Abstract
An enzyme capable of liberating functional tRNALys from Escherichia coli diacetyl-lysyl-tRNALys was purified from the archae Sulfolobus solfataricus. Contrasting with the specificity of peptidyl- tRNA hydrolase (PTH) from E.coli, the S.solfataricus enzyme readily accepts E.coli formyl-methionyl-tRNAfMet as a substrate. N-terminal sequencing of this enzyme identifies a gene that has homologs in the whole archaeal kingdom. Involvement of this gene (SS00175) in the recycling of peptidyl-tRNA is supported by its capacity to complement an E.coli strain lacking PTH activity. The archaeal gene, the product of which appears markedly different from bacterial PTHs, also has homologs in all the available eukaryal genomes. Since most of the eukaryotes already display a bacterial-like PTH gene, this observation suggests the occurrence in many eukaryotes of two distinct PTH activities, either of a bacterial or of an archaeal type. Indeed, the bacterial- and archaeal-like genes encoding the two full-length PTHs of Saccharomyces cerevisiae, YHR189w and YBL057c, respectively, can each rescue the growth of an E.coli strain lacking endogeneous PTH. In vitro assays confirm that the two enzymes ensure the recycling of tRNALys from diacetyl-lysyl-tRNALys. Finally, the growth of yeast cells in which either YHR189w or YBL057c has been disrupted was compared under various culture conditions. Evidence is presented that YHR189w, the gene encoding a bacterial-like PTH, should be involved in mitochondrial function.
INTRODUCTION
Ribosomal translation of mRNA open reading frames (ORFs) normally terminates on a stop codon. At this step, several release factors collaborate with ribosome to liberate a newly synthesized polypeptide [for a review, see Kisselev and Buckingham (1)]. However, during elongation of translation, peptidyl-tRNA molecules may prematurely dissociate from the mRNA template. Such abortive events result in accumulation of peptidyl-tRNAs that are left apart from the translation machinery (2). It was recognized early on that Escherichia coli and yeast cells possessed a hydrolytic activity capable of recycling such peptidyl-tRNA molecules by removing the peptidyl moiety (3–7). This activity was called peptidyl-tRNA hydrolase (PTH). The E.coli PTH was characterized in depth (5,8–12). Provided they are N-blocked, aminoacyl-tRNAs are substrates of the hydrolase. However, formyl-methionyl-tRNAfMet appears to be a relatively poor substrate. Resistance to PTH enables the formylated initiator tRNA to participate in the formation of the ribosomal initiation complex. With elongator tRNAs, the action of the hydrolase is facilitated by the 5′-phosphate at the end of a fully base-paired acceptor stem. Escherichia coli tRNAfMet lacks base pairing at position 1–72. The resulting mismatch at the top of the acceptor helix is believed to twist the position of the phosphate group so that it can no longer trigger the activity of the enzyme.
The gene encoding PTH was first identified in E.coli (13). It is essential to the survival of the bacterium (11,14). Homologs of this gene (pth) were then recognized in the genomes of all bacteria and in those of most eukarya. In Saccharomyces cerevisiae, the homolog, YHR189w, encodes a PTH capable of conferring thermoresistance to a pthts E.coli strain (15). Surprisingly, however, analyses of completed archaeal genome sequences did not reveal any pth homolog. To examine the possibility that archaea use an alternative enzyme to ensure the recycling of peptidyl-tRNA, we took Sulfolobus solfataricus as starting material. A similar search using Methanocaldococcus jannaschii has been reported very recently (16). The latter work resulted in the identification of a PTH activity encoded by the MJ0051 gene. The product of this gene is markedly distinct from the E.coli PTH and the yeast YHR189w gene product. However, its expression in E.coli confers thermoresistance to a pthts strain. Interestingly, the MJ0051 gene has homologs in all archaeal and eukaryal genomes. The product of YBL057c, the ortholog of MJ0051 in S.cerevisiae, cures the thermosensitivity of an E.coli pthts strain. In this case, to obtain complementation, a large part of YBL057c had to be deleted on the N-terminal side of the protein product (16).
Here, we isolate PTH activity from S.solfataricus crude extracts and identify the corresponding gene. This gene, which is homologous to the MJ0051 gene, could be over-expressed in an E.coli context. Comparison of the biochemical properties of purified S.solfataricus PTH with those of E.coli PTH indicates distinct mechanisms of substrate recognition. In particular, S.solfataricus PTH is only weakly sensitive to the presence of a 5′-phosphate at the top of the acceptor helix of tRNA. As a consequence, the enzyme readily accepts E.coli formyl-methionyl-tRNAfMet as a substrate. This behavior can be related to the special feature of translation in archae, where formylation of methionine esterified to initiator tRNA does not happen. In the second part of the present study, we examine various culture conditions in the hope of revealing phenotypes of S.cerevisiae haploid strains defective in either YHR189w or YBL057c. Finally, YHR189w disruption is shown to impair the growth of cells on minimal medium containing a non-fermentable carbon source.
MATERIALS AND METHODS
Isolation and N-terminal sequencing of peptidyl-tRNA hydrolase from S.solfataricus
Cells from S.solfataricus strain P2 (DSM1617) were grown at 78°C essentially as in Zillig et al. (17). The culture (3.5 l) was quickly cooled and centrifuged for 10 min at 4000 g. All buffers used for the purification of PTH contained 10 mM 2-mercaptoethanol and 1 mM EDTA. The cell pellet (21 g, wet weight) was suspended in 100 ml of 50 mM potassium phosphate buffer pH 7.6 containing 0.1 mM dithiothreitol (DTT), 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 µg/ml of each leupeptin, pepstatin A and aprotinin. Cells were disrupted by sonication (10 min, 0°C), and debris removed by centrifugation (60 min, 10 000 g). Nucleic acids were precipitated by addition of streptomycin (30 g/l) to the supernatant, which was then centrifuged for 60 min at 10 000 g. The resulting supernatant was brought to 70% ammonium sulfate saturation. After centrifugation for 30 min at 10 000 g, the protein pellet was dissolved in 15 ml of 20 mM potassium phosphate buffer pH 7.6 containing 0.1 mM PMSF, and dialyzed against 1 l of the same buffer. The resulting solution was applied on a column of Q-Sepharose FastFlow (4.4 × 30 cm; Amersham Biosciences) equilibrated in 20 mM potassium phosphate pH 7.6. The major part of the PTH activity was recovered in the flowthrough and further applied on an SP-Sepharose column (1.1 × 10 cm; Amersham Biosciences) equilibrated in 20 mM potassium phosphate pH 7.6. Elution was carried out at a flow rate of 1 ml/min with a 0.1 l linear gradient of 0–500 mM KCl in the same buffer (200 mM/h). Fractions of 2 ml were collected. PTH activity was recovered at 190 mM KCl. This activity was associated with a material showing a strong light absorbancy at 260 nm. After concentration by ultrafiltration on a Ultrafree 4 centrifugal filter unit 10 K (Millipore), the sample (2 ml) was applied onto a Superdex 75 column (120 ml, 1.6 × 60 cm; Amersham Biosciences) equilibrated in 20 mM potassium phosphate pH 7.6 containing 500 mM KCl and eluted at a flow rate of 0.14 ml/min. PTH activity was recovered at 520 min. Fractions (0.9 ml each) containing activity were pooled and dialyzed against 20 mM potassium phosphate pH 7.6. After addition of 1.2 M ammonium sulfate, the sample (7.4 ml) was applied on a Widepore HI-Propyl column (0.5 × 5.5 cm, particle size 15 µm; Baker) equilibrated in 20 mM potassium phosphate buffer pH 7.6 containing 1.7 M ammonium sulfate. Elution was carried out at a flow rate of 0.15 ml/min with a linear gradient of 1.7–0 M ammonium sulfate (1 M/h). Fractions of 0.2 ml were collected. Enzyme activity free of contaminating nucleic acids was recovered at 95 min. Fractions containing PTH were pooled (1.8 ml) and dialyzed against 20 mM potassium phosphate pH 7.6 containing 60% glycerol, and stored at –20°C. As judged from an SDS–PAGE analysis, the recovered protein (∼10 µg) was at least 90% pure.
An aliquot of purified PTH (1 µg) was electrophoresed on an SDS–polyacrylamide gel. After migration, proteins were transferred on a ProBlot membrane (Applied Biosystems) and stained with amido black. The major protein band, corresponding to an Mr of 15 000 Da, was submitted to 10 cycles of Edman degradation on an Applied Biosystems Procise Sequencer.
Cloning of peptidyl-tRNA hydrolase genes
The SS00175 gene was amplified by PCR using S.solfataricus genomic DNA as template, and oligonucleotides CTGCATCATGCCATGGTTAAGATGGT and CGCGGATCCTCACAGTAATTTT as primers. The resulting DNA fragment was purified using the Qiagen PCR Purification Kit 50, digested by both BamHI and NcoI, and inserted into the corresponding sites of plasmid pTrc99-A to give plasmid pTrc-pthS. The YBL057c gene was amplified from genomic DNA of S.cerevisiae strain YPALS (18), with oligonucleotides CCGAATTCTATGATAACGTCCTTTTTAATGGAAAAGATGACAG and CCAGCCAAGCTTCAATACAA TTTCAAATCACCTGTTATTTGATCC as primers. The resulting DNA fragment was purified, digested by both EcoRI and HindIII, and inserted into the corresponding sites of plasmid pKK223-3 to give plasmid pKKpthY2. The YHR189w gene was amplified from genomic DNA of S.cerevisiae strain YPALS, with oligonucleotides CCATCGATTCTAGAAAGGAGGTACGATCATGTCCGGTAAATGGAGACT and CGCGGATCCCTATGAAATGTAC TGAGTCAGAGCACG as primers. The resulting DNA fragment was purified, digested by both BamHI and ClaI, and inserted into the corresponding sites of plasmid pBluescript(+)KS to give plasmid pBSpthY1. In the three cases, the cloned DNAs were verified by DNA sequencing.
To obtain complementation of an S.cerevisiae strain where the YHR189w gene is disrupted, the HindIII–XbaI polylinker in the expression vector pYES2 (Invitrogen) was replaced by the TCTAGACCCGGGCTCGAGGGTACCCCGCGGGCGGCCGCGTCGAC sequence. The resulting plasmid was called pYES2lpa. Then, the 633 bp XhoI–NotI fragment of plasmid pBSpthY1 was inserted between the corresponding sites of pYES2lpa, to give plasmid pYESpthY1.
Preparation of crude extracts for activity measurements
When PTH activity was assayed in crude E.coli extracts, bacteria were grown overnight in 25 ml of 2× TY (1.6% bacto-tryptone, 1% bacto-yeast extract, 0.5% NaCl) medium containing 10 µg/ml of ampicillin. In the case of strain K37ΔpthTr(pBSpthY1), the growth medium also contained 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After centrifugation at 8000 g for 10 min, bacteria were suspended in 10 mM Tris–HCl pH 7.5 containing 0.1 mM EDTA, 0.1 mM PMSF and 10 mM 2-mercaptoethanol, so that the optical density (OD) of the bacterial suspension was equal to 100 at 650 nm. Cells were disrupted by sonication (3 min, 0°C) and debris removed by centrifugation (15 min, 18 000 g). When the assayed enzyme was the S.solfataricus PTH, the extract was heated at 80°C for 15 min and centrifuged at 18 000 g for 15 min. Total amounts of protein in the extracts were determined by the Bio-Rad protein assay, with bovine serum albumin as standard.
Purification of S.solfataricus peptidyl-tRNA hydrolase using expression in an E.coli strain
Escherichia coli XL1-Blue (Table 1) transformed with plasmid pTrc-pthS was grown at 37°C in 1 l of 2× TY medium containing 80 µg of ampicillin/ml. When the OD of the culture reached 1 at 650 nm, 0.3 mM IPTG was added, and growth was continued for 4 h. Cells were harvested by centrifugation for 30 min at 4000 g. All buffers used for the purification of PTH contained 10 mM 2-mercaptoethanol and 0.1 mM EDTA. The cell pellet was suspended in 50 ml of 50 mM Tris–HCl pH 7.0 containing 0.1 mM PMSF. Cells were disrupted by sonication (3 min, 0°C), and debris removed by centrifugation (60 min, 10 000 g). The resulting extract was centrifuged at 100 000 g for 90 min. The supernatant was heated at 80°C for 10 min and centrifuged at 10 000 g for 30 min. The resulting supernatant was brought to 70% ammonium sulfate saturation. After centrifugation for 30 min at 10 000 g, the pellet was dissolved in 20 mM Tris–HCl buffer pH 7.0 and dialyzed overnight against the same buffer. The resulting sample (1.6 ml) was successively chromatographed on a Superdex 75 column and a HI-Propyl column, as described above in the case of the enzyme purified from S.solfataricus cells. In the elution profile of the HI-Propyl chromatography, the peak of PTH activity was free of contaminating nucleic acids. Active fractions recovered from the HI-Propyl column (1.8 ml) were pooled, dialyzed against 20 mM Tris–HCl buffer pH 7.0 and applied on an SP-Sepharose column (0.6 × 5.5 cm) equilibrated in the same buffer. This column was eluted with a linear gradient from 0 to 500 mM KCl in the buffer of the column (0.3 ml/min, 200 mM/h). Fractions of 0.45 ml were collected. The peak of activity was recovered at 300 mM KCl. In the course of the isolation of PTH from a S.solfataricus extract, the KCl concentration causing elution of activity on the same column was lower. In that case, contaminating nucleic acids may be at the origin of a biased effect of KCl during elution.
Table 1. The E.coli and S.cerevisiae strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| E.coli | ||
| K37 | galK rpsL | (40) |
| K37ΔpthTr | galK rpsL Δpth::kan (pMAK705 pth) | (11) |
| XL1-Blue | endA1 hsdR17 supE thi-1 recA1 gyrA96 relA1 lac (F′ proAB lacIq lacZΔM15 Tn10) | Stratagene |
| S.cerevisiae | ||
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| Y03083 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YBL057c::kanMX4 | Euroscarf |
| Y12883 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YHR189w::kanMX4 | Euroscarf |
The SP-Sepharose column procedure produced 0.26 mg of protein in 5.6 ml. According to an SDS–PAGE analysis, the obtained enzyme was homogeneous. All steps of the purification procedure are summarized in Table 2. Concentration of purified PTH was determined using a light absorption coefficient (1.269 A280 U/mg/ml) and a molecular ratio (2 × 13 139) calculated from the amino acid sequence of the protein.
Table 2. Purification of S.solfataricus PTH from E.coli strain XL1-Blue(pTrc-pthS).
| Purification step | Protein yield (mg) | Total activity (U)a | Specific activity (U/mg) | Yield (%) | Relative purification |
|---|---|---|---|---|---|
| Extract | 170b | 56 000 | 330 | 100 | 1 |
| Supernatant after ultracentrifugation | 140b | 54 000 | 390 | 96 | 1.2 |
| Supernatant after heat treatment | 7.8b | 51 000 | 6500 | 91 | 20 |
| Supernatant after ammonium sulfate precipitation | 5.5b | 45 000 | 8200 | 80 | 25 |
| Superdex 75 | 1.3b | 34 000 | 26 000 | 61 | 79 |
| HI-Propyl | 0.56b | 26 000 | 46 000 | 46 | 140 |
| SP-Sepharose | 0.26c | 18 000 | 69 000 | 32 | 210 |
a1 U is defined as the amount of enzyme capable of deacylating 1 pmol of diacetyl-[14C]lysyl-tRNALys per second in the standard assay conditions (see Materials and Methods).
bProtein analysis by the Bio-Rad protein assay, with bovine serum albumin as standard.
cProtein amount determined from UV absorbancy, assuming that 1 A280 unit corresponds to a protein concentration of 0.788 mg/ml.
Assay of peptidyl-tRNA hydrolase activity
Escherichia coli tRNALys, tRNAHis, tRNAfMet and tRNATyr were prepared as described previously (11,12,19). These tRNAs were fully aminoacylated with l-[14C]lysine (50 or 317 Ci/mol), l-[14C]histidine (50 Ci/mol), l-[14C]methionine (57.9 Ci/mol) and d-[3H]tyrosine (500 Ci/mol), respectively (11,12,19). l-Lysyl-tRNALys and l-histidyl-tRNAHis were then acetylated with acetic anhydride (11). Methionyl-tRNAfMet was formylated in the presence of E.coli methionyl-tRNAfMet formyltransferase and 10-formyltetrahydrofolate (12). Dephosphorylated diacetyl-[14C]lysyl-tRNALys (50 Ci/mol) and formyl-[14C]methionyl-tRNAfMet (57.9 Ci/mol) were obtained as described previously (11,12).
Unless otherwise stated, measurements of S.solfataricus PTH activity were performed at 50°C in 100 µl assays containing 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, 0.75 µM diacetyl-[14C]lysyl-tRNALys (50 Ci/mol) and catalytic amounts of enzyme (11). The reaction was quenched by the addition of 340 µl of ethanol, 14 µl of sodium acetate (3 M, pH 4.8) and 20 µl of carrier RNA from yeast (4 mg/ml). Samples were then centrifuged. Soluble radioactivity in the supernatant was measured by scintillation counting, as described (10). Data were corrected for spontaneous hydrolysis of the substrate in the absence of PTH. One unit corresponded to the enzyme activity capable of deacylating 1 pmol of diacetyl-[14C]lysyl-tRNALys per second in the above conditions.
Km and kcat values were deduced from measurements of the initial rate of hydrolysis in the presence of a variable concentration of diacetyl-lysyl-tRNALys (8–150 nM). To estimate the inhibition constant (KI) associated with tRNALys binding, initial rates of hydrolysis were measured in the presence of variable concentrations of tRNALys (0–20 µM) and diacetyl-[14C]lysyl-tRNALys (0.2–2 µM). The data verified competitive inhibition by tRNALys of diacetyl-lysyl-tRNALys hydrolysis. To obtain Michaelian parameters associated with other N-blocked aminoacyl-tRNAs, initial rates of hydrolysis were measured at a fixed substrate concentration (∼0.75 µM) in the presence of a variable concentration of tRNALys (0–20 µM). Km values for each studied substrate could then be deduced by assuming competitive inhibition by tRNALys. To estimate KI values associated with the other nucleic acids, initial rates of diacetyl-[14C]lysyl-tRNALys (0.2 µM) hydrolysis were assayed in the presence of either E.coli tRNAfMet (0–20 µM), unfractionated E.coli tRNA (0–20 µM), E.coli rRNA (0–0.8 mg/ml; Roche) or herring sperm DNA (0–0.8 mg/ml; Sigma).
Km, kcat and KI values were derived from iterative non-linear fits of the theoretical Michaelis equation to the experimental values, using the Levenberg–Marquardt algorithm (20).
Aminoacylation of tRNALys produced through enzymatic hydrolysis of diacetyl-[3H]lysyl-tRNALys
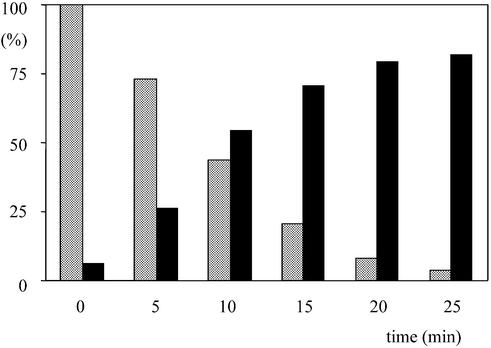
Escherichia coli tRNALys was aminoacylated with l-[3H]lysine (320 Ci/mol), then acetylated with acetic anhydride (11), and treated with sodium periodate as described previously (21). A 130 pmol concentration of the resulting diacetyl-l-[3H]lysyl-tRNALys sample was incubated in 650 µl at 50°C in the presence of 0.12 U of S.solfataricus PTH. At various times (Fig. 1), two 50 µl aliquots were simultaneously withdrawn. The first aliquot was mixed with 1 ml of 5% (w/w) trichloracetic acid (TCA) and 20 µl of carrier RNA from yeast. The second one was diluted in a reaction mixture (50 µl) containing 40 mM Tris–HCl pH 7.6, 19 µM [14C]lysine (317 Ci/mol), 4 mM ATP, 0.1 mM EDTA, 0.1 mM DTT and 2 µM of pure E.coli lysyl-tRNA synthetase (22). After 2 min incubation at 28°C, 1 ml of 5 % (w/w) TCA and 20 µl of carrier RNA from yeast were added to the second sample. Insoluble radioactivity (3H in the first sample, 3H and 14C in the second one) was measured by scintillation counting, as described (23). It was systematically verified that 3H radioactivity was the same in each pair of samples, thus showing that diacetyl-l-[3H]lysyl-tRNALys did not vary upon incubation in the presence of lysyl-tRNA synthetase.
Figure 1.
The SS000175 gene product behaves as a PTH. Diacetyl-l-[3H]lysyl-tRNALys was incubated at 50°C in the presence of catalytic amounts of S.solfataricus PTH. At various times, aliquots were withdrawn and incubated for 2 min with [14C]lysine, ATP and excess pure E.coli lysyl-tRNA synthetase (see Materials and Methods). After quenching of the reaction, TCA-precipitable 3H (open bars) and 14C (closed bars) radioactivities were measured by scintillation counting. Values were normalized to 100% with respect to TCA-precipitable 3H radioactivity at time zero.
Yeast strains and growth media
The S.cerevisiae strains used in this work are summarized in Table 1. Rich media contained 1% peptone and 1% yeast extract (Difco) with either 2% glucose or 2% glycerol. Minimal media contained 0.67% yeast nitrogen base (Difco) with either 2% glucose or 2% glycerol. Required amino acids and uracil were added at final concentrations of 50 µg/ml each. Induction of YHR189w gene expression from the pYESpthY1 plasmid was obtained by adding 0.1% galactose to the medium.
RESULTS AND DISCUSSION
Identification of the SS000175 gene and characterization of its product
Upon assaying PTH activity in an S.solfataricus cell extract with diacetyl-[14C]lysyl-tRNALys as the substrate, dissociation of radioactivity from tRNA could be shown. At 50°C, the reaction occurred at a rate of 8 U/mg of protein in the extract. This assay enabled us to undertake isolation of the archaeal PTH activity through successive chromatographies on Q-Sepharose, SP-Sepharose, Superdex 75 and HI-Propyl columns. At the end of the purification procedure, the PTH specific activity was enriched ∼8500-fold if compared with the activity in the crude extract. A nearly homogeneous protein with a 15 ± 2 kDa molecular weight was obtained, as judged by SDS–PAGE analysis. Edman degradation of this protein indicated an N-terminal sequence, MIKMVIVVRS, that unambiguously designated the ORF SS00175 in the S.solfataricus genome. This ORF encodes a short 120 residue polypeptide with a Mw of 13.1 kDa. It belongs to the family of archaeal PTH homologs, as defined by Rosas-Sandoval et al. on the basis of the MJ0051 gene encoding PTH activity in M.jannaschii (16).
To assess whether the SS00175 ORF actually encoded a protein with a PTH activity, this gene was amplified by PCR and inserted into the expression vector pTrc99-A under control of the trc promoter, to give pTrc-pthS. The E.coli strain K37ΔpthTr was transformed with the resulting plasmid. In this strain, the chromosomal PTH gene (pth) is disrupted while a functional pth gene is added in trans through a plasmid (pMAKpth) harboring a temperature-sensitive replicon (11). At 42°C, the thermosensitive plasmid is lost and cells die. Upon addition of plasmid pTrc-pthS, K37ΔpthTr acquired the capacity to grow at 42°C, in spite of the loss of the thermosensitive plasmid. Transformation with the control plasmid pTrc99-A did not produce thermoresistant clones. This experiment demonstrates that, similarly to the MJ0051 gene, the SS00175 gene can complement for the absence of functional PTH in an E.coli cell.
Furthermore, crude extracts of strain K37ΔpthTr and K37ΔpthTr(pTrc-pthS) were exposed to 80°C for 10 min and PTH activity was assayed. With strain K37ΔpthTr, the E.coli PTH activity was rapidly lost upon heating. With strain K37ΔpthTr(pTrc-pthS), a PTH specific activity of 600 U/mg persisted at the end of the heat treatment. This thermoresistant PTH activity is likely to originate from the S.solfataricus gene introduced in trans in the E.coli recipient strain.
The E.coli strain XL1-Blue transformed with plasmid pTrc-pthS was used to undertake purification of the archaeal PTH. A 0.26 mg aliquot of homogeneous enzyme was obtained from a 1 l culture (Table 2). To determine the molecular mass of the S.solfataricus PTH, a sample of this enzyme was chromatographed on a TSK-GEL G3000-SWXL gel filtration column (0.78 × 30 cm; Tosohaas) equilibrated in 20 mM Tris–HCl buffer pH 7.0 containing 150 mM KCl. In this experiment, marker proteins of known molecular mass included carbonic anhydrase, E.coli PTH and egg white lysozyme. According to the elution times of the various proteins, an Mr of 25 ± 2 kDa could be estimated for the S.solfataricus PTH. Therefore, we concluded that, in contrast to the monomeric E.coli enzyme (7), the S.solfataricus enzyme behaves as a dimer.
Because of their role in tRNA folding, various compounds such as MgCl2, KCl or polyamines were varied in the assay of S.solfataricus PTH activity. Data in Table 3 indicate that, at their optimal concentration, each of these compounds stimulated the rate of diacetyl-lysyl-tRNALys hydrolysis ∼80-fold. We concluded that, similarly to the case of the E.coli enzyme (11), the archaeal PTH requires its substrate to adopt a compact 3D structure.
Table 3. Activity of S.solfataricus PTH under various ionic conditions.
| Compound added | Specific activity (s–1)a |
|---|---|
| None | 0.026 |
| 5 mM MgCl2 | 1.3 |
| 10 mM MgCl2 | 1.8 |
| 40 mM MgCl2 | 2.3 |
| 150 mM KCl | 1.8 |
| 300 mM KCl | 2.0 |
| 500 mM KCl | 1.1 |
| 0.03 mM spermidine-HCl | 1.6 |
| 0.1 mM spermidine-HCl | 1.9 |
aSpecific activities were measured at 50°C in the presence of 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 0.1 mM DTT, 0.75 µM diacetyl-[14C]lysyl-tRNALys and catalytic amounts of enzyme (between 0.1 and 10 nM, depending on the magnitude of the rate to be measured), as described in Materials and Methods. Standard errors on the values shown are <20%.
Substrate specificity of the SS00175 protein product
PTHs purified from bacteria or yeast transform N-acyl-aminoacyl-tRNA into N-acyl-amino acid and tRNA (3,5–7). However, it was reported earlier that rabbit reticulocytes contain a nuclease activity capable of converting N-acyl-aminoacyl-tRNA into N-acyl-aminoacyl-AMP plus truncated tRNA lacking the 3′-AMP terminus (24,25). To examine whether the S.solfataricus enzyme carried such an activity, we determined whether the product of the reaction catalyzed by this protein remained aminoacylatable. A sample of diacetyl-[3H]lysyl-tRNALys was treated with sodium periodate, in order to oxidize, and thus inactivate, any uncharged tRNA molecules. Then, during incubation of this sample in the presence of homogeneous S.solfataricus PTH, pairs of aliquots were withdrawn as a function of time to measure (i) the decline of the diacetyl-[3H]lysyl-tRNALys substrate; and (ii) the appearance of tRNALys aminoacylatable by [14C]lysine upon addition of [14C]lysine, ATP and unlimiting amounts of E.coli lysyl-tRNA synthetase. Along the kinetics, the amount of [14C]lysine that could be esterified with tRNALys exactly paralleled the amount of deacylated diacetyl-[3H]lysyl-tRNALys (Fig. 1). Upon omission of PTH from the reaction mixture, the deacylation rate of diacetyl-[3H]lysyl-tRNALys was strongly reduced (16% deacylation in 25 min), as was the capacity to transfer [14C]lysine onto tRNA (<0.2 pmol of [14C]lysine per pmol of initial diacetyl-[3H]lysyl-tRNALys). These experiments enabled us to conclude that the S.solfataricus enzyme behaves as a hydrolase capable of liberating functional tRNA from an N-blocked aminoacyl-tRNA substrate. Such a hydrolase activity fully accounts for the capacity of the archaeal protein to compensate for the lack of endogeneous PTH activity in an E.coli cell.
In E.coli and in yeast, a tRNA-specific hydrolase distinct from PTH ensures the deacylation of mischarged d-aminoacyl-tRNAs (19,26,27). The genes encoding these d-aminoacyl-tRNA deacylases have homologs in the genomes of all bacteria and eukarya, but not in those of archaea. Therefore, we wondered whether the new PTH isolated from S.solfataricus, which is markedly different from a bacterial PTH, could have a specificity large enough to also catalyze deacylation of d-aminoacyl-tRNAs. Actually, the results shown in Table 4 exclude this possibility. The efficiency of d-aminoacyl-tRNA hydrolysis by the archaeal PTH is poor compared with the transformation of an N-blocked aminoacyl-tRNA. The value of the ratio between the two reaction rates is close to that measured with the E.coli PTH.
Table 4. Hydrolysis of diacetyl-l-[14C]lysyl-tRNALys and d-[3H]tyrosyl-tRNATyr by either S.solfataricus or E.coli PTH.
| Specific activity of deacylation (s–1) | |||
|---|---|---|---|
| Diacetyl-l-[14C]lysyl-tRNALys | d-[3H]tyrosyl-tRNATyr | Ratio value | |
| Purified S.solfataricus PTH (37°C) | 0.53 | 0.0013 | 410 |
| Purified E.coli PTH (28°C) | 0.50 | 0.0025 | 200 |
Specific activities were measured in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 mM EDTA and 0.1 mM DTT, in the presence of either 0.75 µM diacetyl-[14C]lysyl-tRNALys and 1 nM PTH (S.solfataricus or E.coli), or 0.1 µM d-[3H]tyrosyl-tRNATyr and 400 nM PTH. Standard errors on the values shown are <20%.
Escherichia coli PTH has a specificity large enough to recycle any peptidyl-tRNA species (28). However, formyl-methionyl-tRNAfMet escapes hydrolysis by the bacterial enzyme (5,8,10,29). Resistance of formyl-methionyl-tRNAfMet can be accounted for by the absence of a canonical base pair at the top of its acceptor stem. In archaea, formylation of methionyl-tRNAiMet is supposed not to happen, and nucleotides 1 and 72 of initiator tRNA are correctly base paired (30). Therefore, we wondered whether the archaeal PTH exhibited any particular sensitivity to the C1–A72 mismatch in E.coli tRNAfMet. To answer this question, Michaelian parameters of the reaction catalyzed by S.solfataricus PTH were measured in the presence of either E.coli diacetyl-lysyl-tRNALys or E.coli formyl-methionyl-tRNAfMet (Table 5). Comparison of the kcat and Km values obtained clearly shows that the archaeal enzyme does not discriminate between the two tRNAs. With E.coli PTH, under the same experimental conditions, the catalytic efficiencies measured with the two substrates differ by a factor of nearly 20 (12).
Table 5. Catalytic parameters of various N-blocked tRNAs in the hydrolysis reaction catalyzed by S.solfataricus PTH.
| tRNA | kcat (s–1) | Km (nM) | Relative kcat/Km |
|---|---|---|---|
| Diacetyl-lysyl-tRNALys | 1.8 | 11 | 1 |
| Formyl-methionyl-tRNAfMet | 3.0 | 12 | 1.6 |
| Acetyl-histidyl-tRNAHis | 3.4 | 16 | 1.3 |
| Diacetyl-lysyl-tRNALys (dephosphorylated) | 0.86 | 2.8 | 1.9 |
| Formyl-methionyl-tRNAfMet (dephosphorylated) | 3.0 | 30 | 0.62 |
Michaelian kcat and Km parameters were measured as described in Materials and Methods. Relative kcat/Km values were given an arbitrary value of 1 for the measurement in the presence of diacetyl-lysyl-tRNALys. Standard errors on the values shown are <20%.
As explained in the Introduction, the activity of the bacterial PTH towards elongator tRNAs is facilitated by the 5′-terminal phosphate at the end of a fully base-paired acceptor stem. Indeed, the catalytic efficiency of the E.coli enzyme is reduced upon removal of the 5′-phosphate in acetyl-phenylalanyl-tRNAPhe or diacetyl-lysyl-tRNALys (8,11). On the other hand, the activity of the bacterial enzyme does not significantly depend on the dephosphorylation of formyl-methionyl-tRNAfMet (12). Therefore, we also measured the activity of the S.solfataricus PTH in the presence of diacetyl-lysyl-tRNALys or formyl-methionyl-tRNAfMet devoid of 5′-phosphate. As shown in Table 5, upon dephosphorylation, the catalytic efficiencies with the two substrates varied by factors <2.5-fold. The above results force us to conclude that, in an archaeal context, PTH uses neither the 1–72 matching nor the 5′-phosphate as identity elements in the tRNA molecule. We also verified that E.coli acetyl-histidyl-tRNAHis behaved as a substrate of the S.solfataricus PTH (Table 5). Indeed, in all living kingdoms, tRNAHis has the peculiarity of possessing an extra base at its 5′ end. The activity of E.coli PTH is not sensitive to this feature (31).
With E.coli PTH, recognition of the substrate is believed to involve the peptidyl moiety and the acceptor end of the esterified tRNA molecule (9,11). Km values for the various studied substrates are in the micromolar range (9–11). Here, the data in Table 5 show Km values for N-acetyl-aminoacyl-tRNAs in the nanomolar range. Such a difference suggests that the S.solfataricus PTH recognizes additional features in the tRNA structure. In agreement with this view, we observed a strong inhibition of the hydrolytic reaction catalyzed by the archaeal enzyme upon addition of non-esterified tRNALys or tRNAfMet. The KI constants with these tRNAs (50 and 110 nM, respectively) are indicative of markedly stable complexes (Table 6). Formation of these complexes is specific for tRNA. Indeed, unfractionated tRNA from E.coli also inhibited the reaction, with a KI value of ∼100 nM. On the other hand, E.coli rRNA or herring sperm DNA could be added in great excess without significant consequences on the velocity of the reaction. Such a behavior of the S.solfataricus PTH in the presence of tRNA is in contrast to that of E.coli PTH. Indeed, uncharged tRNA had to be added at concentrations in the range of 10 µM to observe inhibition of the bacterial PTH (9).
Table 6. KI values for various nucleic acids in the hydrolysis reaction catalyzed by S.solfataricus PTH.
| Compound | KIa (nM) | (mg/ml) |
|---|---|---|
| Escherichia coli tRNALys | 50 | 1.2 |
| Escherichia coli tRNAfMet | 110 | 2.7 |
| Unfractionated E.coli tRNA | 95 | 2.4 |
| Escherichia coli rRNA | >2000 | |
| Herring sperm DNA | >2000 |
aInitial rates of hydrolysis of 0.2 µM diacetyl-[14C]lysyl-tRNALys in 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA and 0.1 mM DTT were measured at 50°C in the presence of 0.075 nM S.solfataricus PTH. Concentrations of the studied inhibitors were varied as described in Materials and Methods. Standard errors on the values shown are <20%.
Presence of homologs of the SS00175 gene in archaeal and eukaryal genomes
Searches of the sequence databases using the BLAST program (32) indicated the presence of SS00175 homologs in all sequenced archaeal and eukaryal genomes, as well as in the fowlpox virus genome. MJ0051 belongs to this family of homologs. In S.cerevisiae, the homolog is the YBL057c gene. However, homologs could not be found in any of the available bacterial genomes. On the other hand, homologs of the E.coli pth gene are detected in bacteria and eukarya, but not in archaea. Therefore, as recently proposed (16), both bacterial- and archaeal-like PTH enzymes are likely to be encoded in eukarya.
The YHR189w gene product in S.cerevisiae corresponds to a bacterial-like PTH. This gene has already been shown to complement an E.coli thermosensitive mutant deficient in PTH activity (15,16). We could reproduce this experiment with a pth-null mutant, K37ΔpthTr, and a pBluescript derivative carrying YHR189w. To know whether the YBL057c gene product could also function as a PTH, we inserted this gene inside pKK233-3 and transformed the E.coli strain K37ΔpthTr with the resulting plasmid (pKKpthY2). Transformants grew at 42°C despite the loss of the thermosensitive pMAKpth plasmid at this temperature. To confirm that the yeast bacterial-like and archeal-like gene products carry a hydrolytic activity resembling that of the E.coli enzyme, we assayed PTH activity in crude extracts of K37ΔpthTr(pBSpthY1) and K37ΔpthTr(pKKpthY2). Measured rates with diacetyl-lysyl-tRNALys as substrate were 83 and 48 pmol/s/mg, respectively (Table 7). With a crude extract of strain K37, which possesses an intact pth gene, the rate of hydrolysis was only 8 pmol/s/mg. The large discrepancy between the two former and the latter rate values indicates that most if not all activity in K37ΔpthTr(pBSpthY1) and K37ΔpthTr(pKKpthY2) must originate from the YHR189w and YBL057c genes, respectively. We may conclude that yeast cells benefit from two types of PTH at a same time. At first sight, this property explains why inactivation of YHR189w had no effect on yeast growth (15) while a pth gene is essential for E.coli (11,14). Indeed, the presence of the archaeal-like PTH is likely to compensate for deprivation of the bacterial-like enzyme. In fact, the YBL057c and YHR189w genes could both be destroyed without noticeable effect on the growth of S.cerevisiae (16). This finding may reflect the occurrence in yeast of additional PTH-like activities.
Table 7. Comparison of the hydrolysis of E.coli diacetyl-l-[14C]lysyl-tRNALys and formyl-[14C]methionyl-tRNAfMet by the various PTHs (S.solfataricus, S.cerevisiae or E.coli).
| Initial rate of deacylation (pmol/s/mg) | |||
|---|---|---|---|
| Diacetyl-[14C]lysyl-tRNALys | Formyl-[14C]methionyl-tRNAfMet | Ratio value | |
| Crude extract of K37ΔpthTr(pTrc-pthS) | 600 | 1300 | 0.46 |
| Purified S.solfataricus PTH | 69 000 | 140 000 | 0.49 |
| Crude extract of K37ΔpthTr(pKKpthY2) | 48 | 22 | 2.2 |
| Crude extract of K37ΔpthTr(pBSpthY1) | 83 | 480 | 0.17 |
| Purified E.coli PTH | 24 000 | 1200 | 20 |
Initial rates were measured in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 mM EDTA and 0.1 mM DTT, containing catalytic amounts of the PTH assayed (0.02–1 µg of total proteins in the case of the crude extracts, 0.2–0.5 ng of purified S.solfataricus PTH and 1–20 ng of purified E.coli PTH), and either 0.75 µM diacetyl-[14C]lysyl-tRNALys or 0.57 µM formyl-[14C]methionyl-tRNAfMet. Activities were measured at 50°C in the case of the S.solfataricus PTH and at 28°C in all other cases. Plasmids pTrc-pthS, pKKpthY2 and pBSpthY1 harbor the SS000175, YBL057c and YHR189w genes, respectively. Standard errors on the values shown are <20%.
In contrast to our study, Rosas-Sandoval et al. did not succeed in complementing an E.coli pthts strain with YBL057c inserted in the pKQV4 vector (16). To eventually cure the thermosensitive phenotype of the strain, the YBL057c ORF had to be truncated by 258 nucleotides on the 5′ side. To explain this experiment, Rosas-Sandoval et al. considered the possibility that the N-terminal domain of the YBL057c product is a transit signal for mitochondrial localization. The demonstration, in the present study, that the full-length product of YBL057c readily complements an E.coli strain lacking endogeneous PTH forces a rediscussion of this speculation. Actually, we observed that excessive overproduction of S.solfataricus PTH from plasmid pTrc-pthS impairs the growth of E.coli strain XL1-Blue. On this plasmid, the S.solfataricus PTH gene is placed under the control of the inducible trc promoter. Growth of the transformed strain was possible only in the absence of the IPTG inducer. In Table 5, we show that the S.solfataricus enzyme readily accepts E.coli formyl-methionyl-tRNAfMet as a substrate. Consequently, it is tempting to associate the toxicity of SS00175 in an E.coli context with the capacity of the archaeal enzyme to scavenge formyl-methionyl-tRNAfMet. The two yeast PTHs also show efficiency in the hydrolysis of E.coli formyl-methionyl-tRNAfMet (Table 7). Possibly, in the experiments of Rosas-Sandoval et al. (16), truncation of the YBL057c ORF was necessary to reduce the specific activity of the plasmid-encoded yeast PTH to a value supportable by the E.coli translation apparatus.
The capacity of S.solfataricus PTH to use formyl-methionyl-tRNAfMet as substrate in vitro may be explained by the special feature of translation in archaea where formylation of methionyl-tRNAiMet does not occur (30). In the cytoplasm of eukarya, like in an archaeal cell, methionylated initiator tRNA is not formylated. On the other hand, translation initiation in the mitochondrial compartment involves a formylatable tRNAfMet species. In yeast, mitochondrial tRNAfMet displays a mismatch (U1–U72) at the top of its acceptor stem (33). This defect resembles the C1–A72 mispairing in E.coli tRNAfMet. Therefore, in view of their harmful behavior toward E.coli formyl-methionyl-tRNAfMet (Table 7), the two PTHs encoded by YBL057c and YHR189w can be suspected to stay in the cytoplasm of yeast. In agreement with the idea of cytosolic enzymes, the double inactivation of the YBL057c and YHR189w ORFs was reported not to impair the growth of S.cerevisiae on solid medium containing glycerol (16). However, in contrast to this result, another study performed in liquid medium claimed that YHR189w disruption reduces the growth rate on several non-fermentable carbon sources (34).
In this context, one must take care that small variations in the culture conditions may be enough to succeed in revealing mitochondrial dysfunction in response to gene inactivation. For instance, recent reports indicated that the respiratory growth of S.cerevisiae was insensitive to deletion of mitochondrial methionyl-tRNAfMet formyltransferase (35,36). Nevertheless, by changing the growth conditions, a selective advantage could eventually be associated with mitochondrial formylation (36).
To possibly associate phenotypes with YBL057c and YHR189w products, we compared the growths of strains Y12883 (YHR189w::kanMX4) and Y03083 (YBL057c:: kanMX4) with those of their parental strains (Table 1). In agreement with previous studies (15,16), all cells grew normally on rich solid medium containing either glucose or glycerol, or on minimal medium containing glucose. On the other hand, a growth defect of Y12883 became clearly visible on minimal medium containing glycerol (Fig. 2). To establish that this defect was directly caused by the disruption of the YHR189w gene, we cloned this gene into the pYES2 expression vector under the control of the inducible GAL1 promoter and introduced the resulting plasmid (pYESpthY1) in the Y12883 mutant. As shown in Figure 2, the growth defect of the mutant was cured by the transformation. We verified that the parental strain BY4742 was insensitive to the transformation. A residual growth of the strain Y12883 carrying the control plasmid pYES2 can be accounted for by the presence of 0.1% of the fermentable sugar galactose in the medium. Addition of this compound was necessary to induce expression of the plasmid-encoded PTH. These results indicate that an intact YHR189w gene contributes to yeast mitochondrial function provided that the cells are grown on minimal medium.
Figure 2.
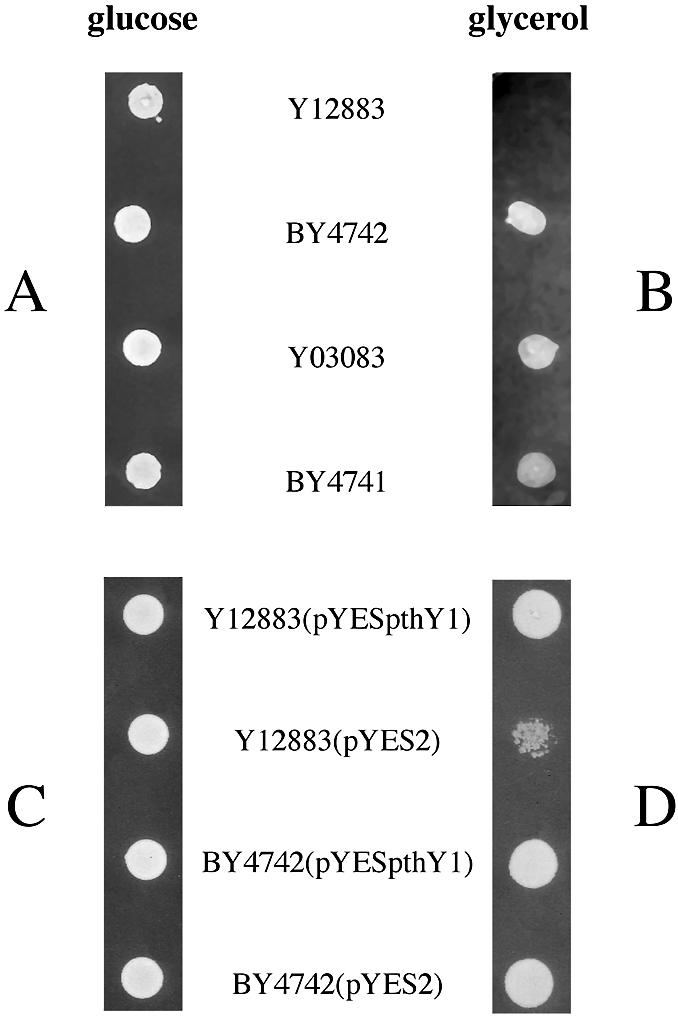
Growth of S.cerevisiae strains BY4742, BY4741, Y12883 (YHR189w::kanMX4), Y03083 (YBL057c::kanMX4), Y12883(pYESpthY1), Y12883(pYES2), BY4742(pYESpthY1) and BY4742(pYES2) on various solid media. Exponentially growing cultures in minimal fermentative medium were diluted in 10 mM Tris–HCl pH 8.0 containing 1 mM EDTA, to obtain suspensions with an OD of 1 at 650 nm. Then, 4 µl aliquots were spotted on minimal medium plates containing either glucose (A and C) or glycerol (B and D) and supplemented with either His, Ura, Lys, Leu and Met (A and B) or His, Lys, Leu and 0.1% galactose (C and D). Plates containing glucose or glycerol were incubated at 30°C for 4 or 7 days, respectively.
By using predictive computational methods [(37), http://mips.gsf.de/cgi-bin/proj/medgen/mitoprot_yeast], the YHR189w product is given only a probability of 0.111 to be mitochondrial. Therefore, the possibility that the YHR189w product remains cytosolic but indirectly contributes to respiration must not be ruled out. Care must be taken, however, in extrapolating this idea to other eukaryotes. For instance, the Arabidospis thaliana genome (http://mips.gsf.de/proj/thal/db/index.html) indicates four bacterial-like PTHs and three archaeal-like PTHs. Using the TargetP algorithm (38), two of the bacterial-like PTHs are predicted to be chloroplastic, whereas one of the bacterial-like PTHs and all three archaeal-like PTHs are predicted to be mitochondrial. Clearly, more work will be necessary to unambiguously assign the cell compartments where the PTH-like products exert activity in eukaryotes. In this search, additional functions will have to be considered. Maize nuclear gene crs2 illustrates this need. This gene has been reported to encode a PTH homolog (39). However, it fails to rescue an E.coli pthts mutant. Instead, the product of crs2 has been proposed to participate in group II intron splicing (39).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jacques d’Alayer and Peggy Chrisment for determining the N-terminal sequence of S.solfataricus PTH, and Marc Nadal for providing us with S.solfataricus strain P2.
REFERENCES
- 1.Kisselev L.L. and Buckingham,R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- 2.Menninger J.R. (1976) Peptidyl transfer RNA dissociates during protein synthesis from ribosomes of Escherichia coli. J. Biol. Chem., 251, 3392–3398. [PubMed] [Google Scholar]
- 3.Cuzin F., Kretchmer,N., Greenberg,R.E., Hurwitz,R. and Chapeville,F. (1967) Enzymatic hydrolysis of N-substituted aminoacyl-tRNA. Proc. Natl Acad. Sci. USA, 58, 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atherly A.G. and Menninger,J.R. (1972) Mutant E.coli strain with temperature sensitive peptidyl-transfer RNA hydrolase. Nature, 240, 245–246. [DOI] [PubMed] [Google Scholar]
- 5.Kössel H. and RajBhandary,U.L. (1968) Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J. Mol. Biol., 35, 539–560. [DOI] [PubMed] [Google Scholar]
- 6.Jost J.P. and Bock,R.M. (1969) Enzymatic hydrolysis of N-substituted aminoacyl transfer ribonucleic acid in yeast. J. Biol. Chem., 244, 5866–5873. [PubMed] [Google Scholar]
- 7.Kössel H. (1970) Purification and properties of peptidyl-tRNA hydrolase from Escherichia coli. Biochim. Biophys Acta, 204, 191–202. [DOI] [PubMed] [Google Scholar]
- 8.Schulman L.H. and Pelka,H. (1975) The structural basis for the resistance of Escherichia coli formylmethionyl transfer ribonucleic acid to cleavage by Escherichia coli peptidyl transfer ribonucleic acid hydrolase. J. Biol. Chem., 250, 542–547. [PubMed] [Google Scholar]
- 9.Shiloach J., Bauer,S., de Groot,N. and Lapidot,Y. (1975) The influence of the peptide chain length on the activity of peptidyl-tRNA hydrolase from E.coli. Nucleic Acids Res., 2, 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutka S., Meinnel,T., Lazennec,C., Mechulam,Y. and Blanquet,S. (1993) Role of the 1–72 base pair in tRNAs for the activity of Escherichia coli peptidyl-tRNA hydrolase. Nucleic Acids Res., 21, 4025–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt E., Mechulam,Y., Fromant,M., Plateau,P. and Blanquet,S. (1997) Crystal structure at 1.2 Å resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase. EMBO J., 16, 4760–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromant M., Plateau,P., Schmitt,E., Mechulam,Y. and Blanquet,S. (1999) Receptor site for the 5′-phosphate of elongator tRNAs governs substrate selection by peptidyl-tRNA hydrolase. Biochemistry, 38, 4982–4987. [DOI] [PubMed] [Google Scholar]
- 13.García-Villegas M.R., De La Vega,F.M., Galindo,J.M., Segura,M., Buckingham,R.H. and Guarneros,G. (1991) Peptidyl-tRNA hydrolase is involved in λ inhibition of host protein synthesis. EMBO J., 10, 3549–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menninger J.R. (1979) Accumulation of peptidyl tRNA is lethal to Escherichia coli. J. Bacteriol., 137, 694–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menez J., Buckingham,R.H., de Zamaroczy,M. and Campelli,C.K. (2002) Peptidyl-tRNA hydrolase in Bacillus subtilis, encoded by spoVC, is essential to vegetative growth, whereas the homologous enzyme in Saccharomyces cerevisiae is dispensable. Mol. Microbiol., 45, 123–129. [DOI] [PubMed] [Google Scholar]
- 16.Rosas-Sandoval G., Ambrogelly,A., Rinehart,J., Wei,D., Cruz-Vera,L.R., Graham,D.E., Stetter,K.O., Guarneros,G. and Söll,D. (2002) Orthologs of a novel archaeal and of the bacterial peptidyl-tRNA hydrolase are nonessential in yeast. Proc. Natl Acad. Sci. USA, 99, 16707–16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zillig W., Kletzin,A., Schleper,C., Holz,I., Janekovic,D., Hain,J., Lanzendorfer,M. and Kristjansson,J.K. (1994) Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol., 16, 609–628. [Google Scholar]
- 18.Plateau P., Fromant,M., Schmitter,J.M. and Blanquet,S. (1990) Catabolism of bis(5′-nucleosidyl) tetraphosphates in Saccharomyces cerevisiae. J. Bacteriol., 172, 6892–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soutourina J., Plateau,P., Delort,F., Peirotes,A. and Blanquet,S. (1999) Functional characterization of the d-Tyr-tRNATyr deacylase from Escherichia coli. J. Biol. Chem., 274, 19109–19114. [DOI] [PubMed] [Google Scholar]
- 20.Dardel F. (1994) MC-Fit: using Monte-Carlo methods to get accurate confidence limits on enzyme parameters. CABIOS, 10, 273–275. [DOI] [PubMed] [Google Scholar]
- 21.Lazard M., Mirande,M. and Waller,J.P. (1987) Expression of the aminoacyl-tRNA synthetase complex in cultured Chinese hamster ovary cells: specific depression of the methionyl-tRNA synthetase component upon methionine restriction. J. Biol. Chem., 262, 3982–3987. [PubMed] [Google Scholar]
- 22.Brevet A., Chen,J., Lévêque,F., Blanquet,S. and Plateau,P. (1995) Comparison of the enzymatic properties of the two Escherichia coli lysyl-tRNA synthetase species. J. Biol. Chem., 270, 14439–14444. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence F., Blanquet,S., Poiret,M., Robert-Gero,M. and Waller,J.P. (1973) The mechanism of action of methionyl-tRNA synthetase. 3. Ion requirements and kinetic parameters of the ATP–PPi exchange and methionine-transfer reactions catalyzed by the native and trypsin-modified enzymes. Eur. J. Biochem., 36, 234–243. [DOI] [PubMed] [Google Scholar]
- 24.Gross M., Starn,T.K., Rundquist,C., Crow,P., White,J., Olin,A. and Wagner,T. (1992) Purification and initial characterization of peptidyl-tRNA hydrolase from rabbit reticulocytes. J. Biol. Chem., 267, 2073–2079. [PubMed] [Google Scholar]
- 25.Gross M., Crow,P. and White,J. (1992) The site of hydrolysis by rabbit reticulocyte peptidyl-tRNA hydrolase is the 3′-AMP terminus of susceptible tRNA substrates. J. Biol. Chem., 267, 2080–2086. [PubMed] [Google Scholar]
- 26.Calendar R. and Berg,P. (1967) d-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. J. Mol. Biol., 26, 39–54. [DOI] [PubMed] [Google Scholar]
- 27.Soutourina J., Plateau,P. and Blanquet,S. (2000) Metabolism of d-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J. Biol. Chem., 275, 32535–32542. [DOI] [PubMed] [Google Scholar]
- 28.Menninger J.R. (1978) The accumulation as peptidyl-transfer RNA of isoaccepting transfer RNA families in Escherichia coli with temperature-sensitive peptidyl-transfer RNA hydrolase. J. Biol. Chem., 253, 6808–6813. [PubMed] [Google Scholar]
- 29.Menninger J.R., Mulholland,M.C. and Stirewalt,W.S. (1970) Peptidyl-tRNA hydrolase and protein chain termination. Biochim. Biophys Acta, 217, 496–511. [DOI] [PubMed] [Google Scholar]
- 30.Bult C.J., White,O., Olsen,G.J., Zhou,L., Fleischmann,R.D., Sutton,G.G., Blake,J.A., FitzGerald,L.M., Clayton,R.A., Gocayne,J.D. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 31.Fromant M., Plateau,P. and Blanquet,S. (2000) Function of the extra 5′-phosphate carried by histidine tRNA. Biochemistry, 39, 4062–4067. [DOI] [PubMed] [Google Scholar]
- 32.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canaday J., Dirheimer,G. and Martin,R.P. (1980) Yeast mitochondrial methionine initiator tRNA: characterization and nucleotide sequence. Nucleic Acids Res., 8, 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinmetz L.M., Scharfe,C., Deutschbauer,A.M., Mokranjac,D., Herman,Z.S., Jones,T., Chu,A.M., Giaever,G., Prokisch,H., Oefner,P.J. et al. (2002) Systematic screen for human disease genes in yeast. Nature Genet., 31, 400–404. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Holmes,W.B., Appling,D.R. and RajBhandary,U.L. (2000) Initiation of protein synthesis in Saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA. J. Bacteriol., 182, 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vial L., Gomez,P., Panvert,M., Schmitt,E., Blanquet,S. and Mechulam,Y. (2003) Mitochondrial methionyl-tRNAfMet formyltransferase from Saccharomyces cerevisiae: gene disruption and tRNA substrate specificity. Biochemistry, 42, 932–939. [DOI] [PubMed] [Google Scholar]
- 37.Claros M.G. and Vincens,P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem., 241, 779–786. [DOI] [PubMed] [Google Scholar]
- 38.Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins B.D. and Barkan,A. (2001) Recruitment of peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J., 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller H.I. and Friedman,D.I. (1980) An E.coli gene product required for lambda site-specific recombination. Cell, 20, 711–719. [DOI] [PubMed] [Google Scholar]