Abstract
The interferon-induced 2′–5′-oligoadenylate synthetases (OAS) are important for the antiviral activity of interferons. The human and murine OAS gene families each contain four genes: OAS1, OAS2, OAS3 and OASL, all having one or more conserved OAS units composed of five translated exons. The OASL gene has both an OAS unit and a C-terminus of two ubiquitin-like repeats. In this study, we demonstrate that murine Oasl1 protein is inactive while murine Oasl2 is active as an OAS. Further more, murine Oasl2 requires double-stranded RNA as co-factor. The affinity of murine Oasl2 for the double-stranded RNA activator is higher than that of human OAS1 (p42 isoform). We propose a model for the evolutionary origin of the murine Oasl1 and Oasl2 genes. The identification of a human orthologue (hOASL2) to the murine Oasl2 gene establishes that the OASL gene was duplicated prior to the radiation of the rodent and primate groups. We suggest that murine Oasl2, which has both enzymatic activity and a ubiquitin-like domain, is a functional intermediate between the active OAS species and the inactive human OASL1/murine Oasl1 proteins. In addition, we propose that murine Oasl1 appears to have gained a hitherto uncharacterized function independent of 2′–5′-linked oligoadenylate synthesis.
INTRODUCTION
Interferons (IFNs) constitute an important part of the mammalian innate immune system (1). They confer resistance to viral infections by regulating the transcription of a large number of genes (2). The 2′–5′-oligoadenylate synthetases (OAS) were among the first characterised IFN-induced antiviral proteins (3–5). The OAS gene family is induced by both type I and type II IFNs. The OAS proteins are expressed as latent enzymes, which require double-stranded RNA (dsRNA) for activation; however, they do not harbour any of the characteristic dsRNA-binding motifs (6). This activation of the OAS enzymes results in the synthesis of 2′–5′-linked oligoadenylates (2–5As) from ATP (6). In turn, these 2–5As can bind to the latent RNase L which subsequently dimerises into the active form. The activated RNase L then degrades viral and cellular RNAs, suppressing protein synthesis and viral growth (7). Most viruses produce dsRNA at some stage in their life cycle, and it is generally believed that the activating dsRNA in infected cells is of viral origin (8). Furthermore, the OAS are involved in the induction of apoptosis and control of cell growth (9,10), and studies show that RNase L and the 2–5A system play a role in tumorigenesis (11).
In humans, the OAS gene family is composed of four genes located on chromosome 12 (6,12,13) The hOAS1, hOAS2 and hOAS3 genes are encoded by a tightly coupled locus on 12q24.1. The additional member of the OAS family in humans is the hOASL gene which is located on 12q24.2. Each OAS gene consists of a conserved OAS unit composed of five translated exons (exons A–E). OAS1 has one unit, whereas OAS2 and OAS3 have two and three units, respectively, and all three genes encode active 2–5A synthetases. The hOASL gene encodes a two-domain protein composed of an OAS unit fused to a 164 amino acid C-terminal domain, homologous to a tandem repeat of ubiquitin. In contrast to the different enzymes encoded by the hOAS1, hOAS2 and hOAS3 genes, no enzymatic activity has been detected for the hOASL protein (14,15).
Another member of the OASL class proteins was identified in chicken (16). The ChOASL protein is active upon induction by poly(I)·poly(C), and the identification of this protein divided the OASL proteins into two classes: the active and the inactive. We have recently described the structure of the murine Oas gene family which included two mOasl genes (mOasl1 and mOasl2) that exist head to tail on chromosome 5F (17,18).
Exons A and B of the OAS genes encode a five-stranded antiparallel β-sheet domain (19, R.Hartmann, unpublished). This domain is structurally homologous to the ‘palm’ domain found in other polymerases and contains a number of amino acids crucial for nucleotide transferase activity (20). The exon B of the OAS family contains a P-loop, which is a short flexible stretch of amino acids forming a helical turn; this motif is present in many nucleoside triphosphate-binding proteins such as kanamycin nucleotidyltransferase and different polymerases (20). The P-loop is involved in the binding of the triphosphate of the nucleotide. Mutational studies have demonstrated the importance of the P-loop in hOAS1 protein for catalytic activity (21). The second motif present in exon B is composed of three conserved aspartic acids in β-strands 2 and 5; these three aspartic acids coordinate the catalytically active magnesium ions (19,21). When considering the amino acid sequence of exon B encoded by mOasl1 and mOasl2 genes, notable differences exist. In mOasl1 protein, two of the three aspartic acids are replaced, and in hOASL1 all three are substituted. Furthermore, in both mOasl1 and hOASL1, several mutations are introduced into the P-loop motif, explaining why the hOASL1 protein is inactive as expected for mOasl1. In contrast, mOasl2 and ChOASL proteins have both motifs conserved, suggesting that the mOasl2 protein belongs to the class of active OASL proteins.
In this study, we have investigated the biochemical properties of mOasl1 and mOasl2 proteins. We purified recombinant protein for examination of the 2–5A synthetase activity and demonstrated that while mOasl1 has no activity, mOasl2 is an active 2–5A synthetase that requires dsRNA. Furthermore, we have determined the affinity of mOasl2 for poly(I)·poly(C) in comparison with hOAS1 (p42 isoform). We propose a model for the evolutionary origin of the mOasl1 and mOasl2 genes. Our data suggest that mOasl2 protein is a functional intermediate between the active hOAS and the inactive hOASL1/mOasl1 proteins. In addition, mOasl1 protein has gained several substitutions in exon B, resulting in loss of 2–5A synthetase activity, suggesting that mOasl1 may have obtained an novel function independent of 2–5A synthesis.
MATERIALS AND METHODS
RT–PCR for determination of IFN induction
Total RNA was purified from L929 (DSMZ: accession 2) cells using the Midi RNEasy purification kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The cells were either untreated, or treated with murine IFN-α (500 U/ml) or IFN-γ (100 U/ml) for 24 h. A 5 µg aliquot of total RNA was reverse-transcribed using the First strand cDNA synthesis kit (Amersham Biosciences, Piscataway, NJ). The PCR was carried out with 30 cycles of 96°C for 30 s, 60°C for 30 s, and 72°C for 2 min, resulting in a 1535 bp PCR product for mOasl1 and a 1526 bp product for mOasl2. The murine β-actin gene (GenBank accession no. M12481) was included as control. The mOasl and control reactions were mixed in equal amounts before gel electrophoresis (1% agarose).
Cloning, expression and purification of recombinant His-tagged protein
The IMAGE clone 1546490 (GenBank accession no. BE13926) was used as a template for mOasl1, and cDNA from IFN-α-induced L929 cells was used as template for mOasl2. The expression vectors (pET30b vector, Novagen, Madison, WI) were introduced into BL21 pRI cells by electroporation using standard conditions. Protein synthesis was induced by addition of 0.5 mM isopropyl-β-d- thiogalactoside (IPTG) overnight at room temperature. Cells were lysed in 25 ml lysis buffer [50 mM Na2HPO4 pH 8.0, 500 mM NaCl, 10% glycerol, 20 mM imidazole, 0.1% (v/v) NP-40, 5 mM β-mercaptoethanol and protease inhibitor cocktail without EDTA (Complete™, Roche Molecular Biochemicals, Indianapolis, IN)] using a French press. The lysate was clarified by centrifugation, and the supernatant was mixed with Ni2+-NTA–agarose beads (Qiagen) and rotated for 1 h. The beads were washed (pH 8.0, 50 mM Na2HPO4, 500 mM NaCl, 10% glycerol and 50 mM imidazole), applied to a column (Bio-Rad, Hercules, CA) and eluted with 15 ml of elution buffer (pH 8.0, 50 mM Na2HPO4, 500 mM NaCl, 10% glycerol, 250 mM imidazole) in 2 ml fractions. The fractions were analysed by 10% SDS–PAGE. The eluted His-tagged protein was loaded on a Heparin HiTrap (5 ml) column (Amersham Biosciences) (buffer A: 20 mM Tris–HCl pH 7.0, 5% glycerol). Bound protein was eluted in a linear gradient of an increasing concentration of NaCl (20–100%) (buffer B: 20 mM Tris–HCl pH 7.0, 5% glycerol, 1 M NaCl). The volume of the gradient was 10.054 ml. The fractions were collected in 0.5 ml and analysed by SDS–PAGE (10%). The eluted protein was dialysed for 2 h (600 mM NaCl, 25 mM HEPES pH 6.8, 5% glycerol), diluted 1:2 with cold 100% glycerol and stored at –20°C.
2′–5′-Oligoadenylate synthetase activity assay by PEI thin-layer chromatography
The activity of the recombinant proteins was determined as previously described (22) with the following overall reaction mixture: 4 µl of 5× OAS buffer, 4 µl of water, 4 µl of recombinant protein, 4 µl of ATP pH 7.5 including 0.025 mCi of [α-32P]ATP/µl, 4 µl of poly(I)·poly(C) (Amersham Biosciences) or water. The 5× OAS buffer contains 20 mM Tris–HCl pH 7.5, 75 mM Mg(OAc)2, 1 mM dithiothreitol, 0.2 mM EDTA, 0.5 mg/ml bovine serum albumin, 10% (v/v) glycerol, 30 mM creatine phosphate (Boehringer Mannheim GmbH) and 0.5 mg/ml creatine kinase (Boehringer Mannheim GmbH). The reactions were stopped with 10 µl of 50 mM EDTA after either 1 or 2 h. Separation of the ATP and the 2′–5′-oligomers was performed by thin-layer chromatography (TLC) as described previously (22). The TLC plate was visualised using a PhosphoImager™ (Molecular Dynamics, Sunnyvale, CA).
Activity assay using Mono Q to detect 2–5As
The incubations to make 2–5As were prepared in a similar way as the activity assay for TLC; however, the total volume was 100 µl. After incubation for 4 h, the OAS enzymes were heat-inactivated, treated with alkaline phosphatase (Boehringer Mannheim GmbH) and applied to a Mono Q HR5/5 column (Amersham Biosciences). The different 2′–5′-oligomers were eluted by a linear gradient of NaCl (0–70%) (buffer B: Tris–HCl pH 7.5, 700 mM NaCl) of 19.64 ml. The 2–5As were detected at 254 nm.
Determination of kinetic parameters
In order to estimate the kinetic parameters of both mOasl2 and hOAS1 (p42 isoform) enzymes, we used the ANEMONA.XLT which is an Excel template (23). This template implements among others the Michaelis–Menten model, and the KM and Vmax parameters are calculated by non-linear regression performed by the least-squares method. The kcat constant defines the number of substrate molecules (ATP) oligomerised per molecule of enzyme per second.
Western blotting
The His-tagged and heparin-purified protein was subjected to 10% SDS–PAGE and blotted onto polyvinylidene difluoride (Immobilon-P, Millipore) membranes. The purified, recombinant proteins were detected using the monoclonal mouse anti-His6 antibody (Boehringer Mannheim GmbH); monoclonal goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Dako) was used. The blots were visualised using the enhanced chemiluminescence method (ECL; Amersham Biosciences) according to the manufacturer’s instructions.
Alignment and phylogenetic analysis
The alignment of sequences was done at the amino acid level using default settings of the ClustalX 1.82 program, followed by manual adjustments. The following sequences were used: hOAS1 (D00068), hOASL1 (AJ225089), mOasl2 (AK010034), mOasl1 (AY089728), ChOASL (AB037592), Geodia cydonium OAS1 (Y18497) and a pseudogene sequence (hOASL2) from the human genome (NT_028327). Phylogenetic trees were reconstructed with and without the partial hOASL2 sequence, using the Geodia sequence as an outgroup, and the minimum evolution criterion for choosing the tree topology. Pairwise distances between sequences were calculated assuming rate heterogeneity in evolutionary rate modelled by a gamma distribution with shape parameter α = 1.5. Only alignment columns without gaps (complete deletion) were used. Standard bootstrap was done using 1000 replicates. For testing for differences in evolutionary rates between human and mouse OASL genes (24), a one-parameter version of the relative rate test was used employing either the hOAS1 sequence or ChOAS1 sequence as outgroup. All analyses were done using MEGA 2.1 (25).
RESULTS
A pseudogene corresponding to mOasl2 in humans
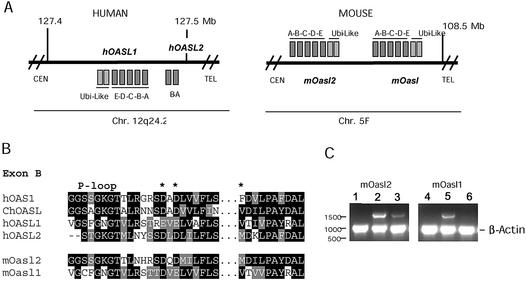
To date, only a single OASL gene has been identified in humans, in contrast to the two genes (mOasl1 and mOasl2) characterised in mice. Therefore, we searched the human genome for a counterpart of the mOasl2 gene. This was done by performing a BLAST search of the translated human genome with the amino acid sequence of mOasl2. Using this approach, we identified two exons (exons A and B) located downstream of the hOASL gene on chromosome 12 position 127.482 kb (exon A) and 127.476 kb (exon B) (Fig. 1A). These exons exhibit 54 and 53% identity to exons A and B in mOasl2, respectively, and their positions correspond to the homologous position of the mOasl2 gene. The N- and C-termini of exon B from human, mouse and chicken OASL proteins are aligned in Figure 1B, showing the important amino acids in the active site. As expected for a pseudogene, several stop codons are present in hOASL2 exon A, but no traces of the remaining sequences of the hOASL2 gene were found. We believe that the identified exons represent an hOASL2 pseudogene, which has not previously been described.
Figure 1.
(A) Schematic drawing of the hOASL and mOasl genes on human chromosome 12 and mouse chromosome 5. Genes with reading frames in the same direction as the annotated nucleotide sequence are indicated by squares above the lines; genes below are in the reverse direction. The genes are not drawn to scale. CEN, centromere; TEL, telomere. (B) Amino acid alignments of hOAS1 p42 (D00068), ChOASL (AB037592), hOASL1 (AJ22089), hOASL2 (NT_028327), mOasl1 (AY089728) and mOasl2 (AK010034) (numbers in parentheses indicate the GenBank accession no. for each protein). Black boxes indicate conserved residues, grey boxes related sequences. Gaps are indicated by –. The middle part of exon B has been cut out as indicated by black dots. The three catalytic aspartate residues are indicated by asterisks. (C) Transcription of mOasl2 (lanes 1–3) and mOasl1 (lanes 4–6) genes. Products of PCRs (30 cycles) with cDNA from untreated (lanes 1 and 4), IFN-α-induced (lanes 2 and 5) and IFN-γ-induced (lanes 3 and 6) mouse L929 cells, analysed by gel electrophoresis (1% agarose). The mOasl reaction and the control reaction (murine β-actin) were mixed in equal amounts before gel electrophoresis.
Interferon induction of mOasl1 and mOasl2
In order to investigate whether the mOasl genes are induced by IFNs, we extracted total RNA from IFN-α- and IFN-γ-induced and untreated mouse L929 cells. By RT–PCR studies, we show that mOasl1 and mOasl2 are induced by IFN-α in murine L929 cells; additionally, mOasl2 is induced by IFN-γ although to a lower extent (Fig. 1C). No transcript was detected in untreated L929 cells with the PCR conditions used here. We have no evidence of the p54/mOasl2 gene identified by Tiefenthaler et al. (26) (GenBank accession no. AF068835) in IFN-induced cells. This p54/mOasl2 variant has a shorter ubiquitin-like domain (1.5 ubiquitin-like repeat). A polymorphism giving allelic variation in the different mouse cells may explain the two forms of mOasl2. The following analysis is based solely on the mOasl2 gene with two ubiquitin-like repeats (GenBank accession no. AK010034).
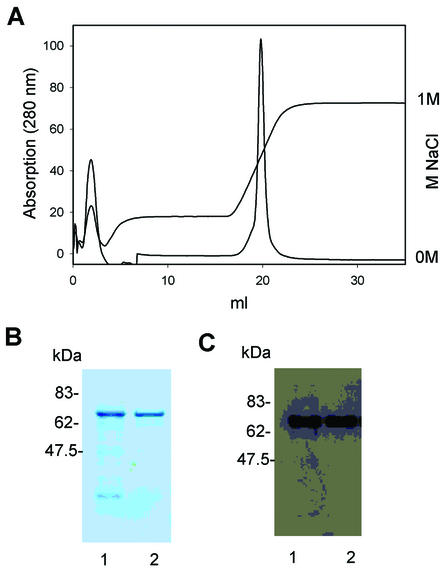
Purification of active murine Oasl2 protein
To explore the biochemical properties of mOasl2, we cloned the cDNA from IFN-α-induced L929 cells. N-terminal His-tagged protein was expressed in Escherichia coli at room temperature to avoid storage of protein in inclusion bodies. The first purification step was His tag purification using Ni2+-NTA beads. mOasl2 was purified further by affinity chromatography using a heparin column to remove several putative degradation products. The elution profile of the heparin column is shown in Figure 2A. A sample of the pooled His tag fractions is shown in Figure 2B (lane 1). This was loaded on the heparin column and then the eluted fractions containing the highest amount of protein were pooled (lane 2). The identity of the protein was verified by western blotting using an anti-His tag antibody (Fig. 2C). mOasl1 protein did not bind to the heparin; therefore, the purification was limited to nickel affinity, followed by dialysis removing the imidazole (data not shown).
Figure 2.
Purification of recombinant mOasl2 protein. (A) The elution profile of mOasl2 using the heparin–Sepharose column for purification. The graphs show the absorption at 280 nm and the NaCl gradient from 0 to 1 M. (B) A 10% SDS–polyacrylamide gel with 5 µM of pooled His tag fractions (lane 1) and 5 µM of pooled fractions from the heparin column (lane 2) was stained with Coomassie brilliant blue. The size of the molecular markers is indicated. (C) Western blot analysis of the samples loaded in (B). They were visualised using an anti-His tag antibody and a secondary goat anti-mouse HRP-conjugated antibody.
mOasl2 possesses true 2′–5′-oligoadenylate synthetase activity
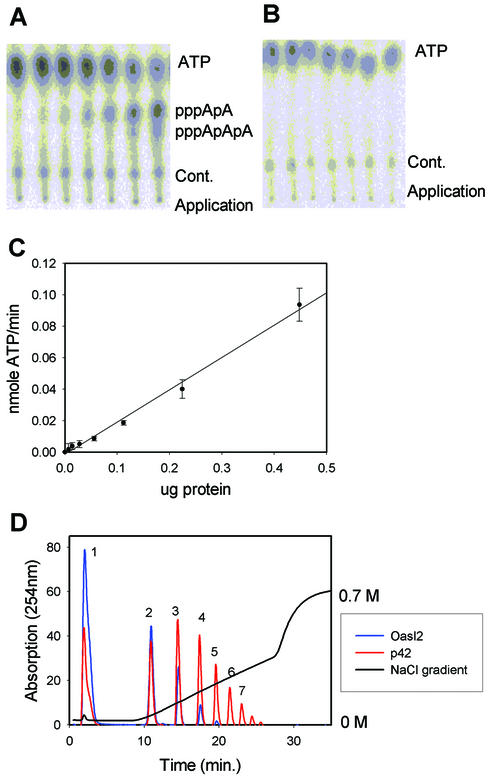
The putative OAS activities of mOasl1 and mOasl2 were tested by assays using synthetic dsRNA [poly(I)·poly(C)] as activator. In the following experiments, the total amount of 2–5A production was measured by TLC. As shown in Figure 3A, mOasl2 is highly active and the protein is enzymatically active after 22 h at 37°C (Fig. 3A and data not shown). As expected for an enzyme, a linear correlation exists between the activity and concentration of mOasl2 enzyme (Fig. 3C). In contrast, mOasl1 was unable to synthesize 2–5A (Fig. 3B) even after longer periods of incubation (22 h at 37°C), hence this protein possesses no 2–5A synthetase activity.
Figure 3.
mOasl2 is active whereas mOasl1 is not. (A) ATP was separated from the produced 2–5A products (pppApA and pppApApA) synthesised by mOasl2 using TLC. Increasing amounts of mOasl2 protein were added (0, 0.0007, 0.0014, 0.0028, 0.0056, 0.0112 and 0.0224 µg/µl) with a final concentration of 100 µg/ml poly(I)·poly(C). Cont., contaminations present in all reactions. (B) 2–5A synthetase activity assay using mOasl1 protein, and nucleotides separated by TLC. The same experimental conditions as in (A). (C) The amount of 2–5A produced by mOasl2 in (A) was quantified and normalised to the ATP spot. A linear correlation exists between the activity (nmol ATP/min) and the concentration of mOasl2 (µg). The experiment was repeated twice. (D) The heat-inactivated 2–5A synthetase reaction [final concentration of 100 µg/ml poly(I)·poly(C)] was treated with alkaline phosphatase and separated using a Mono Q column. The elution profiles of the products produced by mOasl2 (2.5 ng/µl) (blue) and p42 (2.5 ng/µl) (red) are superimposed. 1, adenylate; 2, pppApA; 3, pppA(pA)2; 4, pppA(pA)3; 5, pppA(pA)4; 6, pppA(pA)5; and 7, pppA(pA)6.
In order to verify that the products synthesised by mOasl2 indeed are 2–5A oligomers, we employed a second method. The reaction products were treated with alkaline phosphatase and the oligonucleotides were separated by an anion exchange column (Mono Q). The migration of the mOasl2 products was compared with standard 2–5As produced by hOAS1. The superimposed elution profiles of products synthesised by mOasl2 and hOAS1 were identical (Fig. 3D). mOasl2 is able to synthesise dimeric (pppApA), trimeric (pppApApA) and longer oligomers after an extended incubation time (22 h) (data not shown).
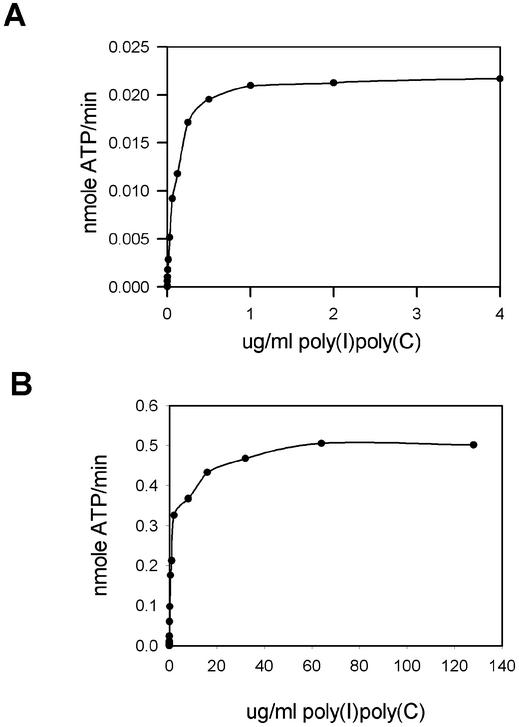
The activity of mOasl2 is dependent on dsRNA
In the previously described experiments, we have used the synthetic dsRNA poly(I)·poly(C) as an activator. Next, we investigated whether mOasl2, like all the human 2–5A synthetases, needs dsRNA as co-factor. Dose–response curves using poly(I)·poly(C) as activator of mOasl2 and hOAS1 are presented in Figure 4A and B, respectively. These curves clearly indicate that the 2–5A synthetase activity of mOasl2 is activated by increasing amounts of dsRNA and that without dsRNA, no 2–5A was produced. hOAS1 (p42 isoform) was included for comparison. Hence mOasl2 needs dsRNA/poly(I)·poly(C) as a cofactor for 2–5A synthetase activity.
Figure 4.
mOasl2 is induced by poly(I)·poly(C). (A) Dose–response curve for poly(I)·poly(C) induction of mOasl2 (2.5 ng/µl). (B) Dose–response curve for poly(I)·poly(C) induction of hOAS1 (2.5 ng/µl).
From the dose–response curves, we have estimated the maximal velocity (Vmax) and the apparent dissociation constant (Kapp) by non-linear curve fit. The Kapp defines the concentration of dsRNA activator, which induces 1/2 Vmax activity. The estimated parameters using either poly(I)· poly(C), poly(A)·poly(U) or poly(GU) are presented in Table 1. mOasl2 has a lower Kapp for poly(I)·poly(C) than hOAS1 (p42 isoform); consequently, our data suggest that mOasl2 has higher affinity for poly(I)·poly(C) than hOAS1. We continued the investigation of the dsRNA binding of mOasl2 by using poly(A)·poly(U) which has been shown to activate hOAS1, although to a lower extent than poly(I)· poly(C) (27). Our experiments show that mOasl2, on the contrary, is not activated by poly(A)·poly(U) to any extent. We also investigated whether poly(GU) can activate mOasl2 even though it does not activate hOAS1. Our studies show no activation of mOasl2 with poly(GU).
Table 1. Kapp for induction by different dsRNA.
| Kappa | mOasl2 | p42 |
|---|---|---|
| Poly(I)·poly(C) | 0.13 µg/ml | 1.40 µg/ml |
| Poly(A)·poly(U) | No activation | 25.00 µg/ml |
| Poly(GU) | No activation | No activation |
aThe apparent dissociation constant (Kapp) was estimated by a non-linear curve fit.
Determination of kinetic parameters [KM(ATP) and kcat]
We continued our biochemical analyses of mOasl2 by determination of the kinetic parameters. We plotted the amount of synthesized 2–5A as a function of the ATP substrate concentration. The Michaelis–Menten equation proposes a model for estimation of substrate affinity of an enzyme. By fitting our data to this equation, we were able to estimate the KM values as shown in Table 2. The KM for mOasl2 with ATP as substrate is 0.66 mM and is 0.31 mM for hOAS1.
Table 2. Kinetic parameters.
| mOasl2 | p42 | |
|---|---|---|
| KM(ATP) | 0.66 mM | 0.31 mM |
| kcat | 0.39 s–1 | 7.20 s–1 |
Phylogenetic analysis
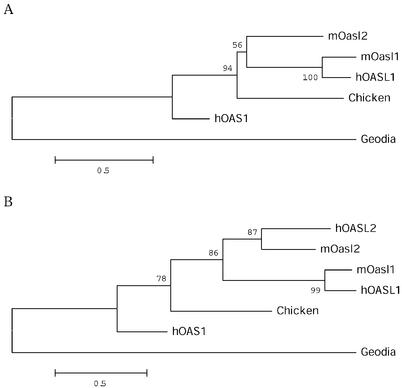
The identification of the hOASL2 pseudogene establishes that the duplication of the OASL gene has occurred prior to the radiation of the rodent and primate groups. However, subsequently during evolution, the hOASL2 gene has evolved into a pseudogene and, during this process, exons C–E and T have been lost and several stop codons have arisen in exon A. We performed a phylogenetic analysis by aligning mOasl1, mOasl2, human p42 (OAS1), hOASL1, hOASL2 (exon B only), ChOASL and the OAS gene from the marine sponge G.cydonium (28). In the subsequent phylogenetic analysis, the Geodia OAS sequence was used as an outgroup to root the trees.
Phylogenetic trees were constructed for the complete gene (Fig. 5A) and for exon B (Fig. 5B) only, using the minimum evolution criterion on a gamma distance matrix (α = 1.5). Analysis on exon B alone allowed us to include the hOASL2 psudogene. Figure 5B shows that mOasl1/hOASL1 and mOasl2/hOASL2 group together with high bootstrap support, suggesting that they indeed represent orthologous pairs of genes and that the duplication occurred prior to the diversification of human and mouse but before the diversification of mammals and birds. Bootstrap support of the shown topology was robust to other choices of α (between 1 and 5) and to other principles of phylogenetic reconstruction (including likelihood methods). The non-parametric relative rates test (24) revealed that mOasl1 has evolved significantly faster than mOasl2 since their divergence (45 versus 28 changes, P < 0.05). This agrees well with the fact that mOasl2 has retained activity and therefore has more common constraints with the ChOASL and hOAS1 sequences. Previous analysis of the OAS family made by Kumar et al. (29) showed that the evolutionary distance between hOASL1 and mOasl2 is longer than expected for two orthologous genes, which led them to suggest that the two genes were paralogues. This was later proven to be true by the identification of the mOasl1 gene. For exon B, the relative rate test suggests not surprisingly that the hOASL2 pseudogene has evolved faster than the mOASL2 sequence since their divergence, though not significantly so (15 versus 7 changes, P = 0.08).
Figure 5.
Phylogenetic analyses. (A) Phylogenetic tree of exons A–E. Bootstrap values are based on 1000 bootstrap replications. Sequences from mOasl2 (AK010034), mOasl1 (AY089725), hOAS1 p42 (D00068), hOASL (AJ22089), hOASL2 (NT_028327), ChOASL (AB037592) and Geodia OAS1 (Y18497) are aligned (numbers in parentheses indicate the GenBank accession no. for each protein). (B) Phylogenetic tree of exon B.
DISCUSSION
This study was performed to understand the biochemical properties of the two mOasl proteins. We have cloned and purified protein encoded by the mOasl1 and mOasl2 genes and characterised their 2–5A synthetase activity. Our results demonstrate that the mOasl2 protein is an active 2–5A synthetase requiring dsRNA as cofactor for activity. As expected from sequence comparisons, mOasl1 is inactive like its human orthologue, the hOASL1 protein (15). Thus it is tempting to speculate that mOasl1 is the functional counterpart of hOASL1.
The OASL proteins consist of an OAS domain and a C-terminal domain of two ubiquitin-like repeats. They have been identified in human, mouse and chicken, and thus we expect this family to exist throughout the mammalian and avian classes. The function of the ubiquitin-like domain of the OASL proteins is unknown. Mice have two mOasl genes, mOasl1 and mOasl2, with striking differences observed in the sequences of exon B of these genes. As described in the Introduction, the P-loop motif and a triad of aspartic acids coordinating the catalytic active magnesium ions are present in exon B. Both these motifs have been altered in the mOasl1 protein, whereas they are retained in the mOasl2 protein (see also Fig. 1B).
In comparison with hOAS1, the specific activity of mOasl2 is ∼20-fold lower; this could at least partly be due to evolution of the mOasl2 protein towards a 2–5A-independent function. The KM we have determined for mOasl2 is marginally higher than for human p42; however, it is still significantly lower than the KM for both human p69 OAS2 and p100 OAS3 proteins (30).
Previous workers have shown that mOasl2 protein only synthesises dimeric 2–5As (pppApA) (18) whereas we report that mOasl2 is able to synthesise up to pentamers, although the major product is dimeric 2–5As. We use creatine kinase and creatine phosphate in our OAS assays to regenerate ATP from ADP and AMP. Since ADP and AMP may be inhibitory to mOasl2, this can explain the lower activity observed in the experiments of Kakuta et al. (18). The activity of mOasl2 is more sensitive towards ADP and AMP inhibition than human p42 under the same experimental conditions (data not shown). Thus it is crucial to avoid degradation of ATP in the reaction when investigating the biochemistry of mOasl2. The assay system used here has the advantage that an eventual degradation of ATP is visualised directly on the chromatogram.
We have measured a higher affinity of mOasl2 for poly(I)·poly(C) compared with hOAS1. This supports published results from other groups showing that the hOAS1 protein in general requires the highest concentrations of dsRNA for activation among the OAS family members (30). The hOAS2 protein requires intermediate concentrations of dsRNA for activation, and the OAS3 proteins are the most sensitive to dsRNA (31). It should be noted that large batch to batch variations of commercially available poly(I)·poly(C) exist, making comparison of the absolute values for poly(I)·poly(C) difficult. In addition, mOasl2 was not activated by poly(A)·poly(U), in contrast to hOAS1 which was activated by this dsRNA although higher concentrations were required. This suggests that the dsRNA recognition mechanism might differ between the mOasl2 and hOAS1 proteins.
Previously published results by Tiefenthaler et al. claimed that the activity of mOasl2 was independent of dsRNA (26). We were unable to detect any activity of the mOasl2 protein in the absence of dsRNA. Furthermore, Tiefenthaler et al. did not observe any increase in the 2–5A synthetase activity upon stimulation with poly(I)·poly(C), whereas we observed a strong and dose-dependent activation of the mOasl2 enzyme by poly(I)·poly(C). The specific activity of the enzyme used by Tiefenthaler et al., although not stated in the paper, appears low, since they use 0.5 µg of protein in contrast to the 0.05 µg used here. The method we utilised for purification of recombinant mOasl2 protein is optimised for expressing the protein at low temperatures, which is essential for recovering active protein. However, at present, we are unable to explain the dsRNA-independent activity observed by Tiefenthaler et al. (26)
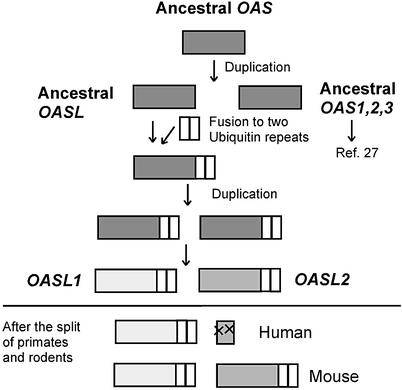
Based on our phylogenetic analyses, we propose a model for the evolutionary origin of the mOasl1 and mOasl2 genes. A duplication of the ancestral OAS gene followed by fusion to the ubiquitin-like domain by exon shuffling resulted in the formation of the original OASL gene (Fig. 6). This gene possesses both the ubiquitin-like domain and a catalytically active OAS domain, as is the case for both mOasl2 and ChOASL genes. Duplication of the ancestral OASL gene led to the precursors of mOasl1 and mOasl2. This duplication occurred either prior to the establishment of the mammalian class or early during mammalian evolution prior to the radiation of the rodent and primate groups, as demonstrated by the identification of an hOASL2 pseudogene. After the duplication of the ancestral OASL gene, mOasl2 has maintained the 2–5A synthetase activity, which is the putative original function of the OASL protein. This is manifested in the conservation of the catalytically important amino acids in the sequence of exon B. Some substitutions of amino acids have occurred outside exon B, explaining why mOasl2 is more closely related to the mOasl1/hOASL1 proteins when considering the entire OAS unit. The hOASL2 gene is not functional in humans, and only exons A and B remain as a pseudogene. Hence, at least in humans, the hOASL2 protein is non-essential. Thus we suggest that mOasl2 having both activity and an ubiquitin-like domain is a functional intermediate between the active hOAS proteins and the inactive hOASL1/mOasl1 proteins.
Figure 6.
Model for the evolutionary origin of the OASL1 and OASL2 genes.
In contrast, accumulations of amino acid substitutions in the mOasl1 gene led to loss of 2–5A synthetase activity. Since both hOASL1 and mOasl1 proteins are inactive, the loss of activity occurred in the common ancestor of man and mouse. Sequence comparisons and functional data demonstrate that the hOASL1 gene is an orthologue to the mOasl1 gene. The most common evolutionary fate of gene duplicates is silencing (32); since this does not seem to be the case for the mOasl1 gene, it appears that mOasl1 has acquired a putative novel function which is independent of 2–5A synthetase activity. Preliminary data suggest that this function requires the ubiquitin-like domain and that hOASL1 has an antiviral activity conferred by the ubiquitin-like domain (33).
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Lisbet Kjeldberg for excellent assistance with enzymatic measurements, Niels Ole Kjeldgaard for stimulating and strong interest, and Robert H. Silverman for critical reading of this manuscript. This work was supported by the Danish Natural Science Research Council, The Novo-Nordisk Foundation and the Danish Cancer Society.
REFERENCES
- 1.Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- 2.Der S.D., Zhou,A., Williams,B.R. and Silverman,R.H. (1998) Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl Acad. Sci. USA, 95, 15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts W.K., Hovanessian,A., Brown,R.E., Clemens,M.J. and Kerr,I.M (1976) Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature, 264, 477–480. [DOI] [PubMed] [Google Scholar]
- 4.Hovanessian A., Brown,R.E. and Kerr,I.M. (1977) Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature, 268, 537–540. [DOI] [PubMed] [Google Scholar]
- 5.Chebath J., Benech,P., Revel,M. and Vigneron,M. (1987) Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature, 330, 587–588. [DOI] [PubMed] [Google Scholar]
- 6.Justesen J., Hartmann,R. and Kjeldgaard,N.O. (2000) Gene structure and function of the 2′–5′-oligoadenylate synthetase family. Cell. Mol. Life Sci., 57, 1593–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong B. and Silverman,R.H. (1995) 2–5A-dependent RNase molecules dimerize during activation by 2–5A. J. Biol. Chem., 270, 4133–4137. [DOI] [PubMed] [Google Scholar]
- 8.Coccia E.M., Romeo,G., Nissim,A., Marziali,G., Albertini,R., Affabris,E., Battistini,A., Fiorucci,G., Orsatti,R. and Rossi,G.B. (1990) A full-length murine 2–5A synthetase cDNA transfected in NIH-3T3 cells impairs EMCV but not VSV replication. Virology, 179, 228–233. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh A., Sarkar,S.N., Rowe,T.M. and Sen,G.C. (2001) A specific isozyme of 2′–5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J. Biol. Chem., 276, 25447–25455. [DOI] [PubMed] [Google Scholar]
- 10.Castelli J.C., Hassel,B.A., Maran,A., Paranjape,J., Hewitt,J.A., Li,X.L., Hsu,Y.T., Silverman,R.H. and Youle,R.J. (1998) The role of 2′–5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ., 5, 313–320. [DOI] [PubMed] [Google Scholar]
- 11.Carpten J., Nupponen,N., Isaacs,S., Sood,R., Robbins,C., Xu,J., Faruque,M., Moses,T., Ewing,C., Gillanders,E. et al. (2002) Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nature Genet., 30, 181–184. [DOI] [PubMed] [Google Scholar]
- 12.Hovnanian A., Rebouillat,D., Mattei,M.G., Levy,E.R., Marie,I., Monaco,A.P. and Hovanessian.A.G. (1998) The human 2′,5′-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69- and 40-kDa forms. Genomics, 52, 267–277. [DOI] [PubMed] [Google Scholar]
- 13.Hovnanian A,, Rebouillat,D,, Levy,E,R,, Mattei,M,G. and Hovanessian,A.G. (1999) The human 2′,5′-oligoadenylate synthetase-like gene (OASL) encoding the interferon-induced 56-kDa protein maps to chromosome 12q24.2 in the proximity of the 2′,5′-OAS locus. Genomics, 56, 362–363. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann R., Olsen,H.S., Widder,S., Jorgensen,R. and Justesen,J. (1998) p59OASL, a 2′–5′ oligoadenylate synthetase like protein: a novel human gene related to the 2′–5′ oligoadenylate synthetase family. Nucleic Acids Res., 26, 4121–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebouillat D., Marie,I. and Hovanessian,A.G. (1998) Molecular cloning and characterization of two related and interferon-induced 56-kDa and 30-kDa proteins highly similar to 2′–5′ oligoadenylate. Eur. J. Biochem., 257, 319–330. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto A., Iwata,A., Koh,Y., Kawai,S., Murayama,S., Hamada,K., Maekawa,S., Ueda,S. and Sokawa,Y. (1998) Two types of chicken 2′,5′-oligoadenylate synthetase mRNA derived from alleles at a single locus. Biochim. Biophys Acta, 1395, 181–191. [DOI] [PubMed] [Google Scholar]
- 17.Eskildsen S., Hartmann,R., Kjeldgaard,N.O. and Justesen,J. (2002) Gene structure of the murine 2′–5′-oligoadenylate synthetase family. Cell. Mol. Life Sci., 59, 1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakuta S., Shibata,S. and Iwakura,Y. (2002) Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res., 22, 981–993. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar S.N., Ghosh,A., Wang,H.W., Sung,S.S. and Sen,G.C. (1999) The nature of the catalytic domain of 2′–5′-oligoadenylate synthetases. J. Biol. Chem., 274, 25535–25542. [DOI] [PubMed] [Google Scholar]
- 20.Holm L. and Sander,C. (1995) DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci., 20, 345–347. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto Y., Sono,D. and Sokawa,Y. (2000) Effects of specific mutations in active site motifs of 2′,5′-oligoadenylate synthetase on enzymatic activity. J. Interferon Cytokine Res., 20, 337–340. [DOI] [PubMed] [Google Scholar]
- 22.Justesen J., Ferbus,D. and Thang,M.N. (1980) 2′5′ oligoadenylate synthetase, an interferon induced enzyme: direct assay methods for the products, 2′5′ oligoadenylates and 2′5′ co-oligonucleotides. Nucleic Acids Res., 8, 3073–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernández A. and Ruiz,M.T. (1998) An EXCEL template for calculation of enzyme kinetic parameters by non-linear regression. Bioinformatics, 14, 227–228. [DOI] [PubMed] [Google Scholar]
- 24.Tajima F. (1993) Simple methods for testing the molecular evolutionary clock hypothesis. Genetics, 135, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S., Tamura,K., Jakobsen,I.B. and Nei,M. (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics, 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 26.Tiefenthaler M., Marksteiner,R., Neyer,S., Koch,F., Hofer,S., Schuler,G., Nussenzweig,M., Schneider,R. and Heufler,C. (1999) M1204, a novel 2′,5′ oligoadenylate synthetase with a ubiquitin-like extension, is induced during maturation of murine dendritic cells. J. Immunol., 163, 760–765. [PubMed] [Google Scholar]
- 27.Baglioni C., Minks,M.A. and De,Clercq (1981) Structural requirements of polynucleotides for the activation of (2′–5′)An polymerase and protein kinase. Nucleic Acids Res., 9, 4939–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiens M., Kuusksalu,A., Kelve,M. and Muller,W.E. (1999) Origin of the interferon-inducible (2′–5′)oligoadenylate synthetases: cloning of the (2′–5′)oligoadenylate synthetase from the marine sponge Geodia cydonium. FEBS Lett., 462, 12–18. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., Mitnik,C., Valente,G. and Smith,F. (2000) Expansion and molecular evolution of the interferon-induced 2′–5′ oligoadenylate synthetase gene family. Mol. Biol. Evol., 17, 738–750. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar S.N. and Sen,G.C. (1998) Production, purification and characterization of recombinant 2′, 5′-oligoadenylate synthetases. Methods, 15, 233–242. [DOI] [PubMed] [Google Scholar]
- 31.Marie I., Blanco,J., Rebouillat,D. and Hovanessian,A.G. (1997) 69-kDa and 100-kDa isoforms of interferon-induced (2′–5′)oligoadenylate synthetase exhibit differential catalytic parameters. Eur. J. Biochem., 248, 558–566. [DOI] [PubMed] [Google Scholar]
- 32.Lynch M. and Conery,J.S. (2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann R., Rebouillat,D., Justesen,J., Sen,S. and Williams,B. (2001) The p59 oligoadenylate synthetase like protein (p59OASL) does not display oligoadenylate synthetase activity but possesses antiviral properties conferred by an ubiquitin-like domain. J. Interferon Cytokine Res., 9 (suppl.), 6–8. [Google Scholar]