Abstract
The process of gene unscrambling in hypotrichous ciliates represents one of nature's ingenious solutions to the problem of gene assembly. With some essential genes scrambled in as many as 51 pieces, these ciliates rely on sequence and structural cues to rebuild their fragmented genes and genomes. Here we report the complex pattern of scrambling in the DNA polymerase α gene of Stylonychia lemnae. The germline (micronuclear) copy of this gene is broken into 48 pieces with 47 dispersed over two loci, with no asymmetry in the placement of coding segments on either strand. Direct repeats present at the boundaries between coding and noncoding sequences provide pointers to help guide assembly of the functional (macronuclear) gene. We investigate the evolution of this complex gene in three hypotrichous species.
Keywords: gene rearrangement, DNA polymerase α, hypotrich, ciliate, DNA computing
All ciliates possess two types of nuclei: an active somatic macronucleus and a germline micronucleus that contributes to sexual reproduction. The macronucleus forms from the micronucleus after cell mating, during the course of development. Prescott and colleagues discovered that the genomic copies of some protein-coding genes in the micronucleus of hypotrichous ciliates are encrypted in three ways (reviewed in ref. 1): (i) intervening non-protein-coding DNA segments [internal eliminated segments (IESs)] interrupt protein-coding DNA segments [macronuclear destined segments (MDSs)] and must be removed from the DNA during macronuclear development (2), (ii) the MDS order in 3 of 10 micronuclear genes is permuted relative to the chronological order in the macronuclear copy (1, 3), and (iii) these scrambled MDSs may be encoded in either orientation on the micronuclear DNA (Fig. 1). Some IESs may be remnants of transposons that lost their transposase and are now excised by a ciliate-specific mechanism for DNA rearrangements (2, 4). The total amount of DNA eliminated from the micronucleus is as great as 98% in Stylonychia (1), a dramatic reduction of noncoding DNA. These acrobatic genome rearrangements therefore present a potentially complicated cellular computational paradigm (5).
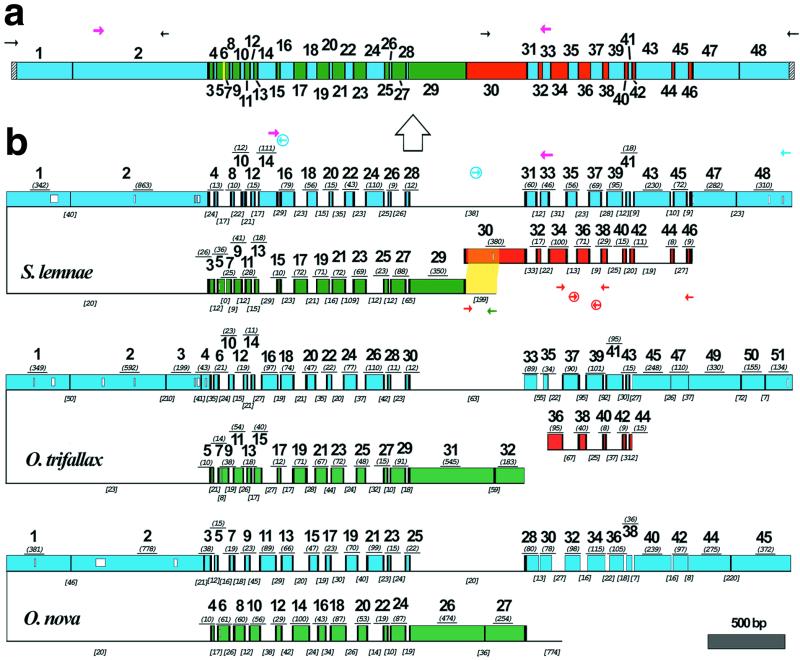
Figure 1.
(a) A schematic map of the macronuclear DNA-polymerase α gene of S. lemnae. The 48 MDS are shown as colored boxes. Green and blue MDSs are both present on the major locus, but in opposite orientation. Red MDSs are derived from the minor locus. The yellow MDS is missing. Pointer sequences are indicated by black vertical bars, telomeres by hatched bars. PCR primers indicated by small arrows; degenerate primers, in magenta, were based on an alignment of this gene in 10 hypotrich species (33). The macronuclear copy is derived from the micronuclear copy (large arrow). (b) A schematic alignment of the micronuclear genes encoding the large catalytic subunit of DNA-polymerase α in S. lemnae, O. trifallax (7), and O. nova (6), with MDSs aligned based on predicted amino acid sequences; open boxes within MDSs indicate gaps in alignment. MDSs are drawn to scale with the length (excluding pointer sequences when available) underlined in italics and parentheses. Green and blue MDSs are encoded on opposite strands of the major locus, indicated by the inversion. Red MDSs are derived from the minor locus. Black vertical bars indicate pointers. IESs are indicated by thin lines, not to scale, but IES length is given in italics and brackets. IES-specific primers are circled. The yellow highlighted region indicates 193/199 bp overlap between the IES that marks the 5′ end of the major locus and MDS 30 in the minor locus of S. lemnae.
Homologous recombination between short repeats at MDS-IES boundaries has been implicated as the likely mechanism of gene unscrambling, as it could simultaneously remove the IESs and reorder the MDSs (1). Typically, a short DNA sequence present at the boundary between MDS n and the downstream IES is repeated between MDS n + 1 and its upstream IES, such that this sequence provides a pointer between MDS n and MDS n + 1 at a distance (refs. 1, 6, and 7; Table 1), with one copy of the repeat retained in the macronucleus. However, the presence of such short direct repeats [average length of 4 bp between nonscrambled MDSs and 9 bp between scrambled MDSs (ref. 8; Table 1)] suggested that, although these pointers are necessary, they cannot be sufficient to direct accurate splicing and may play more of a role in structure than recognition. Otherwise, incorrectly spliced sequences (the results of promiscuous recombination) would dominate. This incorrect hybridization could be a driving force in the production of newly scrambled patterns in evolution; however, only precisely unscrambled genes would encode the canonical protein product and only molecules that acquire telomeres to protect both ends are retained in the macronucleus, ensuring that most promiscuously ordered genes would not be retained.
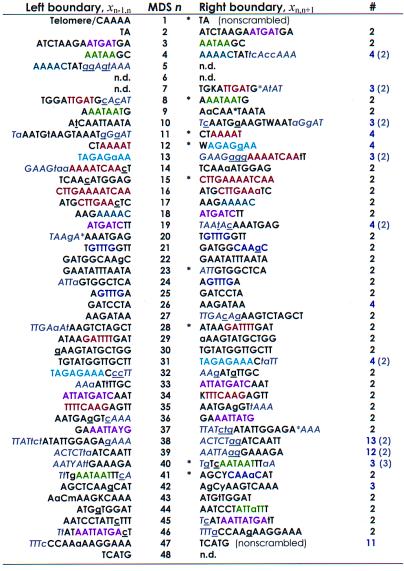
Table 1.
Stylonychia lemnae DNA pol α pointers plus context are unique
This table lists the boundary sequences (pointers xij) at the ends of each MDS in the micronuclear DNA. Flanking sequences in gray and italics provide extended context for pairing. Sequences that are identical between MDS n and n + 1 are in uppercase; asterisks within a sequence represent gaps introduced to maximize alignment (by eye). Where there are mismatches (lowercase), the macronuclear sequence contains the underlined nucleotide. The left boundary of MDS 1 begins with the encoded sequence CAAAA, which serves as a telomere addition site. Pointers marked by an asterisk are the most conserved among S. lemnae, O. nova, and O. trifallax. The right column indicates the multiplicity of each pointer (number of times the identical sequence is found in either strand of the major or minor locus). Numbers in parentheses include an additional gray sequence, which generally reduces the multiplicity of each pointer to two, the expected redundancy at the end of MDS n, and beginning of MDS n + 1. Pointers with multiplicity greater than 2 (even with additional sequences) all follow short (9–28 bp) MDSs that are exchanged for IESs of similar length (MDS 11, 12, 26, 40, 42; Table 2), suggesting that the physical alignment provides the context for pairing. Sequences to the left of MDS 30 and the right of MDS 46 were part of PCR primers and were not sequenced directly. IUPAC symbols specify pairs of nucleotides present at positions that differ between alleles. Sequence motifs (representative “words” or “subwords” that recur on either strand) shared between two or more pointer pairs are indicated by the same color.
Fragmented genes exist in a variety of taxa, including other Alveolates [e.g., split ribosomal RNAs in Plasmodium and Theileria (9, 10)]. However, rarely are they actually “sewn” back together at the level of DNA, to create a contiguous gene from all pieces. A notable exception is V(D)J splicing in the vertebrate immune system. Trans-splicing of RNA molecules (e.g., ref. 11) and even of proteins mediated by inteins (12) provides another approach to merging functional regions located on dispersed elements of a genome. The complexity of DNA splicing events in hypotrichous ciliates even shares some ostensible similarities with the process of gene shuffling in vitro (13), making the scrambled genes one of the most interesting and challenging cases to study. Here, we report that the micronuclear gene encoding the large catalytic subunit of DNA polymerase α in Stylonychia lemnae is broken into 48 MDSs scrambled in an odd/even order with 14 MDSs inverted on the opposite strand of another 24 MDSs, 9 additional MDSs on a separate locus, and 1 MDS not present on either locus, and we compare the evolution of this complex gene with its scrambled homologs in Oxytricha nova (6) and Oxytricha trifallax (7).
Methods
S. lemnae micronuclear and macronuclear DNAs were a gift from H. Lipps (Witten University, Witten, Germany). Micronuclear DNA purified on 0.75% agarose was PCR amplified with degenerate primers from MDS 14→ to 33←, which gave a smaller fragment for micronuclear DNA than contaminating macronuclear DNA (present in high copy number). The micronuclear product was gel-purified, cloned into a Topo-TA vector (Invitrogen), and sequenced, and from the resulting micronuclear sequence we designed IES-specific primers (below) to selectively amplify micronuclear DNA with end primers derived from MDS 30← and 48→. Multiple clones were generally sequenced to detect allelic differences or PCR errors and to derive a consensus. Most point mutations were silent, suggesting that both alleles are expressed and not pseudogenes. The minor locus containing nine MDSs absent from the major locus was recovered by the same strategy, first amplifying a fragment from MDS 34 to 38 that gave a smaller PCR product for micronuclear DNA than macronuclear DNA, and then using the IES sequences to design micronuclear specific primers (below) to amplify to the end of MDS 30→ and 46←, including putative pointers. Example conditions were 40 cycles (94°C, 25 sec; 60°C, 40 sec; 72°C 1–3 min) as in ref. 14, using Taq Extender (Stratagene). Some products were diluted 1:50, and 1 μl of this dilution was reamplified for up to 30 cycles.
The complete macronuclear gene sequence was recovered by PCR with degenerate primers between MDS 2→ and 33← and then by PCR to the ends of the macronuclear chromosomes with nested primers derived from MDS 2← and 30→ in combination with telomere-specific primers (ref 15; and E.A.C. and L.F.L., unpublished work).
All macronuclear primers are indicated in the supplemental alignment (published as supplemental data on the PNAS web site, www.pnas.org). Major locus micronuclear-specific primers were TCGTTACAAATAAAAAATCTGAGGG (IES between MDS 14–16←) and TTGATAAGTTAATTTTGAATTATTCTAG (IES between MDS 28–31→). Minor locus micronuclear-specific primers were CAAGAGTTATATATTTGTTT (IES between MDS 34–36→), CAATATAGAATAACTCATAA (IES between MDS 36–38←).
Results and Discussion
The S. lemnae gene is broken into 48 or more pieces in the germline genome, dispersed across two loci that may even reside on different chromosomes (Fig. 1). Thirty-eight MDSs are on a single 5-kb major locus in the cryptic order 29-27-25-23-21-19-17-15-13-11-9-7-5-3/1-2-4-8-10-12-14-16-18-20-22-24-26-28-31-33-35-37-39-41-43-45-47-48, with one boundary between MDS 3 and 1 inverted such that fourteen odd MDSs 3–29 (italicized) are encoded on the reverse complementary strand with respect to the 24 MDSs downstream. Thus, this gene displays no particular asymmetry in the placement of coding regions on either strand of this chromosome. There are only two IESs placed between nonscrambled MDSs, one at the beginning between MDS 1–2 and one at the end between MDS 47–48; both are present in O. nova and O. trifallax. However, nine even MDSs 30-32-34-36-38-40-42-44-46 are present on an unmapped minor locus separate from the major locus of the micronuclear gene; yet, these pieces are seamlessly woven into the final macronuclear product. Long PCR with primer combinations amplifying away from the ends of both major and minor loci could not detect a micronuclear product that joined them, suggesting that these loci are not near each other, consistent with sequencing 5 kb on either side of the major locus in O. nova (6). The last missing MDS 6 is only 12 bp and has not been found (see discussion below).
Properties of Boundaries.
Pointers are present at MDS-IES junctions that could simultaneously facilitate removal of the IESs and proper reordering of the MDSs (Table 1). There is also a region 15 bp upstream of MDS 29 (on the inverse strand) that contains 193/199 bp identical to a portion of MDS 30 in the minor locus (highlighted in Fig. 1). This may serve as an anchor to bring the two loci into proximity for recombination. The length of overlap may actually be greater because this region was recovered as part of a large PCR fragment, and no sequences further upstream were obtained. Similarly, no sequences were recovered from the minor locus either upstream of MDS 30 or downstream of MDS 46, and so it is possible that there may be extended regions of alignment. This long overlap in MDS 30 also suggests that the piece that became the minor locus probably broke off from the major locus in this region, where it once was linked (Fig. 2), perhaps involving a long staggered cut and repair, producing a long duplicated region that then diverged. Alternatively, the original unsplit version may have coexisted as a polymorphism with the minor locus, which was recruited to supply nine MDSs when a second event led to the truncation of the major locus. Because assembly of the sequences for these loci from overlapping PCR fragments could have led us to believe that pieces such as the major and minor loci were linked, we confirmed the 5-kb size of the major locus between MDS 29 and 47 by Southern blotting (not shown).
Figure 2.
Proposed steps in the evolution of the scrambled DNA polymerase α gene through a series of recombinations between MDSs and IESs. MDSs are shown as boxes (black numbers and color stripes reference cognate regions in the extant S. lemnae gene; Fig. 1); IESs or noncoding DNA are drawn as thin lines; and pointers are shown as black bars. The first general step takes place in a nonscrambled ancestral version of the gene [shown here with one IES between MDS 1–2 because this IES is conserved in all three species and IESs flanked by TA repeats are the most common in other ciliate species (1)]. Reciprocal exchange at the ×s between an MDS and upstream noncoding DNA creates a scrambled hypothetical MDS order (3-1–2-4; new MDSs in purple) and an inversion (up arrow) between 3 and 1; these MDSs are numbered from the 5′ end (… ) and italicized if on the reverse strand. The juxtaposition of MDSs and IESs now promotes homologous recombination at chance matches, perhaps even at favored sites (Table 1), creating the new pattern 5–3-1–2-4–6. Continued exchange at the ×s, in any order (not just the reasonable one shown here), propagates the odd/even MDS splitting: 17–15-13–11-9–7-5–3-1–2-4–6-8–10-12–14-16–18 is shown as one evolutionary intermediate, finally reaching a dense set of 43 contiguous MDSs very similar to O. nova. At any stage in an ancestor of all three species there could be insertion of an IES in the last MDS and finally translocation of a cluster of MDSs at the 5′ end (probably maintained as a polymorphism initially, allowing substantial overlap in the orange region) by reciprocal exchange with a distant DNA fragment. The precise ordering of the previous steps does not matter, but the most recent step is splitting of MDSs 5–7 (by exchange with an unknown fragment) and 11–13 in an ancestor of S. lemnae and O. trifallax, leading to the pattern in S. lemnae.
Correct alignment of all pointers leads to the predicted folded structure in Fig. 1 that could provide a dramatic shortcut for disentangling the micronuclear DNA pol α gene (6, 7). Specifically, Fig. 3 and Table 2 (Table 2 is published as supplemental data) compare the length of DNA exchanged during recombination, excluding pointers, for the scrambled regions that interleave two potentially juxtaposed aligned strands, as in Fig. 1. For MDSs < 30 nt, there is a visible correlation, and we found this generally to be true for other species and other scrambled genes (data not shown). This predicts a zigzag local alignment between these portions of two interleaved micronuclear strands:
Figure 3.
Comparison of MDS versus IES length (nt) in S. lemnae for exchanged regions flanked by the same set of pointers (such as xij and xjk). [Pointer length is not included in the comparison; hence, this graph compares the length of each α and opposing ɛ (see text).] The data are listed in Table 2. Mean MDS length is 49 nt; mean IES length is 23 nt.
… -xij-αj-xjk-ɛb-xkl-αl-xlm-ɛd-…
… -xij-ɛa-xjk-αk-xkl-ɛc-xlm-αm-…
where each xij symbolically represents a pointer between MDS i and j (= i + 1), αj is MDS j (excluding its pointers xij and xjk), and ɛa is the IES between the same two pointers (5, 16). This sequence becomes unscrambled as
… -xij-αj-xjk-αk-xkl-αl-xlm-αm-…
simply by even exchange between aligned MDSs and IESs at matching pointers xij. The question remains, What drives incorporation of MDSs and exclusion of IESs at recombination junctions?
It is noteworthy that the “newest” MDS 12 in S. lemnae is exactly the same length as its exchanged IES, suggesting that it was recently formed by a precise exchange between coding and noncoding DNA. Also, the location and sequence of the pointers flanking this MDS and homologous MDS 14 in O. trifallax are tightly conserved within 1–2 bp on either side. In general, for the region in which comparisons are possible in all three species (Table 2), more than half (13 of 23) of the homologous MDSs between S. lemnae MDS 3–28 preserve the property in two or more species that they recombine with IESs of similar length, differing only between 0 and 13 bp, with 10 of these falling within just 7 bp. The shortest length of DNA exchanged is 8 bp for MDS 44 and 9 bp for several IESs. The longest region exchanged is 230 bp (MDS 43) opposite a 19 bp IES. This is also the greatest difference in length between exchanged DNA in S. lemnae. On the other extreme, a 312-bp IES in O. trifallax exchanges with 15-bp MDS 43. We note that the MDSs with non-unique (often shorter) pointers (see Table 1) generally fall into the class that has no gaps in alignment. We conclude that those IESs that are approximately the same length as their juxtaposed MDSs (which have the same pointers at both ends) have accumulated length changes more slowly since their introduction in an ancestor of these three species, either reflecting their recent introduction or a physical length constraint that facilitates unscrambling.
Overall, pointers are generally shorter when they direct recombination between MDSs and IESs of comparable length, where the physical alignment—pairing at upstream or downstream pointers—will dramatically increase the probability of correct pairing at adjacent pointers. In other cases, additional sequence alignment exists close to the sequences designated as pointers, defined as the direct repeat with no more than a single mismatch. By increasing the effective length of a pointer despite several mismatches, this extended context would provide additional information and alignment, thereby increasing the likelihood of pairing at correct pointers. MDS 38 provides a good example of a pointer (x37,38) which has four of seven nucleotides repeated to the left (between the upstream IES and MDS 37) and three of four nucleotides repeated to the right (Table 1). Although this pointer itself occurs uniquely once in the major locus and once in the minor locus to provide a junction between them, the additional bases extend the context for pairing, promoting exchange in the correctly matched region. Both pointers flanking MDS 39 (x38,39 and x39,40), however, require the additional context to provide specificity (Table 1). MDS 39 is 70 bp longer than the IES between MDSs 38 and 40, which introduces long gaps in the physical alignment. Alternatively, one could just relax the definition of pointer, allowing two or even three mismatches in the “direct repeat,” especially because many pointers in Table 1 do contain single mismatches.
Our model for the guided homologous recombinations that take place during gene unscrambling relies on the presence of specific contexts to influence splicing (refs. 5 and 16; see discussion below). The two independent factors, the physical alignment anchored by pointers and conservative spacing between MDSs and IESs, and the local sequence alignment influenced by the broader context surrounding pointers make the context of each designated pointer “unique” within this locus. Taken together, every “redundant” pointer between scrambled MDSs (one with multiplicity >2 in Table 1) lies within a region of either precise spacing, occurring after short (9–28 bp) MDSs that exchange with IESs of similar length (Fig. 3), or additional DNA sequence alignment, driving correct pairing during unscrambling.
In general, IESs, pointers, and MDS-IES boundaries are labile and accumulate differences rapidly in sequence and position (17). But although most pointers have shifted their location by 1–20 nucleotides in at least one of the three species, a few pointers are actually conserved, and several overlap by a few nucleotides (see annotated alignment in supplemental material on the PNAS web site). Accordingly, there may be less “sliding” of pointers either in conserved regions of the protein, because these regions have fewer replacement substitutions that contribute to mismatches between pointers, or in regions that constrain MDSs and IESs to be of comparable length, to facilitate their alignment and exchange.
The pattern of mismatches in the middle of pointers (Table 1) suggests their stochastic incorporation into macronuclear sequence; there is no apparent bias for the presence of the sequence derived from the left or the right end in the final macronuclear copy. We occasionally see evidence of two distinct alleles in both the micronuclear and resulting macronuclear copies, with most substitutions occurring at silent positions, confirming the expression of both micronuclear alleles in the macronucleus. Although some of the apparent mismatches in pointers could be caused by PCR mixtures of alleles, or even PCR error, most mismatches probably reflect the ongoing process of point mutation that leads to sliding of the boundaries between IES and MDS as the alignment increases at one end and subsequently deteriorates at the other end by drift. The matching sequences that currently provide additional context for pointers may simply be the footprints of former repeats. A similar sliding process has occurred between guide RNAs and mRNAs that undergo RNA editing in kinetoplastid mitochondria, leading to shifting of the boundaries of interactions between these molecules (18). Thus, both processes probably reflect a more general mutational trend among overlapping nucleic acid molecules that drive the assembly of protein-coding genes from their constituent pieces.
Evolution of the Reading Frame.
Previous published reports (6, 7) of the DNA pol α gene in O. nova and O. trifallax assigned the first AUG codon downstream of a conserved UGA stop as the putative start of the reading frame, but both of these AUG codons are absent from MDS 1 in S. lemnae, as well as the UGA stop, with the next AUG codon over 300 bp downstream in MDS 2. Close inspection of upstream sequences revealed the presence of a conserved AUG codon in all three species that would introduce another 45 amino acids in the O. trifallax sequence and 52 in O. nova, with several synonymous and conservative replacement substitutions across all three species as evidence for initiation at this AUG codon (see supplemental aligned data). The S. lemnae sequence, however, has a different complication. A single A insertion 40 bp downstream of the conserved AUG in both the macronuclear and micronuclear sequence places this AUG out-of-frame, with the sequence AUG AGU G now parsed as A UGA GUG, with a new in-frame UGA stop overlapping the conserved AUG. Evidence for translation from the conserved AUG—requiring a mechanism to merge the two reading frames—is the conservation of 13/14 predicted amino acids after this AUG, with five silent third position substitutions in the S. lemnae sequence. There is, however, an in-frame upstream AUG that would introduce only 17 amino acids if it initiates translation, again requiring a mechanism to compensate for the UGA codon overlapping the conserved AUG.
These in-frame UGA codons, two of which are conserved in O. trifallax and O. nova, could be sites of selenocysteine incorporation (19) or leaky stop codons, either naturally suppressed nonsense mutations or a case of context-dependent stop codon read-through, both phenomena known in eukaryotes (20, 21). Indeed the UGA codon—the only stop codon used in these hypotrichs (22)—has been described as particularly leaky (20). Alternatively, in S. lemnae, either ribosomal frameshifting soon after initiation at the conserved AUG, initiation by the GUG codon after the in-frame UGA, or RNA editing (23) could restore the reading frame.
Evolutionary Dynamics and the Origin of Scrambled Genes.
The observation that S. lemnae and O. trifallax share a similar pattern of accumulated MDSs (e.g., the presence of a missing MDS in the same region and MDS 11–13 in S. lemnae, homologous to MDS 13–15 in O. trifallax) suggests that these two species may have shared a common ancestor more recently than either has with O. nova; however, O. nova and O. trifallax share the presence of an IES between nonscrambled MDSs at a similar position (13 bp apart) just before MDS 3 in O. nova. It is possible that this IES was either lost from S. lemnae or added independently in the two other lineages, especially as the position of the other shared nonscrambled IES between MDS 1 and 2 is precisely conserved in all three species. Alternatively, the presence of this IES may have been a polymorphism when these species diverged. We can, however, infer that the last common ancestor of these three species had at least 44 MDSs for this gene.
Fig. 2 is a general model for the origin and accumulation of scrambled MDSs in a gene with a nonrandomly scrambled order. Although Prescott (3) proposed that creation of the inversion was the last step in the origin of the scrambled DNA pol α gene, we conjecture that the appearance of an inverted segment instead behaved as a “catalyst” for the fragmentation of MDSs and the creation of new nonrandomly scrambled patterns during evolution. This can be tested by recovery of the homologous scrambled gene in several outgroup lineages. By stabilizing the hairpin alignment that juxtaposes coding and noncoding sequences, an inversion would promote germline recombination that breaks apart MDSs. This could occur at short arbitrary repeats in S. lemnae's AT-rich DNA or at potentially favored “words” that are enriched in the set of pointers in Table 1. These junctions would then become the pointers, and selection would constrain the pointers to co-evolve to guide unscrambling at these junctions. For example, MDS 6 and 10 in O. nova could have given rise to MDS 7–9 and 13–15 in O. trifallax (refs. 7 and 24) and 5–7 and 11–13 in S. lemnae (Figs. 1 and 2). It is suggestive that the pointer x12,13 flanking new MDS 12 in S. lemnae recurs in x31,32. This model suggests that most IESs between scrambled MDSs are not degenerate transposons, and that pointers between many scrambled MDSs derive from recombination between chance occurrences of repeat sequences in the genome.
Given the presence of an “unsolicited capacity” (25) for gene rearrangement, alleles with increasing degrees of scrambling may be effectively neutral, as long as there is the sorting machinery available to unscramble them. We have proposed analogously that the extent of RNA editing in kinetoplastid mitochondria may have increased by accumulation of mutations that could be easily repaired by RNA editing, given a mechanism for U insertion (26) and the presence of guide RNAs to direct them. In both cases, the gene editing or unscrambling event acts as a suppressor. The probability of reversion to “wild-type” is expected to be very low because it requires site-specific repair of the germline DNA. Such a bias would lead to continued fragmentation of these genes and accumulation of excessive “neutral” mutations in a ratchet-like fashion. However, as ciliates reproduce asexually for many generations (given a plentiful food source), undergoing sexual reproduction to produce a new macronucleus only in conditions of starvation, and as kinetoplastid mitochondria may effectively be asexual populations as well with only one mitochondrion per cell, the influence of Muller's ratchet could lead to the exaggeration of these two degenerative processes—the accumulation of scrambled MDSs and extent of RNA editing in the respective lineages. In the case of ciliates, the ratchet would act on the relatively quiescent micronucleus during asexual reproduction, and in kinetoplastids on the organelle genome (27). In particular, in the absence of selection to preserve individuals with the most conservative pattern of MDSs, the accumulation of scrambled alleles would be expected by random loss of the least mutated class during the asexual stage of reproduction (28). It remains to be tested whether individuals with more scrambled or edited alleles do have reduced fitness.
The missing MDS 6 in S. lemnae provides an interesting clue in support of our model, as it provides an example of a functional gene that has simply “lost” a short segment in the middle! Only 12 nt flanked by a TC repeat, and shifted 15 nt downstream of the missing MDS 8 in O. trifallax, this MDS encodes four conserved amino acids in the macronuclear copy of the gene (see supplemental alignment). Its closest match in the two loci is 11/12 nt in the IES between MDS 15 and 17 but without additional flanking pointers, making it unlikely that this is the actual MDS. Instead, the parent MDS (a homolog of MDS 6 in O. nova) in an ancestor of both O. trifallax and S. lemnae probably participated in a pair of rogue recombination events with some distant part of the genome, perhaps at matches of preferred words (Table 1). The actin II gene in O. nova is similarly missing 121 bp at the 5′ end that have not been identified anywhere (D. M. Prescott, personal communication), which further supports the model. Moreover, it suggests that such a recombination mechanism can lead to dramatic genome-wide rearrangements, affecting multiple, now interacting loci.
Computational Aspects of Gene Unscrambling.
The enormous plasticity of both gene scrambling and editing mechanisms over evolutionary time may be one reason for the survival of such a wide range of biological systems driven by sequence matching rules. Furthermore, the recent proof that a model that describes this process of gene unscrambling has the computational power of a universal Turing machine (16)—the most widely accepted model for electronic computation—suggests that these hypotrichous ciliates may in principle possess the capacity to perform any formal computation carried out by an electronic computer. This hints at both the capacity of these cells to perform daring feats of biological computation (16, 29) and the creativity of evolution to form complex, potentially nonadaptive genetic systems.
Computationally, we ask, what is the problem that is actually being solved? The first step, alignment, involves a series of basic combinatorial pattern matching steps (Fig. 4, available as supplemental data), perhaps resulting in an alignment like Fig. 1. There are many opportunities to deviate from this alignment because of the redundancy of some pointers (Table 1 and Fig. 4), and it is plausible that the DNA in the developing macronucleus searches through several possible matches and their resulting conformations, via either intramolecular or intermolecular strand associations. We have speculated that this route could be similar to the solution of a directed Hamiltonian path problem, like the “classic” DNA computing experiments in vitro (4, 30, 31). Alternatively, the second step, homologous recombination at aligned pointers, also involves choices at each junction of whether to retain either the IES or the MDS between two pointers. This decision process could then be analogous to solving an n-bit instance of a satisfiability problem, where n is the number of scrambled MDSs. At each MDS-IES junction, there is a choice of which of two possible sequences to discard or to keep in the surviving strand, marked by telomere addition at both ends. This choice may be governed by the epigenetic influence of the old copy of the gene in the degraded macronucleus, which regulates IES splicing in the ciliate Paramecium (32). Both of these hypotheses about the computational nature of gene unscrambling make specific testable predictions about the types of errors or intermediates that may be identified during gene rearrangement, such as linking of segments at incorrect pointers or retention of noncoding instead of coding sequence.
Supplementary Material
Acknowledgments
We thank Hans Lipps and members of his lab for the generous gift of S. lemnae DNA; David Ardell for suggesting the broader alignment of pointers; Lila Kari for theoretical modeling; David Prescott, Erik Winfree, Andrew Goodman, Grzegorz Rozenberg, Charles Kurland, and Tamara Horton for discussions, Vernadette Simon for technical assistance, and three anonymous reviewers for comments. This work was supported by National Institute of General Medical Sciences Grant GM59708 to L.F.L.
Abbreviations
- MDS
macronuclear destined segment
- IES
internal eliminated segment
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF194336, AF194337, and AF194338).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040574697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040574697
References
- 1.Prescott D M. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klobutcher L A, Jahn C L, Prescott D M. Cell. 1984;36:1045–1055. doi: 10.1016/0092-8674(84)90054-0. [DOI] [PubMed] [Google Scholar]
- 3.Prescott D M. Nucleic Acids Res. 1999;27:1243–1250. doi: 10.1093/nar/27.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klobutcher L A, Herrick G. Prog Nucleic Acid Res Mol Biol. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- 5.Landweber L F, Kari L. BioSystems. 1999;52:3–13. doi: 10.1016/s0303-2647(99)00027-1. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman D C, Prescott D M. Nucleic Acids Res. 1996;24:3337–3340. doi: 10.1093/nar/24.17.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman D C, Prescott D M. Nucleic Acids Res. 1997;25:1883–1889. doi: 10.1093/nar/25.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prescott D M, Dubois M L. J Eukaryotic Microbiol. 1996;43:432–441. doi: 10.1111/j.1550-7408.1996.tb04502.x. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie D E, Salazar N A, Rehkopf D H, Feagin J E. Nucleic Acids Res. 1999;27:2416–2422. doi: 10.1093/nar/27.11.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kairo A, Fairlamb A H, Gobright E, Nene V. EMBO J. 1994;13:898–905. doi: 10.1002/j.1460-2075.1994.tb06333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malek O, Brennicke A, Knoop V. Proc Natl Acad Sci USA. 1997;94:553–558. doi: 10.1073/pnas.94.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Hu Z, Liu X Q. Proc Natl Acad Sci USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stemmer W P C. Science. 1995;270:1510. doi: 10.1126/science.270.5241.1510. [DOI] [PubMed] [Google Scholar]
- 14.Landweber L F, Kreitman M. Methods Enzymol. 1993;218:17–26. doi: 10.1016/0076-6879(93)18004-v. [DOI] [PubMed] [Google Scholar]
- 15.Curtis E A, Landweber L F. Ann NY Acad Sci. 1999;870:349–350. doi: 10.1111/j.1749-6632.1999.tb08900.x. [DOI] [PubMed] [Google Scholar]
- 16.Landweber L F, Kari L. In: Evolution as Computation. Landweber L F, Winfree E, editors. Berlin: Springer; 2000. , in press. [Google Scholar]
- 17.DuBois M, Prescott D M. Mol Cell Biol. 1997;17:326–337. doi: 10.1128/mcb.17.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landweber L F, Fiks A G, Gilbert W. Proc Natl Acad Sci USA. 1993;90:9242–9246. doi: 10.1073/pnas.90.20.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Commans S, Bock A. FEMS Microbiol Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 20.Parker J. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valle R P C, Morch M-D. FEBS Lett. 1988;235:1–15. doi: 10.1016/0014-5793(88)81225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helftenbein E. Nucleic Acids Res. 1985;13:415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landweber L F, Gilbert W. Nature (London) 1993;363:179–182. doi: 10.1038/363179a0. [DOI] [PubMed] [Google Scholar]
- 24.Landweber L F. Biol Bull (Woods Hole, Mass) 1999;196:324–326. doi: 10.2307/1542961. [DOI] [PubMed] [Google Scholar]
- 25.Stoltzfus A. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- 26.Landweber L F. BioSystems. 1992;28:41–45. doi: 10.1016/0303-2647(92)90006-k. [DOI] [PubMed] [Google Scholar]
- 27.Lynch M. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- 28.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 29.Ehrenfeucht A, Prescott D M, Rozenberg G. In: Evolution as Computation. Landweber L F, Winfree E, editors. Berlin: Springer; 2000. , in press. [Google Scholar]
- 30.Adleman L M. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 31.Kari L, Landweber L F. Methods Mol Biol. 1999;132:413–430. doi: 10.1385/1-59259-192-2:413. [DOI] [PubMed] [Google Scholar]
- 32.Meyer E, Duharcourt S. Cell. 1996;87:9–12. doi: 10.1016/s0092-8674(00)81317-3. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman D C, Prescott D M. J Mol Evol. 1997;45:301–310. doi: 10.1007/pl00006234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.