Abstract
Ume6p plays essential roles in the regulation of early meiotic genes in Saccharomyces cerevisiae. Ume6p exerts repression via recruitment of the Sin3p-Rpd3p histone deacetylase and Isw2p chromatin remodeling complexes. The transcriptional step that is ultimately inhibited by Ume6p is unknown. Here, in vivo footprinting shows that transcriptional activators Hap1p and Abf1p occupy upstream sites in repressed and derepressed promoters. In contrast, chromatin immunoprecipitation shows that TATA box-binding protein (TBP)- promoter binding is reduced upon repression of HOP1. Fusion of TBP to a zinc cluster DNA binding domain relieves repression at a HOP1 promoter modified to include the zinc cluster target site. We suggest that TBP binding is inhibited through chromatin modification by the Sin3p-Rpd3p and Isw2p complexes recruited by Ume6p.
INTRODUCTION
In the yeast Saccharomyces cerevisiae, repression of early meiotic genes depends on histone deacetylation and chromatin remodeling activities. These genes are repressed during mitotic growth by the binding of Ume6p to the URS1 site in their 5′ regions (reviewed in 1). Ume6p recruits the histone deacetylase (HDAC) complex Sin3p-Rpd3p to exert repression (2,3). It is well established that the recruitment of Rpd3p generates a localized histone deacetylation domain over a range of one to two nucleosomes in the targeted promoter (4–7). Ume6p also recruits the Isw2p chromatin remodeling complex, which promotes the formation of a nuclease-inaccessible chromatin structure proximal to the URS1 site at target promoters (8,9). Both Sin3p-Rpd3p and Isw2p are required for full repression by Ume6p, thus supporting the idea that nucleosome modification and position together govern promoter function (8–10).
Though effects of the Ume6p complex on local chromatin structure are well documented (8,9), the transcription factors that are ultimate targets of repression are uncertain. Histone deacetylation in a TATA-proximal nucleosome inhibits TATA box-binding protein (TBP) binding at the human pS2 promoter (11), but inhibits activator Adr1p binding at the yeast ADH2 promoter (12). Histone deacetylation at Polycomb-repressed Drosophila promoters does not affect TBP binding, but may inhibit RNA polymerase II recruitment (13). SIR-generated repressive chromatin is permissive for both activator and TBP binding (14). Based on these precedents, Ume6p may repress transcription through inhibition of activator or TBP binding or by inhibition of a later step in transcription initiation.
Here we report analysis of Ume6p-repressed promoters through in vivo footprinting and chromatin immunoprecipitation (ChIP) assays. Our findings argue that inhibition of TBP binding is a critical step in Ume6p-dependent repression.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains used were SK1 derivatives, have genetic markers ura3 trp1 leu2 lys2 ho::LYS2 and are isogenic except as noted as follows: AMP107 (MATa), AMP1779 (MATα hap1::ZC-TBP-TRP1), AMP1780 (MATα hap1::LEU2), MHS21 (MATα hap1::ZC-TBP-TRP1 ume6Δ2), MHS22 (MATα hap1::LEU2 ume6Δ2), MHS24 (MATa ume6Δ2), yx268 (MATa ume6Δ1:: TRP1 gal80::LEU2 his3Δ), yx423 (MATa gal80::LEU2 his3Δ). MHS21, 22 and 24, which are ume6Δ2 strains, carry a ume6Δ allele lacking codons 159–836. Construction of the ume6Δ1:: TRP1 mutation in yx268 was described previously (15).
ZC-TBP is an in-frame fusion protein in which the zinc cluster DNA binding domain of Hap1p (amino acids 1–247) (16) is fused to the N-terminus of TBP. It was created through several steps in the genome of strain AMP1779 as follows. A DNA fragment containing the TBP open reading frame (ORF) (called SPT15 in S.cerevisiae) and 300 bp of its 3′-untranslated region was cloned between the SpeI and XhoI sites of pRS304, forming plasmid pAD6. PCR was then performed with pAD6 as template using a 5′-primer (5′-AGT AAC GGA ACC ATC CAC TTA GGT GCC ACC CAC TGG TTG TCT ATC ATG AAA GGT GAC CCG ATG GCC GAT GAG GAA CGT TTA) which corresponds to nucleotide positions 682–741 of the HAP1 coding strand and positions 1–21 of the SPT15 coding strand and a 3′-primer (5′-GTT AGA CAC GTC CTG GCT GGT TGC TGG AAT GGT AGC GTT TAA TTG AGG AAA ATT ATC AGG CGG CAT CAG AGC AGA TTG T-3′) which corresponds to positions 2160–2100 of the HAP1 non-coding strand and positions 151–169 of pRS306. The amplified fragment was integrated into the hap1::LEU2 locus of AMP1780 to express the Hap1p DNA binding domain fused with the N-terminus of TBP (referred to as ZC-TBP).
Epitope-tagged TBP was constructed in a TRP1 centromeric plasmid pRS314 as follows. To express HA epitope-tagged TBP (HA3-TBP) from its natural promoter, the SPT15/TBP1 gene with 5′-flanking sequences (–840 to –1) and 3′-flanking sequences (the stop codon to 300 bp downstream) was cloned between the KpnI and SpeI sites of pRS314 and a NotI site was created after the first codon of TBP. Then, a DNA fragment that encodes three copies of HA epitope was obtained by digestion of pGTEP1 with NotI (17) and was inserted into the NotI site after the first codon of TBP, in-frame with the TBP ORF.
CYC1-lacZ plasmids pKB112, pKB143 and pLGΔ312ΔRS were described previously (18,19). Plasmid pKT5-1 was constructed from pAV79B (generously provided by A. Vershon; 20) as follows: the region –131 to –114 of the HOP1 promoter in pAV79B was replaced by the CYC1 UAS1 sequence (5′-GGC CGG GGT TTA CGG ACG-3′), forming pKT5-1.
All the expression and reporter plasmids constructed here were verified by DNA sequencing.
Culture conditions, media, strain construction, transformations and the β-galactosidase assay followed standard recipes and protocols as described (21).
In vivo footprinting
In vivo UV and DMS footprintings by primer extension were performed as described (22,23). Primer sequences are available upon request.
Chromatin immunoprecipitation assay
Chromatin-containing whole cell extract was prepared from 100 ml of log phase culture as described (24–26). Chromatin corresponding to 2 × 108 cell equivalents of the whole cell extract (an average size of ∼500 bp) was combined in a final volume of 0.2 ml with 10 µl of monoclonal anti-HA antibody (5 mg/ml 12CA5; Roche). Immunoprecipitated DNA was analyzed by quantitative PCR using primer sets for specific regions, which were designed to 24–28mers. Primer sequences are available on request. PCR was first performed with decreasing amounts of DNA templates to determine the linear range. The PCR conditions were 94°C for 5 min, followed by 27 cycles of 94°C for 30 s, 40°C for 30 s and 72°C for 30 s, then 72°C for 5 min. PCR products were resolved by 2% agarose gel electrophoresis, stained with SYBR-Green, visualized and quantitated with a FujiFilm Luminescent Image Analyzer LAS-1000plus.
RESULTS AND DISCUSSION
The Ume6p repressor complex does not affect activator binding in the CYC1 and HOP1 promoters
To understand the mechanism of Ume6p-dependent repression, we compared binding of transcriptional activators to repressed and derepressed promoters in vivo. We first used the well characterized CYC1-lacZ promoter region. This hybrid gene is activated by the transcription factor Hap1p, which binds to the UAS1 site. Binding of Hap1p results in UV hypersensitivity within UAS1 (23) and is thus detectable by UV photofootprinting. CYC1-lacZ is not normally repressed by Ume6p, so we created the hybrid CYC1-URS1-lacZ gene, which has the Ume6p binding site (URS1) inserted between UAS1 and the TATA region (19). We verified that CYC1-URS1-lacZ expression was repressed about 20-fold by Ume6p (Fig. 1A), in agreement with previous reports (2,19). We then analyzed activator Hap1p binding in the CYC1-URS1-lacZ promoter by in vivo UV photofootprinting (Fig. 1B). Hap1p-UAS1 binding was readily detectable at CYC1-URS1-lacZ in the presence or absence of Ume6p. Therefore, repression of this hybrid promoter does not result from inhibition of activator binding.
Figure 1.
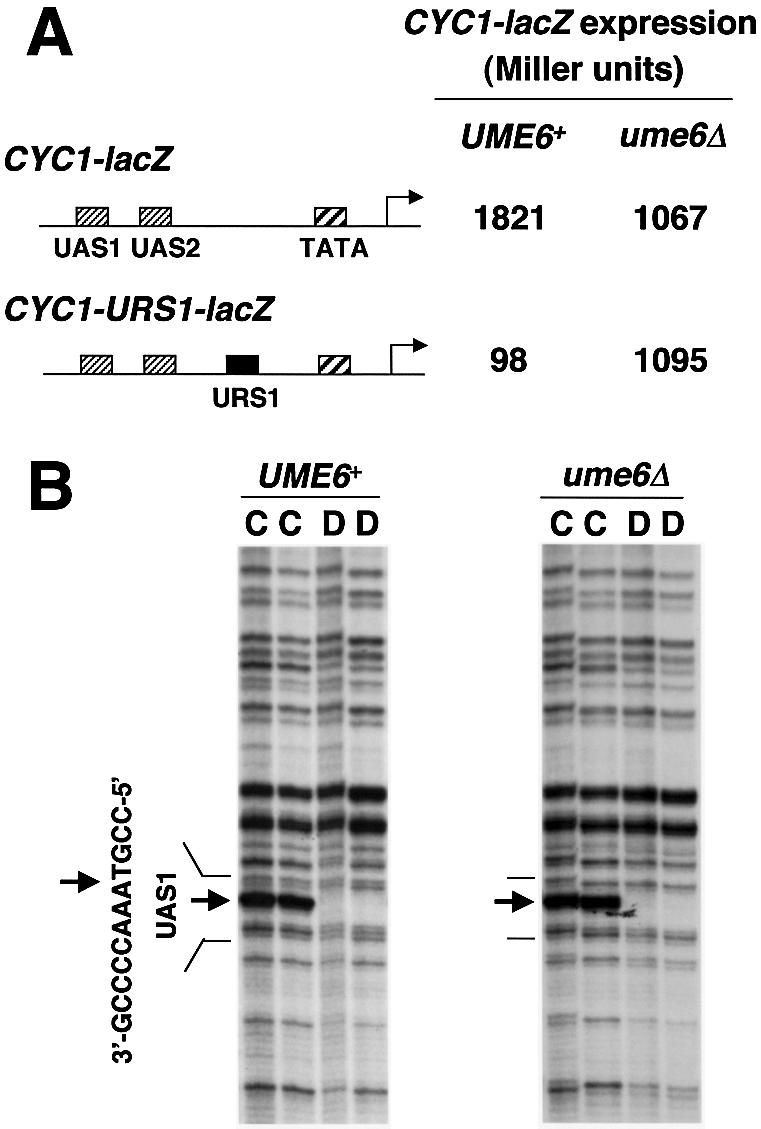
(A) Repression of CYC1-lacZ reporter gene through URS1, a Ume6p binding site, in high copy plasmids [CYC1-lacZ, pKB112; CYC1-URS1-lacZ, pKB143 (17,18)] in UME6 and ume6Δ strains (yx423 and yx268, respectively). CYC1-lacZ expression is indicated in Miller units of β-galactosidase. Location of UAS1, UAS2, TATA and regions are diagrammed. The standard deviations were <20% for the triplicate determinations. (B) In vivo UV photofootprinting of the bottom strand of the UAS1 region in a CYC1-URS1-lacZ plasmid in UME6 and ume6Δ strains (yx423 and yx268, respectively). Lanes marked C are samples from intact cells irradiated with UV light (500 and 750 mJ/cm2); lanes marked D are samples from purified DNA irradiated with UV light (120 and 240 mJ/cm2). The UAS1 sequence is shown to the left of the gels. Arrows indicate sites of enhancement of UV photoproducts in irradiated cells (lane C) compared with irradiated purified DNA (lanes D).
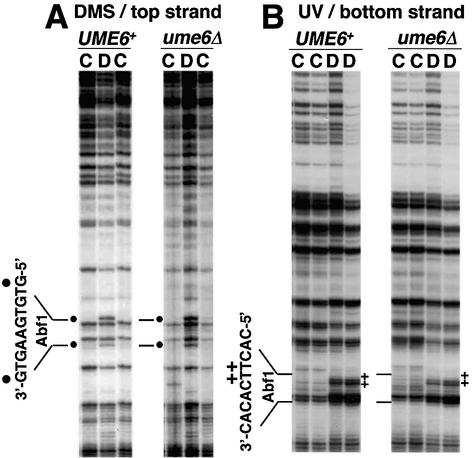
We also examined the HOP1 promoter, a natural Ume6p repression target. The HOP1 promoter is activated by binding of the transcription factor Abf1p to an upstream site (27). HOP1 is normally repressed in mitotic cells by Ume6p (20) and expressed only in meiotic cells when Ume6p repression is lifted (1,20). We verified that HOP1-lacZ expression was repressed about 800-fold by Ume6p in mitotic cells (data not shown). We carried out in vivo DMS footprinting of the Abf1p site in mitotic UME6 and ume6Δ cells to determine whether repression affected Abf1p-DNA binding. We detected protection of two guanine residues in DNA treated with DMS in whole cells (Fig. 2A, lanes C), compared to DNA treated with DMS after purification (lanes D). The protected guanine residues coincide with in vitro carboxymethylation interference footprints of Abf1p (28). We also observed a UV photofootprint at this Abf1p site: a comparison of UV photofootprinting of DNA in whole cells or after purification revealed protection from pyrimidine dimerization in vivo in the center of the Abf1p binding site (Fig. 2B). By both measures, Abf1p binding to the HOP1 promoter was comparable in both UME6 and ume6Δ strains. These findings demonstrate that at both the natural HOP1 promoter and the hybrid CYC1-URS1-lacZ promoter, transcriptional activators bind to their target sites in promoters repressed by Ume6p.
Figure 2.
In vivo footprinting of Abf1p in a high copy HOP1-lacZ plasmid, pAV79B (20), in UME6 and ume6Δ strains (yx423 and yx268, respectively). The Abf1p binding site is shown to the left of the gels. (A) DMS footprints of the top strand of the HOP1 promoter. Lanes marked C are samples from intact cells treated with 0.12 and 0.06% DMS; lanes marked D are samples from purified DNA treated with 0.1% DMS. Dots indicate sites protected from DMS modification in cells. (B) UV photofootprints of the bottom strand of the HOP1 promoter. Lanes marked C are samples from intact cells irradiated with UV light (500 and 750 mJ/cm2); lanes marked D are samples from purified DNA irradiated with UV light (120 and 240 mJ/cm2). + indicates sites protected from UV irradiation in cells.
Analysis of TBP occupancy at the HOP1 promoter by chromatin immunoprecipitation assay
We considered the hypothesis that repression results from inhibition of TFIID binding to the TATA region. This idea was tested through a ChIP assay for TBP-DNA binding. An epitope-tagged derivative of TBP (HA3-TBP) was expressed from the natural TBP1/SPT15 promoter in a TRP1 centromeric plasmid. The HA3-TBP construct was required for efficient recovery of promoter regions in anti-HA ChIP experiments (data not shown). Binding of HA3-TBP to the HOP1 and control ACT1 TATA regions was compared after anti-HA immunoprecipitation (Fig. 3). Recovery of the HOP1 promoter relative to ACT1 from the repressed UME6 strain was ∼40%, compared to that from the derepressed ume6Δ strain. Therefore, repression by Ume6p is associated with reduced binding of TBP to the HOP1 promoter.
Figure 3.
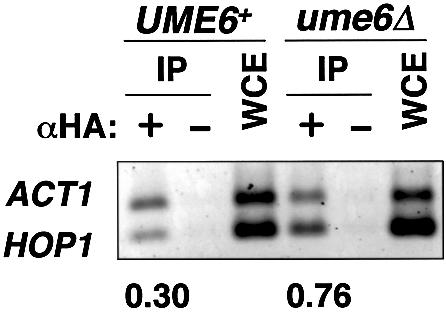
ChIP assay for HA3-TBP binding in UME6 and ume6Δ strains (AMP107 and MHS24, respectively) expressing HA3-TBP. IP indicates chromatin immunoprecipitated DNA with or without monoclonal anti-HA antibody (lanes marked with αHA: + and –, respectively). WCE indicates DNA isolated from whole cell extract. Multiplex PCR reactions were performed with primer sets for promoters of ACT1 and HOP1. PCR amplified regions were: ACT1, –300 to –56; HOP1, –179 to –1. Agarose gel was stained with SYBR-Green, visualized and quantitated by a Luminescent Image Analyzer. The ratios HOP1/ACT1 in IP were normalized by dividing by HOP1/ACT1 in WCE, and are shown at the bottom of the gel. The ChIP experiment was carried out with three independent sets of samples, and the results shown here are typical.
Artificial recruitment of TBP relieves repression by Ume6p at the HOP1 promoter
Inhibition of TBP binding may be the cause of repression by Ume6p at HOP1 or it may be an indirect consequence of the repressed state. The former model predicts that expression of a TBP derivative which is able to bind to the HOP1 TATA region, independently of Ume6p, will relieve repression. To create such a derivative, we made use of the observation above that the activator Hap1p binds to CYC1 UAS1 independently of Ume6p (Fig. 1B). We fused the zinc-cluster DNA binding domain of Hap1p (16) to the N-terminus of TBP to create ZC-TBP (in strains lacking intact Hap1p). We also introduced the Hap1p binding site, CYC1 UAS1, upstream of the HOP1 TATA region to create the HOP1-UAS1 promoter. We reasoned that ZC-TBP would bind to HOP1-UAS1 with increased affinity because it can make both TBP-TATA and ZC-UAS1 protein–DNA contacts. In vivo UV photofootprinting verified that ZC-TBP is bound to the HOP1-UAS1 promoter in both UME6 and ume6Δ strains (data not shown).
We then examined expression of HOP1 and HOP1-UAS1 in the presence and absence of ZC-TBP (Fig. 4A). In the absence of ZC-TBP, the HOP1 and HOP1-UAS1 promoters were expressed at equivalent levels; Ume6p caused several hundred-fold repression. In the presence of ZC-TBP, the two promoters were expressed at similar derepressed levels (ume6Δ strain). However, whereas the HOP1 promoter (WT TATA) was repressed over 600-fold, the HOP1-UAS1 promoter (UAS1+WT TATA) was repressed only 2.5-fold. As a control, we prepared an additional strain expressing ZC alone and found that Ume6p repression was not relieved by the expression of ZC (data not shown). Thus artificial recruitment of TBP by ZC at the HOP1 TATA region relieves repression by Ume6p (Fig. 4B). This finding supports the model that Ume6p causes repression through inhibition of TBP binding at the HOP1 promoter.
Figure 4.
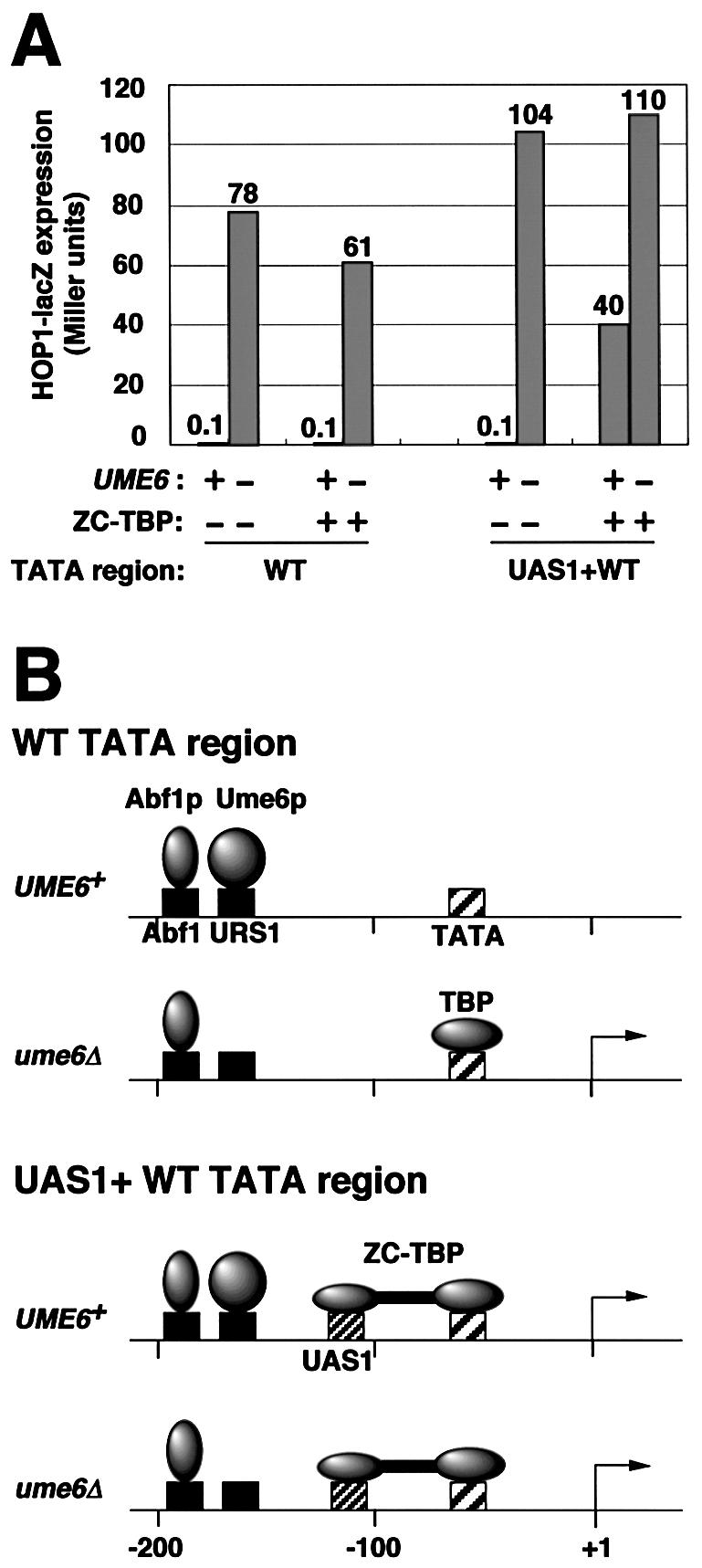
Effect of artificial recruitment of TBP on repression of the HOP1 promoter by Ume6p. (A) HOP1-lacZ expression in Miller units was measured in pAV79B (wild-type HOP1 promoter, referred to as WT TATA region) (20) and pKT5-1 [HOP1 promoter region in which –131 to –114 was replaced by the ZC binding site (CYC1 UAS1), referred to as UAS1+WT TATA region] in UME6 and ume6Δ strains with or without ZC-TBP expression. Strains used were AMP1780 (MATα hap1::LEU2), MHS22 (MATα hap1::LEU2 ume6Δ2), AMP1779 (MATα hap1::ZC-TBP-TRP1) and MHS21 (MATα hap1::ZC-TBP-TRP1 ume6Δ2). All strains lack Hap1p, the activator that binds to CYC1 UAS1. The standard deviations were <20% for the triplicate determinations. (B) Model for occupancy of Abf1p, TBP and ZC-TBP in wild-type HOP1 (WT TATA) and HOP1-UAS1 (UAS1+WT TATA) promoters. TBP binds to the derepressed wild-type HOP1 promoter, but does not bind to the repressed one. ZC-TBP, which is artificially recruited by ZC (zinc cluster DNA binding domain of Hap1p) to the CYC1 UAS1, binds to both repressed and derepressed HOP1-UAS1 promoter.
Ume6p repression: mechanistic implications
The localized histone deacetylation domain established by the Ume6p-Sin3p-Rpd3p complex (4–7) has been proposed to inhibit the binding of activators and/or TFIID to their cognate sites (4,5,29,30). This model is based upon a prevailing idea for transcriptional regulation by histone acetylation: acetylation weakens histone–DNA interaction and allows trans-acting factors access to cis-elements; deacetylation prevents access (30). Our findings indicate that Ume6p does not inhibit access of activators to CYC1-URS1 and HOP1, but that it impairs TBP binding at HOP1. Furthermore, at the HOP1 promoter, the relief of repression by tethered TBP argues that reduced TBP binding is the major cause of repression.
One simple model for repression is that the Ume6p repressor complex interferes with TBP binding through deacetylation of histones in a nucleosome which occludes the TATA region. This explanation is consistent with the finding that a deacetylated nucleosome can block TBP binding in vitro and in vivo (11,31,32). It is also consistent with the finding that Isw2p complex recruitment by Ume6p generates a local nuclease-inaccessible chromatin structure (8–10). These precedents argue that Ume6p-dependent chromatin modification can inhibit TBP binding directly.
A second possibility is that Ume6p affects TBP-promoter binding indirectly. For example, inhibition of the SAGA complex or another coactivator would lead to reduced TBP recruitment (33,34). According to this model, inhibition of activator function would then reduce TBP binding (25,35) and may be bypassed by tethering of TBP (36–39). Although we cannot rule out indirect models, our study narrows the target of Ume6p repression to an event between post-activator binding and TBP recruitment at the HOP1 promoter.
Another possibility is that Ume6p has multiple repression targets, since Ume6p interacts with both the Sin3p-Rpd3p and Isw2p complexes. This may explain the observation that the difference in TBP occupancy is not as great as the difference in repression we observed.
Very recently, similar results were published for artificial his3 promoters with or without IME2 URS1 sites (40). Deckert and Struhl (40) showed that binding of activators to their cognate sites, which were introduced to the his3 promoter, was unaffected by Rpd3p recruitment, whereas TBP occupancy was reduced upon Rpd3p recruitment in the range 2- to 6-fold, both in the artificial promoters and in the natural promoters INO1, CAR1, CAR2, SPO11 and SPO13. They also showed that Rpd3 repression at the his3 promoter can be bypassed by artificial recruitment of TFIID components (40). Our results for the natural HOP1 and artificial CYC1 promoters agree with the earlier study (40), though promoter structures and properties are different, indicating that blocking of TBP binding but not activator binding is a common mechanism for repression of Ume6p-regulated genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. P. Davis, Y. Xiao and W. Xu for materials and advice, N. Tomita and M. Hara for technical assistance, A. Vershon for plasmid pAV79B, T. Ohta for guidance with the ChIP assay and T. Bestor and C. Schindler for comments on this manuscript. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan to M.S., by NIH grant GM39531 to A.P.M. and by the Japan–US Cooperative Science Program, from JSPS and NSF, to M.S. and A.P.M.
REFERENCES
- 1.Mitchell A.P. (1994) Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev., 58, 56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadosh D. and Struhl,K. (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell, 89, 365–371. [DOI] [PubMed] [Google Scholar]
- 3.Kadosh D. and Struhl,K. (1998) Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev., 12, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadosh D. and Struhl,K. (1998) Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rundlett S.E., Carmen,A.A., Suka,N., Turner,B.M. and Grunstein,M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- 6.Suka N., Suka,Y., Carmen,A.A., Wu,J. and Grunstein,M. (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell, 8, 473–479. [DOI] [PubMed] [Google Scholar]
- 7.Deckert J. and Struhl,K. (2001) Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol., 21, 2726–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldmark J.P., Fazzio,T.G., Estep,P.W., Church,G.M. and Tsukiyama,T. (2000) The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- 9.Kent N.A. Karabetsou,N., Politis,P.K. and Mellor,J. (2001) In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev., 15, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazzio T.G., Kooperberg,C., Goldmark,J.P., Neal,C., Basom,R., Delrow,J. and Tsukiyama,T. (2001) Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol., 21, 6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sewack G.F., Ellis,T.W. and Hansen,U. (2001) Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol., 21, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdone L., Wu,J., van Riper,K., Kacherovsky,N., Vogelauer,M., Young,E.T., Grunstein,M., Di Mauro,E. and Caserta,M. (2002) Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J., 21, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breiling A., Turner,B.M., Bianchi,M.E. and Orlando,V. (2001) General transcription factors bind promoters repressed by Polycomb group proteins. Nature, 412, 651–655. [DOI] [PubMed] [Google Scholar]
- 14.Sekinger E.A. and Gross,D.S. (2001) Silenced chromatin is permissive to activator binding and PIC recruitment. Cell, 105, 403–414. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Y. and Mitchell,A.P. (2000) Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol. Cell. Biol., 20, 5447–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeifer K., Kim,K.S., Kogan,S. and Guarente,L. (1989) Functional dissection and sequence of yeast HAP1 activator. Cell, 56, 291–301. [DOI] [PubMed] [Google Scholar]
- 17.Schneider B.L., Seufert,W., Steiner,B., Yang,Q.H. and Futcher,A.B. (1995) Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast, 11, 1265–1274. [DOI] [PubMed] [Google Scholar]
- 18.Bowdish K.S. and Mitchell,A.P. (1993) Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowdish K.S., Yuan,H.E. and Mitchell,A.P. (1995) Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol., 15, 2955–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vershon A.K., Hollingsworth,N.M. and Johnson,A.D. (1992) Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol., 12, 3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Shimizu M., Li,W., Covitz,P.A., Hara,M., Shindo,H. and Mitchell,A.P. (1998) Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res., 26, 2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu M., Li,W., Shindo,H. and Mitchell,A.P. (1997) Transcriptional repression at a distance through exclusion of activator binding in vivo. Proc. Natl Acad. Sci. USA, 94, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- 25.Kuras L. and Struhl,K. (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature, 399, 609–613. [DOI] [PubMed] [Google Scholar]
- 26.Davie J.K., Trumbly,R.J. and Dent,S.Y.R. (2002) Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol., 22, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gailus-Durner V., Xie,J., Chintamaneni,C. and Vershon,A.K. (1996) Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol. Cell. Biol., 16, 2777–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prinz S., Klein,F., Auer,H., Schweizer,D. and Primig,M. (1995) A DNA binding factor (UBF) interacts with a positive regulatory element in the promoters of genes expressed during meiosis and vegetative growth in yeast. Nucleic Acids Res., 23, 3449–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 30.Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- 31.Imbalzano A.N., Kwon,H., Green,M.R. and Kingston,R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481–485. [DOI] [PubMed] [Google Scholar]
- 32.Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 33.Dudley A.M., Rougeulle,C. and Winston,F. (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev., 13, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belotserkovskaya R., Sterner,D.E., Deng,M., Sayre,M.H., Lieberman,P.M. and Berger,S.L. (2000) Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol., 20, 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X.Y., Virbasius,A., Zhu,X. and Green,M.R. (1999) Enhancement of TBP binding by activators and general transcription factors. Nature, 399, 605–609. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee S. and Struhl,K. (1995) Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature, 374, 820–822. [DOI] [PubMed] [Google Scholar]
- 37.Klages N. and Strubin,M. (1995) Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature, 374, 822–823. [DOI] [PubMed] [Google Scholar]
- 38.Xiao H., Friesen,J.D. and Lis,J.T. (1995) Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. Cell. Biol., 15, 5757–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keaveney M. and Struhl,K. (1998) Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell, 1, 917–924. [DOI] [PubMed] [Google Scholar]
- 40.Deckert J. and Struhl,K. (2002) Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA and TATA binding protein. Mol. Cell. Biol., 22, 6458–6470. [DOI] [PMC free article] [PubMed] [Google Scholar]