC | |||
| 34kb f | AAT TCA ATA ATG GCT TCT CAC TCC | 476 | 58.1 | 76.4 |
| 34kb r | CAT TCT TTC CCA TGT ATT TTC AAA C | |||
| Con f | TGG GGG AGG TGG GGA CTT A | 432 | 61.5 | 80.1 |
| Con r | TTG CAG ATG GCG GAC TTC TC | |||
| D f | TTT AGT GGG GTA GTT ACT CCT | 540 | 60.4 | 77.5 |
| D r | TTA TTA TTT ATT GGG TAG TGT GAT A | |||
| E f | CAG AAT CTC AGG CAC ATC ATC | 385 | 60.2 | 76.1 |
| E r | TAT AAC CCC TTT GGA AAA CCT T | |||
| F f | AAT AAT CCT TTT GTC TCT CCA C | 472 | 60.2 | 75.5 |
| F r | TTC CCA TCA GCA TAA ATA AGT A | |||
| G f | GGG CAG GAT TCA GGA TGA C | 421 | 61 | 79.6 |
| G r | GTG TCT GGG GGA ACA AAA AG | |||
| IN2 f | TTT CCC CTT CTT TTC TAT GGT TAA G | 820 | 55 | 81.5 |
| IN2 r | GTG GGA GGA AGA TAA GAG GTA TGA | |||
| H f | GCC CAC AAG TAT CAC TAA GC | 529 | 60 | 78.3 |
| H r | GGG GAA AGG TGG TAT CTC TA | |||
| I f | GGA GAA TCA GGA AAC TAT TAC TC | 475 | 56.2 | 80.8 |
| I r | ATT AAC ATA CCC ATT ATC TCA CAT | |||
| β-actin f | AAC ACC CCA GCC ATG TAC G | 254 | 60 | 87 |
| β-actin r | ATG TCA CGC ACG ATT TCC C | |||
| PRM2 f | AGG CAA GGC AAG GGG GTC TGT | 440 | 60 | 89.3 |
| PRM2 r | TGG CCA TGG GTC CTC TCG TAG A |
Ta, annealing temperature; Optical read temperature, temperature selected to eliminate spurious PCR products prior to plate read.
The initial amount of target and the amplification efficiency were calculated from the fluorescent amplification curves generated by the Opticon Monitor using the TAQ software (http://vortex.cs.wayne.edu/Projects.html). The conjectural product doubling of PCR (26,27) is illustrated by equation 1, where N is the number of amplified molecules, N0 is the initial amount of target and n is the number of amplification cycles.
N = N0 (2n)1
This equation expresses the exponential relationship between the original amount of target and the final quantity of product. A log-linear relationship can be obtained between N and n, by expressing equation 1 as a function of its log, as shown in equation 2, where E represents amplification efficiency.
ln(N) = ln(N0) + n ln(1 + E)2
In this example, E assumes a value between 0 and 1. Using identical primer sets in different parallel reactions E typically ranges from 0.80 to 0.99 (35). In comparison, using different primer sets E can range anywhere from 0.46 to 0.99 (28–30). The TAQ software employs a linear regression of the log transformed fluorescence intensity data to quantify the reaction efficiency and the initial number of target molecules.
RESULTS AND DISCUSSION
It is tempting to utilize the variation in maximum or 50% of the maximum fluorescence as a gauge to estimate differences in target molecules with real-time PCR. However, PCR reactions saturate for different reasons, including the disparity of primer concentration, competition between the denatured product and primer for primer sites during annealing, the premature inactivation of enzyme, insufficient indicator and limitations in detection devices (36). For these reasons, a comparison of starting template concentrations using measurements dependent on maximum fluorescence is deemed inappropriate. To overcome this problem, most quantitative real-time PCR methods employ an arbitrarily selected fluorescence threshold (31,36). To be valid, these methods require that the amplification efficiencies between test and reference samples be equivalent, as differences in starting template estimates are easily changed by shifting the arbitrarily selected threshold up or down. However, PCR amplification efficiencies are seldom equivalent (28–30).
TAQ software
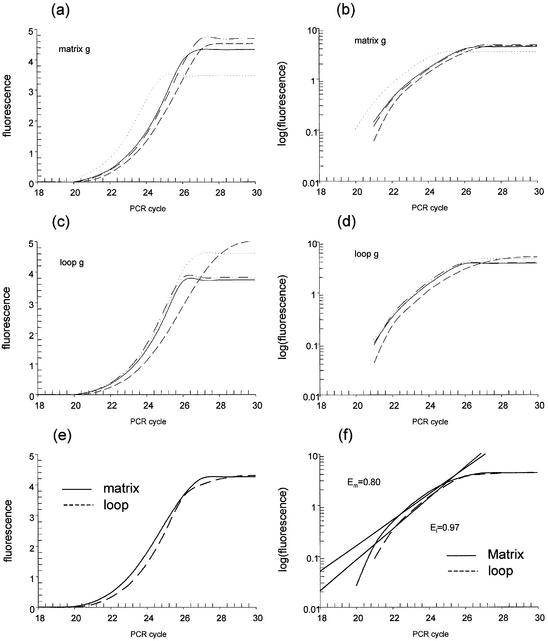
To increase the accuracy in estimating the number of target molecules, we have implemented a log-linear regression analysis of real-time PCR data in the TAQ software (http://vortex.cs.wayne.edu/Projects.html). As shown in Figure 1, the relative amount of initial template is automatically calculated as a function of amplification efficiency, independent of an arbitrarily selected fluorescent threshold. Figure 1a and c shows the original fluorescence intensity data for the G amplicon in the GM00468 cells for both the MA and LE DNA samples. The TAQ software log transforms the data as shown in Figure 1b and d, then fits a line to the best portion of the curve. For illustrative purposes only, the average intensity across the samples at each cycle was taken, as shown in Figure 1e, and the corresponding best fit lines as determined by the TAQ software are shown in Figure 1f. An exhaustive search strategy has been implemented to identify the interval that best fits the model. In effect, the TAQ software fits multiple linear equations to the PCR amplification data then, using the coefficient of determination (r2) and a maximum number of data points, selects the best model of the data. The equation of this line is solved for the y-intercept, to estimate the fluorescence intensity of the 0th cycle of PCR. This value is directly proportional to the initial amount of template. The slope of the fitted line reflects the PCR reaction efficiency.
Figure 1.
Computational method utilized by the TAQ software. Real-time PCR estimates the amount of product by measuring the intensity of a DNA-specific fluorescent dye after each cycle of amplification. (a and c) The original fluorescence intensity data obtained from the Opticon monitor for the G amplicon in the GM00468 cells for both the MA enriched and LE DNA samples. The TAQ software log transforms the data as shown in (b) and (d). (e) The intensity average at each cycle for quadruplicate PCR reactions, clearly demonstrating differences in amplification efficiencies. After transforming the data, the TAQ software fits a line to the best portion of each curve submitted, as shown in (f). An exhaustive search strategy has been implemented to identify the interval that best fits the model. The equations are solved for their y-intercepts, which estimate the fluorescence intensity associated with the 0th cycle of PCR. The slope of the fitted line directly reflects the PCR reaction efficiency and for the average fluorescence intensities across cycles in MA is 0.80, and 0.97 for the LE fraction.
When the TAQ software is launched, a graphical user interface is provided. The user specifies a file containing tab delimited fluorescence intensity data in the appropriate text field. The default format is that of the MJ Research Opticon Monitor. This format has a single top row containing column headers. The first three columns contain ‘Read’, ‘Cycle’ and ‘Step’, followed by any number of columns of fluorescence intensity data. Prior to analysis, the minimum acceptable regression efficiency can be set. This allows the user to independently constrain the sample estimates to the ‘best’ portion of the log-linear phase. Upon selecting the ‘compute’ button, the software parses several iterations of the log-linear regression to yield the best model that minimizes the residual and maximizes the number of points used. As a general rule, only four or five cycles are within the ‘log-linear’ portion of the curve (37). Together, these constraints eliminate poor or failed reactions and the software demarcates these reactions in the final data set as ‘N/A’. When the computations are complete, the ‘save results’ button is activated. This must be selected to specify an output file name.
As shown in Table 3, the output generated from the data shown in Figure 1 is a simple tab delimited text file containing eight columns. The first column, labeled N, lists the reaction name as indicated in the first row of the input file. The second column, Init_amount, estimates the initial amount of fluorescence, which is directly proportional to the initial quantity of target. The third and fourth columns, labeled Efficiency and Residual, report the PCR reaction efficiency and its respective standard error. The fifth column, Position_Start, reflects the specific cycle number from which the data extraction began the computation. Column six, Interval, reports the number of PCR cycles used in the regression analysis. The last two columns report the t and P value estimates of significance for the linear regression models. Clearly, even though there are discrepancies in reaction efficiencies as observed in Figure 1 and measured in Table 3, the TAQ software reliably estimates initial template quantities within the quadruplicates of the reactions. They were essentially the same. Only differences of greater than 10-fold represent a significant deviation since the data is collected as an exponential function yet for calculative purposes is expressed as a logarithmic function.
Table 3. TAQ software output.
| N | Init_amout | Efficiency | Residual | Position_Start | Interval | t | P |
|---|---|---|---|---|---|---|---|
| MA G | 4.64E-07 | 0.869900 | 0.019313 | 22 | 4 | 14.24186 | <0.05 |
| MA G | 2.08E-07 | 0.984040 | 0.024831 | 21 | 4 | 13.74928 | <0.05 |
| MA G | 1.12E-07 | 0.965715 | 0.034472 | 22 | 4 | 11.51121 | <0.05 |
| MA G | 3.60E-07 | 0.885257 | 0.014498 | 22 | 4 | 16.65232 | <0.05 |
| Average MA G | 1.22E-06 | 0.801287 | 0.025852 | 22 | 4 | 11.57435 | <0.05 |
| LE G | 1.29E-07 | 0.963167 | 0.021076 | 22 | 4 | 14.69342 | <0.05 |
| LE G | 2.06E-07 | 0.929908 | 0.015444 | 22 | 4 | 16.73029 | <0.05 |
| LE G | 2.39E-07 | 0.882500 | 0.081501 | 22 | 5 | 12.13693 | <0.05 |
| LE G | 1.37E-07 | 0.965879 | 0.022374 | 22 | 4 | 14.29009 | <0.05 |
| Average LE G | 1.21E-07 | 0.966171 | 0.02063 | 22 | 4 | 14.88531 | <0.05 |
MA G, matrix-associated enriched DNA sample G amplicon; LE G, loop enriched DNA sample G amplicon; average, data generated using fluorescence intensity average at each cycle for quadruplicate PCR reactions.
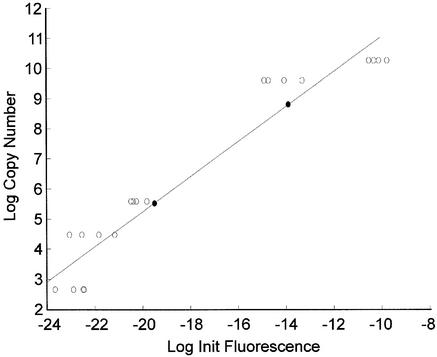
As an initial test of the reliability of the TAQ software, serially diluted human genomic DNA was subjected to real-time PCR. Amplification of β-actin was carried out in quadruplicate for each dilution. The raw data were processed by the TAQ software and the initial fluorescence calculated. These values were compared to initial template concentrations calculated via UV spectrophotometric quantification using linear regression (y = 0.58x + 16.87, r2 = 0.900, P < 0.05). Two test reactions containing 5714 and 143 copies of the initial template were also run. Using the TAQ software and the generated standard curve, it was estimated that the test samples contained 6200 ± 1714 and 249 ± 31 (mean ± SE of triplicates) initial target molecules, respectively. The difference observed in the second test sample likely reflects hardware detection limits. This is supported by the substantial change in fluorescence intensity associated with the lower target ranges, shown in Figure 2. The PCR amplification efficiency of each reaction was compared to its initial fluorescence estimate using linear regression. No relationship was observed (y = –0.01x + 0.62, r2 = 0.096, P > 0.05). This verifies that differences in initial template are not simply a reflection of variations in amplification efficiency and supports the premise that PCR amplification efficiencies are indeed considered by the TAQ software when the number of initial target molecules is estimated. Together, these results show that the TAQ software reliably estimates relative differences in the initial amount of template.
Figure 2.
TAQ software estimates of serial dilutions. Serial dilutions of human genomic DNA were subjected to real-time PCR using β-actin- specific primers. The fluorescence data obtained were analyzed using the TAQ software and the estimates of initial fluorescence were compared to known concentrations using linear regression. The regression equation obtained was then used to estimate the concentration of two test samples shown as black circles.
Matrix attachment region (MAR) analysis of the β-globin locus using the TAQ software
Understanding the mechanism of gene potentiation is one of today’s greatest scientific challenges. It is known that chromatin structure provides a key component to the spatial and temporal control of gene expression (6,38,39). It has been proposed that genes are organized into functional domains by DNA anchors, known as MARs, and anchor to a proteinacious nuclear scaffold. This facilitates genes assuming a transcriptionally permissive state (7,10) while insulating the loci from neighboring enhancers and the silencing effects of heterochromatin (11,12). Even though the precise components of the nuclear matrix have not been clearly defined, several in silico and wet bench methods have been developed to facilitate their detection.
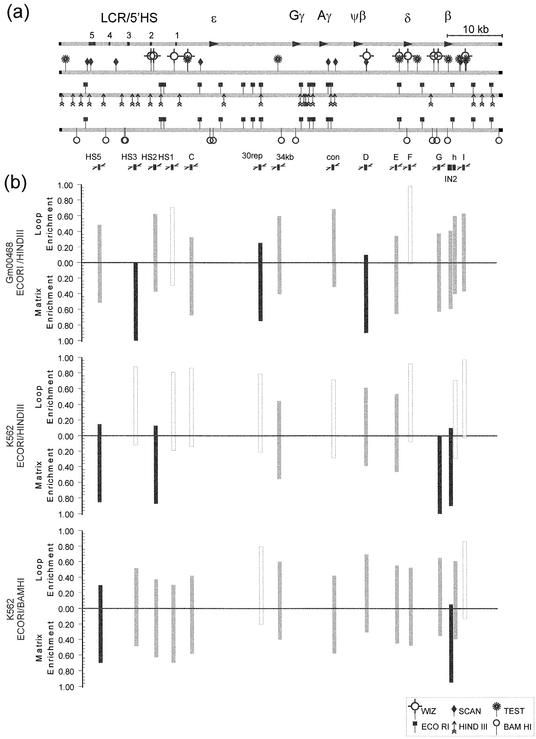
Utilizing an in silico approach, a mechanism mediated by alterations in gene-specific nuclear matrix attachment has been offered to support the looping model for spatial and temporal regulation of the β-globin locus (22). This model envisages that the β-globin LCR is consistently located at the nuclear matrix and that specific genes are either recruited to or away from the nuclear matrix-bound LCR to either enhance or diminish transcription. To build upon this model, the MAR prediction map shown in Figure 3a was developed using three publicly available pattern matching programs (32,33,40). MARFINDER, now known as MARWIZ, (32) utilizes a predefined series of sequence-specific patterns that have previously been suggested as containing MAR activity. Using a sliding widow approach, the representation of each pattern is calculated and weighted as a function of the probability that each pattern occurred by chance. Similarly, SMARTest (33) uses a series of precalculated weight matrices derived from the S/MAR database (41). Once a threshold value of the co-occurrence of weighted matrix patterns is exceeded, a candidate MAR is reported. In contrast, MARSCAN identifies the 8–16 bp bipartite candidate MAR sequence element pairs that occur within a 200 bp span (34). Interestingly, only MARWIZ and MARSCAN predict that the LCR/5′HS region of the β-globin locus is matrix attached. In contrast, all three programs predicted multiple attachment sites in close proximity to the δ- and β-globin genes.
Figure 3.
Identification of MARs using real-time PCR. (a) Strategy graph for the human β-globin locus. A strategy graph of the human β-globin locus was prepared as an initial step in determining in vivo nuclear matrix attachment. The first line illustrates the location of the LCR/5′HS and the five genes of the locus. The second line highlights those regions suspected to have MAR activity based on the three different in silico MAR prediction programs MARWIZ (WIZ), MARSCAN (SCAN) and MARTest (TEST). The third and fourth lines show the specific restriction fragments generated by EcoRI/HindIII and EcoRI/BamHI digestion, respectively. The interrogated PCR amplicons are shown as black boxes in the fifth row. (b) Target amplicon quantification. Real-time PCR, using equal amounts of MA enriched and LE DNA fractions from GM00468 and K562 cells, was performed in quadruplicate. The data were analyzed using the TAQ software to estimate the initial amount of template. The percent of each amplicon found in the MA fraction was calculated assuming 100% when the median of the MA and LE fractions are combined. Those amplicons having at least 75% in the MA fraction were considered matrix bound and are shown using black columns. Those having less than 75% but more than 25% in the MA fraction were considered matrix associated and are shown using gray columns. Those amplicons having less than 25% in the MA fraction were considered non-associated and are designated with white columns. The globin non-expressing GM00468 cells show matrix attachment in the LCR, 30rep and D amplicons. The globin expressing K562 cells show matrix attachment in the O, A, G and IN2 amplicons. These attachment sites were verified and narrowed to the O and IN2 amplicons using a second set of endonculease restriction enzymes. These data provide the first evidence that the looping transcriptional control of this locus is mediated through nuclear matrix association.
To verify or reject the predicted MARs in vivo, human fetal fibroblasts (GM00468), which do not express any elements of the β-globin locus, and cells from the globin expressing K562 cell line were fractionated into MA and LE DNA. Successful partitioning and analysis was verified by the absence of nuclear matrix association of the male germ cell-specific expressed gene PRM2. Assuming that 100% of the amplicon is contained within the MA plus LE fraction, the preparations utilized in these assays showed that 65 (GM00468), 89 (K562, EcoRI/HindIII) and 68% (K562, EcoRI/BamHIII) of the PRM2 amplicon was contained within the LE fraction, whereas 35, 11 and 32% was contained within their respective MA fractions. This is in accord with previous work, showing that PRM2 is consistently found in the LE fraction of somatic cells (10,25). As determined by either the PCR assay (10) or by fluorescent in situ hybridization (42), it is only bound in cells of the spermatogenic lineage.
Fifteen different regions spanning the human β-globin locus, as shown in Figure 3a, were assessed using quantitative real-time PCR. Again, assuming that 100% of each amplicon would be found if the MA and LE fractions were combined, the y-intercept medians of the quadruplicates were used to determine the percent of amplicon enrichment within the MA fraction. As illustrated in Figure 3b, matrix attachment was shown in the HS3, 30rep and D amplicons for the non-expressing GM00468 cells. The 596 bp HS3 amplicon maps to a downstream region flanking the 5′HS3. In this case nuclear matrix association may silence the expression of the globin locus, similar to the mode of regulation of the Ig-µH and IL-2Rα genes (7,43) observed during T cell differentiation. In the latter, SATB1 is bound to the enhancer and promoter regions forming a network that recruits chromatin silencers, including Mi-NURD, ISWI, pARP and histone deacetyltransferases, to the loci (7). The 434 bp segment 30rep maps to a LINE1 repeat sequence upstream of the Gγ gene. The 540 bp D amplicon localizes to a region ∼200 bp downstream of the ψβ gene and corresponds to a previously determined site of nuclear matrix attachment (13). We propose that tethering of these two innocuous regions to the nuclear matrix prevents the globin genes from looping back to the LCR/5′HS region. The adoption of this nuclear architecture would augment the maintenance of the non-potentiated closed chromatin state that provides the primary means of silencing a locus. The absence of specific erythropoietic trans-acting factors would further ensure silencing. The GM00468 cell line appears to uphold the classic ‘looping’ model of regulation as the LCR/5′HS region is matrix attached while regions surrounding the various non-expressed globin genes are loop enriched. If the model is correct, actively expressing cells should maintain nuclear matrix attachment at the LCR/5′HS region and should then recruit those genes being expressed to the nuclear matrix.
To challenge the role of nuclear matrix attachment in the spatial and temporal regulation of the β-globin locus, the same restriction fragments were interrogated using MA and LE DNA fractionated from human erythromyeloid cells (K562). This cell line expresses the ε, Gγ and Aγ genes of the β-gobin locus (44). As shown in Figure 3b, the 30rep and D regions were released, the β-globin gene was brought to the nuclear matrix and there was an intriguing shift within the LCR/5′HS region. Within the LCR/5′HS region, the HS3 amplicon itself was no longer attached to the nuclear matrix. Instead, the LCR/5′HS region was attached by the 393 bp HS5 amplicon and the 514 bp HS2 amplicon. Attachment of the HS5 amplicon corroborates the Jarman and Higgs study (13), while attachment of the HS2 amplicon is novel. These regions correspond to the 5′HS5 and 5′HS2 of the LCR/5′HS region, respectively, and have been implicated in insulation (13,45,46) and recruitment/enhancing (21,47). Similarly, the 421 bp G and 820 bp IN2 regions were attached to the nuclear matrix. These amplicons map to regions ∼800 bp upstream of the first exon and within the second intron of the β-globin gene. In K562 cells, the locus appears anchored at each end. Together this supports the view that the chromatin domain achieves a malleable structure. In turn this enables the ‘active genes’ to loop back and interact with the LCR/5′HS region that is attached to the nuclear matrix.
To substantiate these findings, a second fractionation using an EcoRI/BamHI digest was employed. As shown in Figure 3b, the data confirmed matrix attachment by the HS5 amplicon and refined the region of attachment to the IN2 amplicon. Surprisingly, with this digest the HS2 amplicon showed more of an association than nuclear matrix attachment. This may reflect the substantial change in restriction fragment size. The larger 6.6 kb fragment generated by the EcoRI/BamHI digestion may have been partially removed from the nuclear matrix by inadvertent mechanical forces that were incurred during the various extraction procedures. The smaller restriction fragments produced by the EcoRI/HindIII digestion localized the site of nuclear matrix attachment within 5′HS2. Attachment of intron 2 of the β-globin gene again corroborated previous studies (13). These data clearly demonstrate that the LCR/5′HS region and that the β-globin gene is attached to the nuclear matrix in K562 cells. Such changes in the attachment sites show that the β-globin locus displays a dynamic association with the nuclear matrix that is dependent on expression status. Together this supports the view that nuclear matrix association helps mediate the looping transcriptional control of the β-globin locus.
Utilizing a unique approach to real-time PCR analysis, we were able to investigate nuclear matrix association in a variety of regions spanning the β-globin locus. Without the TAQ software, the determination of amplicon enrichment would have required considerable PCR reaction optimization. The TAQ software method offers several advantages when compared to other analysis routines commonly used for quantitative real-time PCR. First, it eliminates the deceptive discrepancies introduced by different amplification efficiencies that are often observed between experiments. Second, the user is not required to select an arbitrary threshold. The threshold is predetermined and thus consistent among experiments. Third, the amplification efficiency is calculated for each reaction, providing the user with invaluable information regarding the ‘quality’ of the PCR and facilitating the optimization of laboratory procedures. Armed with the TAQ software, investigators can now ponder the biological implications of their studies rather than conditionally optimizing the technical logistics of PCR quantification.
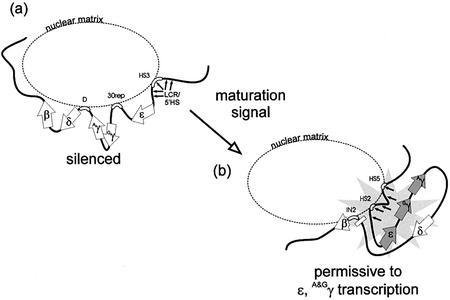
The results presented in this communication provide the first evidence that nuclear matrix association dynamically mediates the looping transcriptional control of the β-globin locus. As summarized in Figure 4, we provide evidence that the LCR/5′HS region of the human β-globin locus is attached to the nuclear matrix in β-globin locus non-expressing cells. This may facilitate the silencing of the enhancer capacity attributed to the LCR/5′HS region, thus providing another example of negative regulation of a gene locus through nuclear matrix association. Two additional segments between the LCR/5′HS region and the β-globin gene were also tethered to the nuclear matrix in these cells. These regions likely provide a means to spatially separate the LCR/5′HS region from the globin genes, ensuring its suppression. In an erythroid cell line that expresses the genes within the β-globin locus, the tethered regions were released from the matrix. The chromatin domain was remodeled with distinct matrix attachment sites appearing within the LCR/5′HS region and the β-globin gene. This dynamic release of the central tether would enable each gene destined for transcription to loop back and interact with the matrix-attached LCR/5′HS region.
Figure 4.
MARs and the regulation of the β-globin locus. (a) β-globin non-expressing cells. The β-globin locus is comprised of the ε→Gγ→Aγ→δ→β globin genes. These genes are expressed in a time- and spatial-dependent manner. Attachment is indicated by outlined segments adjacent to the nuclear matrix. Attachment of the LCR/5′HS region of the human globin locus in non-erythroid cell lines is proposed to play a role in quenching expression. Two additional regions are tethered to the nuclear matrix between the LCR and β-globin gene to spatially separate and silence the locus. (b) Cells permissive for β-globin locus expression. In response to a maturation signal, the previously tethered regions between the LCR/5′HS and the β-globin gene are released from the nuclear matrix and the chromatin domain is remodeled. With new nuclear matrix tethers, a chromatin conformation is then established that facilitates looping of the active genes back to the LCR/5′HS region.
Acknowledgments
ACKNOWLEDGEMENTS
A special thank you is extended to Susan Wykes for her help in naming the software and Carol Rubin from University of California–Davis for her help with cell culture. This work was funded in part by NIH grant HD36512 to S.A.K. G.C.O. is funded in part by Wayne State University’s School of Medicine Dean’s Post-Doctoral Fellowship Award and the NICHD Contraception and Infertility Loan Repayment Program.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Bode J., Benham,C., Knopp,A. and Mielke,C. (2000) Transcriptional augmentation: modulation of gene expression by Scaffold/Matrix-Attached Regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr., 10, 73–90. [PubMed] [Google Scholar]
- 4.Dundr M. and Misteli,T. (2001) Functional architecture in the cell nucleus. Biochem. J., 356, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mielke C., Christensen,M.O., Westergaard,O., Bode,J., Benham,C.J. and Breindl,M. (2002) Multiple collagen I gene regulatory elements have sites of stress-induced DNA duplex destabilization and nuclear scaffold/matrix association potential. J. Cell. Biochem., 84, 484–496. [PubMed] [Google Scholar]
- 6.Orphanides G. and Reinberg,D. (2002) A unified theory of gene expression. Cell, 108, 439–451. [DOI] [PubMed] [Google Scholar]
- 7.Yasui D., Miyano,M., Cai,S.T., Varga-Weisz,P. and Kohwi-Shigematsu,T. (2002) SATB1 targets chromatin remodelling to regulate genes over long distances. Nature, 419, 641–645. [DOI] [PubMed] [Google Scholar]
- 8.Wolffe A.P. and Hansen,J.C. (2001) Nuclear visions: functional flexibility from structural instability. Cell, 104, 631–634. [DOI] [PubMed] [Google Scholar]
- 9.Jackson D.A. (2000) Features of nuclear architecture that influence gene expression in higher eukaryotes: confronting the enigma of epigenetics. J. Cell. Biochem., 35 (suppl.), 69–77. [DOI] [PubMed] [Google Scholar]
- 10.Kramer J.A. and Krawetz,S.A. (1996) Nuclear matrix interactions within the sperm genome. J. Biol. Chem., 271, 11619–11622. [DOI] [PubMed] [Google Scholar]
- 11.McKnight R.A., Shamay,A., Lakshmanan,S., Wall,R.J. and Hennighausen,L. (1992) Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 6943–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Namciu S.J., Blochlinger,K.B. and Fournier,R.E. (1998) Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol., 18, 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarman A.P. and Higgs,D.R. (1988) Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J., 7, 3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bender M.A., Reik,A., Close,J., Telling,A., Epner,E., Fiering,S., Hardison,R. and Groudine,M. (1998) Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse beta-globin locus control region. Blood, 92, 4394–4403. [PubMed] [Google Scholar]
- 15.Cao A. and Moi,P. (2002) Regulation of the globin genes. Pediatr. Res., 51, 415–421. [DOI] [PubMed] [Google Scholar]
- 16.Reik A., Telling,A., Zitnik,G., Cimbora,D., Epner,E. and Groudine,M. (1998) The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol., 18, 5992–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter D., Chakalova,L., Osborne,C.S., Dai,Y.F. and Fraser,P. (2002) Long-range chromatin regulatory interactions in vivo. Nature Genet., 32, 623–626. [DOI] [PubMed] [Google Scholar]
- 18.Felsenfeld G. and Groudine,M. (2003) Controlling the double helix. Nature, 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 19.Tolhuis B., Palstra,R.J., Splinter,E., Grosveld,F. and de Laat,W. (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell, 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- 20.Dillon N., Antoniou,M., Berry,M., DeBoer,E., Drabek,D., Ellis,J., Fraser,P., Hanscombe,O., Imam,A., Koken,M. et al. (1993) Regulation of the human β-globin expression domain. In Heslop-Harrison,J.S. and Flavell,R.B. (eds), The Chromosome. BIOS Scientific Publishers, Oxford, UK, pp. 149–160.
- 21.Harju S., McQueen,K.J. and Peterson,K.R. (2002) Chromatin structure and control of beta-like globin gene switching. Exp. Biol. Med., 227, 683–700. [DOI] [PubMed] [Google Scholar]
- 22.Walter W.R., Singh,G.B. and Krawetz,S.A. (1998) MARs mission update. Biochem. Biophys. Res. Commun., 242, 419–422. [DOI] [PubMed] [Google Scholar]
- 23.Lauber A.H., Barrett,T.J., Subramaniam,M., Schuchard,M. and Spelsberg,T.C. (1997) A DNA-binding element for a steroid receptor-binding factor is flanked by dual nuclear matrix DNA attachment sites in the c-myc gene promoter. J. Biol. Chem., 272, 24657–24665. [DOI] [PubMed] [Google Scholar]
- 24.Mirkovitch J., Mirault,M.E. and Laemmli,U.K. (1984) Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell, 39, 223–232. [DOI] [PubMed] [Google Scholar]
- 25.Kramer J.A. and Krawetz,S.A. (1997) PCR-based assay to determine nuclear matrix association. Biotechniques, 22, 826–828. [DOI] [PubMed] [Google Scholar]
- 26.Arnheim N. and Erlich,H. (1992) Polymerase chain reaction strategy. Annu. Rev. Biochem., 61, 131–156. [DOI] [PubMed] [Google Scholar]
- 27.Tyrrell D.A. (1997) Polymerase chain reaction. Br. Med. J., 314, 5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang A.M., Doyle,M.V. and Mark,D.F. (1989) Quantitation of mRNA by the polymerase chain reaction. Proc. Natl Acad. Sci. USA, 86, 9717–9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chelly J., Kaplan,J.C., Maire,P., Gautron,S. and Kahn,A. (1988) Transcription of the dystrophin gene in human muscle and non-muscle tissue. Nature, 333, 858–860. [DOI] [PubMed] [Google Scholar]
- 30.Choi Y.W., Kotzin,B., Herron,L., Callahan,J., Marrack,P. and Kappler,J. (1989) Interaction of Staphylococcus aureus toxin “superantigens” with human T cells. Proc. Natl Acad. Sci. USA, 86, 8941–8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak K.J. and Schmittgen,T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 32.Singh G.B., Kramer,J.A. and Krawetz,S.A. (1997) Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res., 25, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frisch M., Frech,K., Klingenhoff,A., Cartharius,K., Liebich,I. and Werner,T. (2002) In silico prediction of scaffold/matrix attachment regions in large genomic sequences. Genome Res., 12, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Drunen C.M., Sewalt,R., Oosterling,R.W., Weisbeek,P.J., Smeekens,S.C.M. and van Driel,R. (1999) A bipartite sequence element associated with matrix/scaffold attachment regions. Nucleic Acids Res., 27, 2924–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiesner R.J. (1992) Direct quantification of picomolar concentrations of mRNAs by mathematical analysis of a reverse transcription/exponential polymerase chain reaction assay. Nucleic Acids Res., 20, 5863–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen R., Morrison,T., Herrmann,M. and Wittwer,C. (1998) Quantitative PCR by continuous fluorescence monitoring of a double strand DNA specific binding dye (Rhoche Applied Science). Biochemica, 2, 8–11. [Google Scholar]
- 38.Ito T. (2003) Nucleosome assembly and remodeling. Curr. Top. Microbiol. Immunol., 274, 1–22. [DOI] [PubMed] [Google Scholar]
- 39.Farkas G., Leibovitch,B.A. and Elgin,S.C. (2000) Chromatin organization and transcriptional control of gene expression in Drosophila. Gene, 253, 117–136. [DOI] [PubMed] [Google Scholar]
- 40.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European molecular biology open software suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 41.Liebich I., Bode,J., Reuter,I. and Wingender,E. (2002) Evaluation of sequence motifs found in scaffold/matrix-attached regions (S/MARs). Nucleic Acids Res., 30, 3433–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid C., Heng,H.H.Q., Rubin,C., Ye,C.J. and Krawetz,S.A. (2001) Sperm nuclear matrix association of the PRMI→PRM2→TNP2 domain is independent of Alu methylation. Mol. Hum. Reprod., 7, 903–911. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez J.D., Yasui,D.H., Niida,H., Joh,T., Loh,D.Y. and Kohwi-Shigematsu,T. (2000) The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev., 14, 521–535. [PMC free article] [PubMed] [Google Scholar]
- 44.Groudine M., Kohwi-Shigematsu,T., Gelinas,R., Stamatoyannopoulos,G. and Papayannopoulou,T. (1983) Human fetal to adult hemoglobin switching: changes in chromatin structure of the beta-globin gene locus. Proc. Natl Acad. Sci. USA, 80, 7551–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanimoto K., Liu,Q., Bungert,J. and Engel,J.D. (1999) Effects of altered gene order or orientation of the locus control region on human beta-globin gene expression in mice. Nature, 398, 344–348. [DOI] [PubMed] [Google Scholar]
- 46.Tanimoto K., Liu,Q., Bungert,J. and Engel,J.D. (1999) The polyoma virus enhancer cannot substitute for DNase I core hypersensitive sites 2–4 in the human beta-globin LCR. Nucleic Acids Res., 27, 3130–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bungert J., Tanimoto,K., Patel,S., Liu,Q., Fear,M. and Engel,J.D. (1999) Hypersensitive site 2 specifies a unique function within the human beta-globin locus control region to stimulate globin gene transcription. Mol. Cell. Biol., 19, 3062–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramchandran R., Bengra,C., Whitney,B., Lanclos,K. and Tuan,D. (2000) A (GATA)(7) motif located in the 5′ boundary area of the human beta-globin locus control region exhibits silencer activity in erythroid cells. Am. J. Hematol., 65, 14–24. [DOI] [PubMed] [Google Scholar]
- 49.Milot E., Strouboulis,J., Trimborn,T., Wijgerde,M., de Boer,E., Langeveld,A., Tan-Un,K., Vergeer,W., Yannoutsos,N., Grosveld,F. et al. (1996) Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell, 87, 105–114. [DOI] [PubMed] [Google Scholar]
- 50.Iler N., Goodwin,A.J., McInerney,J., Nemeth,M.J., Pomerantz,O., Layon,M.E. and Lowrey,C.H. (1999) Targeted remodeling of human beta-globin promoter chromatin structure produces increased expression and decreased silencing. Blood Cells Mol. Dis., 25, 47–60. [DOI] [PubMed] [Google Scholar]
- 51.Ellis J., Tan-Un,K.C., Harper,A., Michalovich,D., Yannoutsos,N., Philipsen,S. and Grosveld,F. (1996) A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO J., 15, 562–568. [PMC free article] [PubMed] [Google Scholar]
- 52.Epner E., Reik,A., Cimbora,D., Telling,A., Bender,M.A., Fiering,S., Enver,T., Martin,D.I., Kennedy,M., Keller,G. et al. (1998) The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol. Cell, 2, 447–455. [DOI] [PubMed] [Google Scholar]
- 53.Pasceri P., Pannell,D., Wu,X. and Ellis,J. (1998) Full activity from human beta-globin locus control region transgenes requires 5′HS1, distal beta-globin promoter and 3′ beta-globin sequences. Blood, 92, 653–663. [PubMed] [Google Scholar]
- 54.Bender M.A., Roach,J.N., Halow,J., Close,J., Alami,R., Bouhassira,E.E., Groudine,M. and Fiering,S.N. (2001) Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood, 98, 2022–2027. [DOI] [PubMed] [Google Scholar]