Abstract
There is a substantial interest in implementing bioinformatics technologies that allow the design of oligonucleotides to support the development of microarrays made from short synthetic DNA fragments spotted or in situ synthesized on slides. Ideally, such oligonucleotides should be totally specific to their respective targets to avoid any cross-hybridization and should not form stable secondary structures that may interfere with the labeled probes during hybridization. We have developed OligoArray 2.0, a program that designs specific oligonucleotides at the genomic scale. It uses a thermodynamic approach to predict secondary structures and to calculate the specificity of targets on chips for a unique probe in a mixture of labeled probes. Furthermore, OligoArray 2.0 can adjust the oligonucleotide length, according to user input, to fit a narrow Tm range compatible with hybridization requirements. Combined with on chip oligonucleotide synthesis, this program makes it feasible to perform expression analysis on a genomic scale for any organism for which the genome sequence is known. This is without relying on cDNA or oligonucleotide libraries. OligoArray 2.0 was used to design 75 764 oligonucleotides representing 26 140 transcripts from Arabidopsis thaliana. Among this set, we provide at least one specific oligonucleotide for 93% of these transcripts.
INTRODUCTION
DNA microarrays enable parallel expression monitoring of thousands of genes. For the production of microarrays, DNA can either be synthesized on a solid support (1–3) or can be deposited in a pre-synthesized form onto a suitable surface. In this case the DNA can be in the form of PCR products (4) or oligonucleotides (5,6). Recent work has shown that microarrays of 60mer oligonucleotides can reach a sensitivity level close to one copy of mRNA per human cell and that a single oligonucleotide per gene is sufficient to monitor gene expression (2). These authors and others (7) have also studied the specificity limit of short oligonucleotides in terms of sequence identity with mRNAs other than their target and have shown greater than 66% identity can lead to cross-hybridization. These results demonstrate that oligonucleotide microarrays compare well with cDNA microarrays. When short sequences are considered for hybridization targets, it is important to avoid regions that can fold to form stable secondary structures.
The emergence of new flexible technologies in microarray fabrication that require only sequence data (2,3) and the availability of an increasing number of sequenced genomes (http://www.ncbi.nlm.nih.gov/Genomes/index.html) have prompted us to develop a program to design specific oligonucleotides for microarrays. In a previous work (8), we have described OligoArray, now known as OligoArray 1.0, a program that computes gene-specific and secondary structure-free oligonucleotides for genome-scale oligonucleotide microarray construction. In this first version, the oligonucleotide specificity was based on a comparison of sequence similarity between the specific target and putative non-specific targets. Here, we present OligoArray 2.0, an improved version where the computation of the specificity is based on the thermodynamics of the hybridization.
MATERIALS AND METHODS
Arabidopsis thaliana sequences
All described transcript sequences were extracted from the A.thaliana chromosome sequences (GenBank accession nos NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076; http://www.ncbi.nlm.nih.gov/) and saved in a single file.
Design of an oligonucleotide set representing the whole A.thaliana transcriptome
We have set the OligoArray 2.0 parameters as follows. The oligonucleotide length range was set to 45–47 nt, the melting temperature (Tm) range to 82–90°C and the GC content range to 35–50% according to the low GC content of the A.thaliana genome. The search was restricted to the last 1500 nt of the input sequences. This limitation was chosen to minimize the bias toward the 3′ end generated by abortive reverse transcription when probes are labeled using oligo(dT) to anchor the reaction. The thresholds to reject oligonucleotides that can fold to form stable secondary structures and to start to consider putative cross-hybridizations were both set to 65°C. Furthermore, oligonucleotides containing either AAAAA, TTTTT, GGGGG, CCCCC or longer homopolymers were rejected.
RESULTS
Algorithm
Prior to running OligoArray 2.0, all transcribed sequences for an organism are saved in a file in FASTA format. These sequences can be mRNA sequences, CDSs or exon sequences, depending on which part of the sequence the search will be restricted to, but they should not be redundant. This file is used as an input file for the design and to format the BLAST database used by OligoArray 2.0 to compute oligonucleotide specificity. It is also possible to use only a subset of these sequences for design.
For each entry in the input file, the sequence length is measured. If this length is longer than the maximum distance accepted between the 5′ end of an oligonucleotide and the 3′ end of the input sequence, the sequence is shrunk from its 5′ part to get a final length equal to this maximum distance. Then, this sequence is masked for the presence of prohibited sequences, such as stretches of the same nucleotide, that may interfere with synthesis chemistry. All bases corresponding to these prohibited sequences are replaced by ‘N’ and, in the case of longer stretches, this substitution is extended to the end. Di- and tri-nucleotide repeats spanning more than 10 nt are also automatically masked.
To ensure the specificity of the oligonucleotide for its target, our approach consists first of detection of sequence similarity between the target and other sequences. The masked sequence is compared to all other sequences using the BLAST program (9) specially tuned for this task as follows. The ‘DUST’ filter is inactivated using the –F F option in order to consider all sequences excepted the one previously masked. The –S option is set to 1 to restrict the search to the plus strand only, the one that will produce labeled probes during reverse transcription. The word size for the BLAST search is set to the smaller value allowed (–W 7) to detect a maximum of sequence similarities. The options controlling the output from BLAST are modified to reduce the number of one-line descriptions (–v 5) and to increase the number of reported alignments (–b 10 000). Before starting to process any sequence, OligoArray 2.0 will compute the Expectation value necessary to report alignments longer than 13 nt as a function of the length of the query and the database (see the OligoArray 2.0 web site for more information: http://berry.engin.umich.edu/oligoarray2). Thus, for each sequence to be processed, this parameter (–e) will be set regarding the length of the query to ensure the reporting of short alignments. The BLAST output is parsed and a matrix will keep a record of the possible similarity between each position of the input sequence and other sequences.
The input sequence is read backwards from the 3′ end by using a moving window length equal to the minimal length of the oligonucleotide. This window sequence is first examined for the absence of prohibited sequences. Then, the percentage of G and C is compared to the range chosen by the user. If one of these two tests fails, the sequence window is moved iteratively by 5 nt to the 5′ end of the sequence. The next step consists of verifying the Tm of the oligonucleotide. This Tm is computed using the nearest neighbor (NN) model (10) with Na+ and DNA concentrations set to 1 M and 1 µM, respectively. If the Tm of the current oligonucleotide is outside the selected range, the program will try to adjust the size of the oligonucleotide to fit both the Tm and size ranges. If there is no successful combination, the sequence window will be moved backward by 1 nt to test the next oligonucleotide. Once a sequence fulfills these criteria, it is tested for the absence of secondary structure. The minimum free energies of all possible secondary structures are computed by using the MFOLD program (11) and thermodynamic parameters from SantaLucia (10), a Na+ concentration of 1 M and the temperature set by the user as a threshold. An oligonucleotide will be rejected if it presents a structure with a negative free energy at this temperature.
If all the previous tests are successfully passed, OligoArray 2.0 will compute the oligonucleotide specificity. If they exist, all similarities between the current oligonucleotide sequence and other sequences are retrieved from the similarity matrix and used to compute the thermodynamic values (Tm, free energy, enthalpy and entropy) of all possible hybridizations between the oligonucleotide sequence and the complementary strand of similar sequences. This computation is done by using the thermodynamic parameters from SantaLucia (10) included in the MFOLD package. It can either process perfect matches or mismatches between the two sequences. If there is no possible cross-hybridization with a Tm above the specificity threshold set by the user, the oligonucleotide is considered to be specific for its target and saved to the output file. If there is possible cross-hybridization, this data is saved in memory for further usage. If the number of specific oligonucleotides found is lower than the number of oligonucleotides required by the user or if none has been found, non-specific ones will be considered. The oligonucleotides with a lower number of putative cross-hybridizations will be reported first. In order to avoid any overlap between sequences and thus minimize competition between two oligonucleotides from the same gene during hybridization, OligoArray 2.0 can be tuned to have at least a minimum specified length separating two adjacent oligonucleotides. By default, this length is set to half the mean length of the oligonucleotide.
Implementation
OligoArray 2.0 is written in Java and was developed under Linux Red Hat 7.2 (http://www.redhat.com/software/linux/) using Java development kit 1.4 from Sun Microsystems (http://java.sun.com/j2se/1.4/index.html). It relies on two other programs, BLAST (9) (ftp://ftp.ncbi.nih.gov/blast/executables/) and MFOLD (11) (http://www.bioinfo.rpi.edu/~zukerm/rna/mfold-3.1.html), to achieve specificity computation and runs under Unix operating systems. The program is started from the command prompt and can be tuned by using 19 options. An extensive description of these options can be obtained with the –h option or from the web site (see the OligoArray 2.0 web site for more information). They control input and output file names (–d, i, o, r and R options), the maximum number of oligonucleotides to design per input sequence (–n), the ranges of length (–l and –L), of GC content (–p and –P) and of melting temperature (–t and –T) of these oligonucleotides, a threshold to reject oligonucleotide sequences that can fold to form stable secondary structures (–s), a threshold to start to consider cross-hybridization (–x), the number of sequences to process in parallel if more than one processor is available (–N) and the minimum distance between the 5′ ends of two adjacent oligonucleotides (–g). It is also possible to define a list of prohibited sequences that should not appear in the oligonucleotide sequence (–m). Such sequences can be stretches of the same nucleotide or any other kind of sequence, such as a restriction site pattern. Another option allows the user to set the maximum distance (–D) that can be accepted between the 5′ end of the oligonucleotide and the 3′ end of the input sequence. This last option is useful for a user who wants to prepare labeled probes using oligo(dT) priming and so design oligonucleotides close to mRNA poly(A) tails.
OligoArray 2.0 generates three output files. One is a log file, named OligoArray.log by default. It contains program status and a step by step analysis of the design process. This file can be used to understand why the design failed for a sequence or which parameter leads to the rejection of most of the oligonucleotides. The second file, named rejected.fas, contains all sequences for which OligoArray 2.0 was not able to design any oligonucleotides. These sequences are saved in a FastA format and are ready to be processed using more permissive parameters if necessary. The oligonucleotide data are saved in a third file named oligo.txt by default. Data are in a TAB delimited format easy to parse or to import into spreadsheet programs. For each oligonucleotide, the gene identifier is given first, followed by the position of the 5′ end of this oligonucleotide on the input sequence. Then its length, the free energy of the hybridization to its target at 37°C (kcal/mol), the enthalpy (kcal/mol), entropy (cal/kmol) and Tm (°C) of the duplex are reported. These thermodynamic data are followed by a list of target(s) for this oligonucleotide. If there is only one target, the oligonucleotide is considered to be specific and the gene identifier is reported alone. In other cases, the program will report first the identifier of the sequence it comes from and then a list of possible non-specific targets. For each cross-hybridization, we also report the free energy, the enthalpy, the entropy and the Tm of the hybridization between the current oligonucleotide and the non-specific target. The free energy is given for the temperature used as threshold for the specificity computation (–x option). The sequence of the oligonucleotide will close the line. The program reports the same strand as the one provided in the input file. So if the input file contains mRNA sequences, the sequences reported in the output file are the oligonucleotide sequences that should be on the microarray if a hybridization is planned with labeled cDNA.
The computation time will obviously depend on the number of sequences to be processed, the number of oligonucleotides required per sequence and the number of sequences in the BLAST database. To give an example, it takes from 4 to 12 h to design up to three 45mers per gene for most of the bacterial genomes on 1.2 GHz dual Xeon processors. For larger genomes, around 100 sequences can be processed per hour. The program binaries, but not yet the source codes, are available for non-profit use upon request from the authors.
Relation between oligonucleotide length, GC content and Tm
In the NN model, the Tm is computed using knowledge of the nucleotide sequence. This model assumes that the stability of a given base pair depends on the identity and orientation of neighboring base pairs. Thus, it is necessary to consider dimers of base pairs instead of a single base pair. Nevertheless, if we consider the average free energies of dimers (10) containing no GC pair (–0.82 ± 0.22 kcal/mol), one GC pair (–1.37 ± 0.09 kcal/mol) or two GC pairs (–2.08 ± 0.21 kcal/mol), we can approximate that the stability of a duplex will depend on the GC content of the sequence.
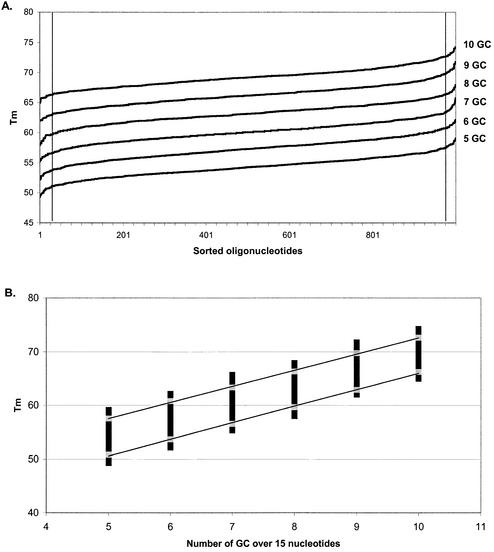
We have only considered the relationship between Tm and GC content. For a given oligonucleotide length and GC content, the melting temperatures will belong to a narrow range of values depending on the exact sequence composition. Since OligoArray 2.0 provides filters based on GC content, Tm and oligonucleotide length, it is very important to not use mutually exclusive parameters. In order to provide a tool to help OligoArray 2.0 users to choose appropriate parameters, we have investigated the relationship between the Tm and both the GC content and length of the oligonucleotide. For each oligonucleotide length comprising between 15 and 70 nt (increasing steps of 5 bases), we have generated random oligonucleotide sequences with a GC content in the 30–70% range. As an example, for a 15mer we have generated 1000 sequences containing five G or C (33% GC), 1000 sequences containing six G or C, up to 10 G or C (66% GC). In the same way, we have generated 29 000 70mer sequences with a number of G or C between 21 and 49. The Tm of each sequence was computed using the NN model for DNA and Na+ concentrations of 1 µM and 1 M, respectively. For each GC content, Tm values were sorted. As represented in Figure 1A for 15mer oligonucleotides, the Tm distribution is not linear at the two extremities. We have observed the same behavior for every oligonucleotide length and GC content. Since these low and high values are poorly representative of the set, we have chosen to filter out the 2.5% extreme temperatures in every set of Tm. These thresholds are represented by the black vertical line in Figure 1A.
Figure 1.
Distribution of melting temperature of 15mer oligonucleotides with various GC contents. For each GC content (from 5 to 10 G or C over 15 nt), 1000 random sequences have been generated and their Tm calculated and sorted. (A) For each GC content, the Tm distribution has been plotted. The two vertical lines visualize the position of the 26th and 974th Tm values and define the 2.5% of data excluded at each extremity of the curves (total 5%). (B) The Tm values from the same data set have been plotted as a function of the GC content of the oligonucleotide (number of GC over 15 nt). The gray squares represent the 26th and 974th Tm values from the distribution.
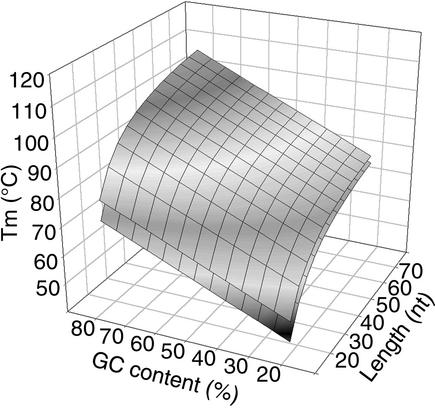
Figure 1B represents the melting temperatures of 15mers as a function of the number of GC present in the sequences. The gray squares represent the two extremes of the 95% of the Tm values remaining after the previously described filter was applied. For each oligonucleotide length we have fitted these values to a linear regression curve, for lower and higher Tm values separately as shown on the graph (r2 = 0.998 for each curve). The equations allow us to compute a theoretical Tm for each GC content between 20 and 80%, using an increment of 5%. We have plotted these theoretical Tm values as a function of the GC content and the oligonucleotide length (Fig. 2). In this way, we obtained two planes that define the upper and lower Tm that can be expected for a given oligonucleotide. The distance between the planes is only a function of the oligonucleotide length and does not vary with the GC content (data not shown). Any value in the space between these two planes is valid. We can use these data in a Java applet (see the OligoArray 2.0 web site for more information) to predict a Tm range as a function of the GC content and the oligonucleotide length. For given GC content and length ranges, this tool will report the narrowest and the widest expected Tm ranges. The narrowest Tm range is defined by the upper expected Tm for the lower content and length values and the lower expected Tm for the larger content and length values. In the same way, the widest Tm range is extended to the lower and higher expected Tm.
Figure 2.
Minimum and maximum melting temperatures as a function of the length and GC content of oligonucleotides. For each oligonucleotide length between 15 and 70, with an increment of 5 nt, and for each GC content between 20 and 80% of GC, the minimum and maximum valid Tm have been plotted. The upper plane defines the maximum Tm that can be computed for a given length and GC content. The lower plane defines the minimum Tm.
Design of an oligonucleotide set representing the whole A.thaliana transcriptome
To test the program, we used it to select oligonucleotides from the genome of the plant A.thaliana (26 178 transcripts; see Materials and Methods). Prior to running OligoArray 2.0, we have determined that the mean GC content of the input sequences is 42%, with a minimum and a maximum of 24 and 69%, respectively, and that 90% of these sequences have a GC content between 38 and 49%. We have used these data to set the OligoArray 2.0 parameters as described in Materials and Methods.
We have successfully designed 75 764 oligonucleotides representing 26 140 transcripts (2.9 oligonucleotides per input sequence on average). Among these 75 764 oligonucleotides, 69 122 are considered to be fully specific for their targets according to the design parameters and there is at least one specific oligonucleotide for 24 502 transcripts (93% of all input sequences). The design failed for 38 transcripts (0.15% of all input sequences). By analyzing the OligoArray.log file, we have determined that these failures were mostly due to short sequences containing prohibited sequences and/or a GC content >50%. These rejected sequences can be further processed using less stringent filters. All these data are available from the OligoArray 2.0 web site.
DISCUSSION
Although the algorithm used by OligoArray 2.0 is similar in some points to the previously described version of OligoArray (8), it shows major improvements. The oligonucleotide specificity is now computed by considering the sequence itself and the thermodynamic properties of its hybridization to targets. We expect, based on theoretical grounds, that this should be much more accurate than when this computation was only based on percentage and length of sequence similarities. This can particularly take into account short and highly GC-rich sequences leading to stable cross-hybridization at temperatures commonly used during hybridization and that may have improperly been selected by OligoArray 1.0. Another improvement concerns the oligonucleotide length. To achieve a better uniformity during hybridization, it is more important to have a narrow Tm distribution rather than a uniform oligonucleotide length. The new algorithm presented here gives more flexibility to adjust the sequence length by one or a few nucleotides to fit the narrowest Tm range.
We use the NN model to calculate the Tm of DNA duplexes using thermodynamic parameters obtained from DNA in solution. In microarray experiments, one strand of DNA is linked to the surface of the support. Such an interaction may modify the thermodynamics of the hybridization as previously suggested from hybridization experiments performed on microarrays of gel pads (12) and may lead to some error in Tm prediction. We are currently investigating the influence of microarray supports on the thermodynamics of the hybridization to determine if a correction need be applied to the NN model to predict hybridization Tm on chips.
The NN method is well adapted to compute the Tm of short sequences, but may lead to an overestimate of the Tm of sequences longer than 50 nt. On such long sequences we can expect some cross-hybridization involving only a shorter part of the sequence that will have a more accurately predicted Tm due to its shorter length. In the end, the difference in terms of real Tm between a long and perfect hybrid and a short non-perfect one may be lower than expected from predicted Tm and will lead to a lower specificity. Thus, we do not recommend the use of OligoArray 2.0 to design oligonucleotides longer than 50mer. This is not a major limitation. Indeed, Hughes et al. have reported that the optimal length for an in situ synthesized oligonucleotide on a chip is around 60 (2). Furthermore, Shchepinov et al. have shown that a spacer of length equivalent to 10mer–15mer is required to place the sequence sufficiently far from the microarray support to be fully available during hybridization (13). Taken together, these data suggest that of a 60mer oligonucleotide, no more than 50 bases are really involved in hybridization. In this case, the design can be restricted to a 45mer–50mer sequence than can be further elongated with a spacer. Reducing the oligonucleotide length will also enhance the oligonucleotide specificity by conferring a larger destabilizing effect on mismatches between the target and non-specific probes.
One of the first steps of sequence processing is to mask prohibited sequences selected by users. Masking is done by replacing unwanted bases by the same number of N. Then all oligonucleotides containing at least one N will be filtered out. One side-effect is that sequences containing some N before the masking step will also be filtered out. This may not be problematic since degenerate sequences are not desired for specific design.
Since OligoArray 2.0 selects only oligonucleotides that fulfill the user’s parameters, it may happen that the end of the input sequence is reached before finding the expected number of oligonucleotides. If some non-specific oligonucleotides exist, they are reported, but in the case of a short input sequence it may be impossible to design more than one or two oligonucleotides respecting the minimum distance allowed between two adjacent ones. In some cases, none is found and the corresponding input sequence is saved in a file. By searching for the identification number of that sequence in the log file and analyzing why each single tested oligonucleotide was rejected, it is possible to determine what the major origin of rejection was and then to run the software against this sequence using more adapted parameters.
There is no ranking of the oligonucleotides during the design, so the first reported is the closer one to the 3′ end of the input sequence. Further improvements will focus on output ranking correlated to experimental data. It is important to bear in mind that when random oligonucleotides are considered for anchoring reverse transcription, there is a better representation of the mRNA 5′ end. In this case, it will be better to pick oligonucleotides relatively far from the poly(A) tail. On the other hand, 3′ located oligonucleotides will be preferred if oligo(dT) priming is envisaged.
Preliminary data obtained from a set of 2500 oligonucleotides (45mer–47mer) designed with OligoArray 2.0 and representing 500 Saccharomyces cerevisiae genes show that we can successfully and reproducibly detect genes known to be expressed under the experimental conditions used. We have also obtained similar results with in vitro transcribed polyadenylated RNA spiked in endogenous total RNA before reverse transcription and probe labeling (Rouillard et al., in preparation).
There are few other algorithms described to design oligonucleotides for microarrays. ProbeSel (14) uses a suffix tree to search for sequence similarity to compute the thermodynamic parameters of the alignments, but this program lacks oligonucleotide self-folding computation. ProbeSelect (15) uses a suffix tree to search for sequence similarity and the myersgrep program to search for matching sequences with few mismatches. The possible secondary structures formed by oligonucleotides are predicted by searching for self-complementarity within the sequence. Relógio et al. propose a modified version of Gene Skipper (16). They exclude oligonucleotides showing only a perfect sequence identity with non-specific targets and do not consider possible mismatches. Wright and Church (17) propose an algorithm based on a BLAST search to define oligonucleotide specificity. They use a thermodynamic prediction of secondary structures based on RNA parameters. They also introduce an interesting concept to define oligonucleotide sequence complexity based on the Lempel-Ziv (LZ) compression algorithm. ArrayOligoSelector (18) uses a BLAST approach to search for sequence similarity and will compute the thermodynamic properties for only the most probable non-specific hybridization. This program also uses the LZ sequence complexity criteria. The last two programs are restricted to the design of 70mer oligonucleotides. Another strategy also focuses on the optimization of an oligonucleotide set for parallel oligonucleotide synthesis on chips (19).
OligoArray 2.0 only allows the design of oligonucleotides for use on microarrays for gene expression profiling. It can manage neither polymorphism nor splicing variants, except by considering each variant as a different input sequence. We are currently implementing new software specially devoted to the design of oligonucleotides for the detection of splicing variants of the same gene and for the identification of single nucleotide polymorphisms. We are also implementing an oligonucleotide database (Rouillard et al., in preparation) to provide pre-designed sets of oligonucleotides for transcriptome analysis for every sequenced genome available from public databases.
Acknowledgments
ACKNOWLEDGEMENTS
Financial support of this research by the Michigan Life Sciences Corridor funding grant no. MEDC-GR171 and DARPA contract no. N39998-01-C-7071 (Xeotron Prime Contractor) is gratefully acknowledged. M.Z. is supported, in part, by a grant from NIH, no. GM54250.
REFERENCES
- 1.Lipshutz R.J., Fodor,S.P., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nature Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 2.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 3.Gao X., LeProust,E., Zhang,H., Srivannavit,O., Gulari,E., Yu,P., Nishiguchi,C., Xiang,Q. and Zhou,X. (2001) A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res., 29, 4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto T., Suzuki,T. and Yamamoto,N. (2000) Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat. Biotechnol., 18, 438–441. [DOI] [PubMed] [Google Scholar]
- 6.Zammatteo N., Jeanmart,L., Hamels,S., Courtois,S., Louette,P., Hevesi,L. and Remacle,J. (2000) Comparison between different strategies of covalent attachment of DNA to glass surfaces to build DNA microarrays. Anal. Biochem., 280, 143–150. [DOI] [PubMed] [Google Scholar]
- 7.Kane M.D., Jatkoe,T.A., Stumpf,C.R., Lu,J., Thomas,J.D. and Madore,S.J. (2000) Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res., 28, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouillard J.M., Herbert,C.J. and Zuker,M. (2002) OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics, 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SantaLucia J. Jr, (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide, NATO ASI Series. Kluwer Academic Publishers, Dordrecht, NL.
- 12.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shchepinov M.S., Case-Green,S.C. and Southern,E.M. (1997) Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res., 25, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaderali L. and Schliep,A. (2002) Selecting signature oligonucleotides to identify organisms using DNA arrays. Bioinformatics, 18, 1340–1349. [DOI] [PubMed] [Google Scholar]
- 15.Li F. and Stormo,G.D. (2001) Selection of optimal DNA oligos for gene expression arrays. Bioinformatics, 17, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 16.Relogio A., Schwager,C., Richter,A., Ansorge,W. and Valcarcel,J. (2002) Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res., 30, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright M.A. and Church,G.M. (2002) An open-source oligomicroarray standard for human and mouse. Nat. Biotechnol., 20, 1082–1083. [DOI] [PubMed] [Google Scholar]
- 18.Bozdech Z., Zhu,J., Joachimiak,M.P., Cohen,F.E., Pulliam,B. and DeRisi,J.L. (2003) Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol., 4, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tolonen A.C., Albeanu,D.F., Corbett,J.F., Handley,H., Henson,C. and Malik,P. (2002) Optimized in situ construction of oligomers on an array surface. Nucleic Acids Res. 30, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]