Abstract
Translation of Xenopus laevis Connexin41 mRNA is strongly controlled by the three upstream open reading frames (uORFs) in its 5′ untranslated region. Mutation of uAUG1 into AAG induced a 100-fold increase in translation of a green fluorescent protein (GFP) reporter ORF. The termination codon of uORF1 was mutated and the uORF was linked in-frame with the GFP ORF, enabling visualisation of initiation at uAUG1 by synthesis of an elongated GFP form. Unexpectedly, hardly any elongated GFP was made, suggesting that translation of uORF1 in wild-type mRNA causes constraining of the entry of 40S ribosomal subunits upstream of uORF1. A rare leucine codon, the third codon of uORF1, contributed to the slow translation and thus to slow scanning. Replacement of the rare leucine codon in uORF1 with a common leucine codon stimulated GFP translation. Remarkably, the rare leucine codon, the termination codon of uORF1, uAUG2 and uAUG3 all improved recognition of uAUG1. Apparently, the block formed by a stalled ribosome on any element in uORF1 prevented the landing of new ribosomal subunits next to the cap and therefore downregulated GFP translation.
INTRODUCTION
The binding of the 40S ribosomal subunit to mRNA is mediated by eukaryotic initiation factors (eIFs). After landing of the 40S subunit next to the m7G cap structure, the mRNA is scanned from 5′ to 3′ for initiation codons, in general AUG (1). Recognition of the initiation codon is mediated by eIF1, eIF2 and base pairing of the anticodon of Met-tRNAi with the AUG codon (2), and is dependent on sequences surrounding the AUG codon. The most frequently found nucleotides surrounding the AUG start codon in Xenopus laevis mRNAs are MMAMMAUGR (M = A or C, R = A or G) (http://uther.otago.ac.nz/Transterm.html). The most frequent nucleotides (in bold) are A at –3 and A or G at +4 (the A of the AUG codon is designated as +1). These two nucleotides are considered the most important nucleotides next to the AUG codon involved in recognition. A good initiation site contains both these nucleotides. When only one of these nucleotides is present, the sequence is said to be adequate for initiation. Whether these most frequent nucleotides indeed correspond to the most active initiation sequence, as has been shown for higher eukaryotes (3), remains to be confirmed.
Many mRNAs have 5′ untranslated regions (UTRs) containing at least one AUG codon (uAUG) upstream of the initiation codon preceding the main open reading frame (ORF) (4–7). These uAUGs, especially those in a good context, will generally inhibit translation of the main ORF. However, several examples are described of uAUGs controlling the expression of the main ORF in a flexible way, dependent on the circumstances (6). When an uAUG is not recognised, the ribosome continues scanning, a process named leaky scanning (8). Constraining scanning could be a powerful mechanism to prevent harmful overproduction of potent regulatory proteins (7). A few examples exist in which uORFs stimulate expression of downstream ORFs (9–13).
Some mRNAs have complex 5′ UTRs, containing multiple uAUGs and sometimes overlapping uORFs, which seem to be crucial for correct expression. Xenopus laevis Connexin41 (Cx41) mRNA contains such a complex 5′ UTR. Expression of this mRNA is controlled by a nested set of three uORFs in the 174 nt 5′ UTR (14; Fig. 1). All three uAUGs are recognised by at least some ribosomes, since mutation of each of the individual uAUGs resulted in an increased translation of the reporter gene. The presence of uAUG1 had the largest effect on downstream initiation (14). The sequence around uAUG1 forms an adequate initiation site (U at –3 and G at +4, Fig. 1A). Although the 5′ UTR of Cx41 is at least partially translated (14), the designation 5′ UTR will be used throughout this study to indicate the sequence from cap site to initiation codon of the connexin ORF.
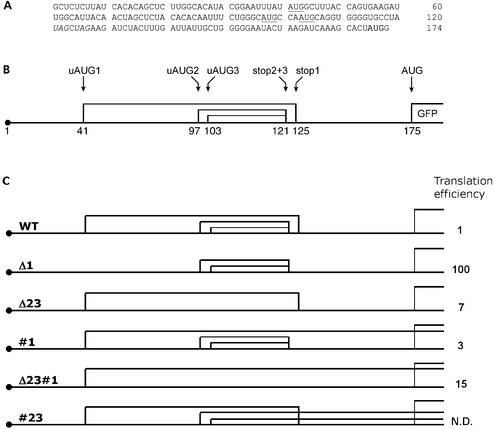
Figure 1.
Translation of Cx41 is controlled by three uORFs. (A) Sequence of the X.laevis Cx41 5′ UTR. (GenBank™ accession no. AF238222). uAUG codons are underlined, termination codons of the uORFs are in italics, and the GFP AUG is in bold (nucleotides 171–173). (B) Schematic representation of the three overlapping uORFs in the Cx41 5′ UTR. uORF2 and uORF3 are in the same reading frame and share a termination codon, they are also in the same reading frame as the main ORF. uORF1 is in a different reading frame. (C) Schematic representations of the ORFs present in the different mutants used in this study. For a detailed description of these and other mutants, see Table 1. The relative translation efficiencies of the different mutants after injection into one cell-stage Xenopus embryos are based on the results presented in Figure 1. The #23 mutant was not injected and was only used for in vitro assays. N.D.: not done.
The strong increase of translation after mutation of uAUG1 suggested that the majority of ribosomes that bound to the wild-type mRNA recognised this uAUG and dissociated from the mRNA after termination at uORF1. Translation of the main ORF was therefore expected to be dependent on the relatively few ribosomes that skipped the uAUGs. We tested this suggestion in this study by making constructs in which the termination codon of uORF1 was mutated. An extra nucleotide was inserted to link this uORF in-frame to the green fluorescent protein (GFP) ORF. These mutations allowed detection of initiation at uAUG1, in the form of an N-terminal extended GFP. The results show that a surprisingly low number of ribosomes initiate at uAUG1. Apparently, the translation of uORF1 poses a severe block for scanning ribosomes. One of the contributing elements is a rare leucine codon, at the third position of uORF1. The results show a remarkably complex mechanism to control the frequency at which ribosomes reach the main ORF.
MATERIALS AND METHODS
Plasmid construction
All plasmids were derived from pT7TS and contain the (mutated) Cx41 5′ UTR, the GFP ORF, the X.laevis β-globin 3′ UTR, and a track of 30 A and 30 C residues. mRNAs with a poly(AC) tail have the same stability and are translated with the same efficiency as mRNAs with a poly(A) tail of 75 nt (15). The mutants used for translation analyses (Fig. 1C and Table 1) were based on the plasmid containing the wild-type Cx41 5′ UTR (WT). Construction of WT, Δ1 (mutation of uATG1) and Δ23 (mutation of uATG2+3) has been described previously (14). The symbol Δ indicates mutation of the uATG codon to AAG.
Table 1. Mutants used for translation analysis of Cx41 5′ UTR.
| Name | Template PCR | Primers PCRa | Mutationsb | Position | Effect |
|---|---|---|---|---|---|
| WT | – | 5F1 | CTATGG→ | 174 | Introduction NcoI site |
| 5R1 | CCATGG | ||||
| Δ1 | WT | 5F2 | uATG1→AAG | 42 | Annihilation of uAUG1 |
| 5F1 | |||||
| 5R1 | |||||
| Δ23 | WT | 5F3 | uATG2→AAG | 98 | Annihilation of uAUG2 and uAUG3 |
| 5F1 | uATG3→AAG | 104 | Mutation of two cysteine residues in uORF1 into serine | ||
| 5R1 | |||||
| #1 | WT | 5F1 | TAG1→TTG | 126 | uORF1 connected to ORF GFP |
| 5R4 | A insertion | 164 | |||
| Δ23#1 | Δ23 | 5F1 | uATG2→AAG | 98 | Annihilation of uAUG2 and uAUG3 |
| 5R4 | uATG3→AAG | 104 | uORF1 connected to ORF GFP | ||
| TAG1→TTG | 126 | ||||
| A insertion | 164 | ||||
| #23 | WT | 5F1 | TAG2+3→AAG | 121 | uORF2+3 connected to ORF GFP |
| 5R5 | TGA→AGA | 139 | uORF1 6 nt shorter | ||
| TAA→AAA | 160 | ||||
| CB– | WT | 5F5 | GCT→GCG | 46 | Creation of rare codons for ALPVc in uORF1 |
| 5R1 | CCA→CCG | 52 | |||
| GTG→GTA | 55 | ||||
| CB+ | WT | 5F6 | TTA→CTG | 47 + 49 | Creation of common codons for ALPVc in uORF1 |
| 5R1 | |||||
| #1CB– | #1 | 5F5 | GCT→GCG | 46 | Creation of rare codons for ALPVc in uORF1 |
| 5R4 | CCA→CCG | 52 | uORF1 connected to ORF GFP | ||
| GTG→GTA | 55 | ||||
| TAG1→TTG | 126 | ||||
| A insertion | 164 | ||||
| #1CB+ | #1 | 5F6 | TTA→CTG | 47 + 49 | Creation of common codons for ALPVc in uORF1 |
| 5R4 | TAG1→TTG | 126 | uORF1 connected to ORF GFP | ||
| A insertion | 164 | ||||
| Δ23CB– | Δ23 | 5F5 | GCT→GCG | 46 | Creation of rare codons for ALPVc in uORF1 |
| 5R1 | CCA→CCG | 52 | Annihilation of uAUG2 and uAUG3 | ||
| GTG→GTA | 55 | ||||
| uATG2→AAG | 98 | ||||
| uATG3→AAG | 104 | ||||
| Δ23CB+ | Δ23 | 5F6 | TTA→CTG | 47 + 49 | Creation of common codons for ALPVc in uORF1 |
| 5R1 | uATG2→AAG | 98 | Annihilation of uAUG2 and uAUG3 | ||
| uATG3→AAG | 104 | ||||
| Δ23#1CB– | Δ23#1 | 5F5 | GCT→GCG | 46 | Creation of rare codons for ALPVc in uORF1 |
| 5R4 | CCA→CCG | 52 | uORF1 connected to ORF GFP | ||
| GTG→GTA | 55 | Annihilation of uAUG2 and uAUG3 | |||
| uATG2→AAG | 98 | ||||
| uATG3→AAG | 104 | ||||
| TAG1→TTG | 126 | ||||
| A insertion | 164 | ||||
| Δ23#1CB+ | Δ23#1 | 5F6 | TTA→CTG | 47 + 49 | Creation of common codons for ALPVc in uORF1 |
| 5R4 | uATG2→AAG | 98 | uORF1 connected to ORF GFP | ||
| uATG3→AAG | 104 | Annihilation of uAUG2 and uAUG3 | |||
| TAG1→TTG | 126 | ||||
| A insertion | 164 |
aPrimers are listed in Table 2.
bAll mutants are based on the WT Cx41 5′ UTR containing the NcoI restriction site.
cALPV = alanine, leucine, proline, valine, amino acid residues 2–5 of uORF1.
The stop codon of uORF1 was mutated in a PCR with primers 5F1 and 5R4 (Table 2) and the WT or Δ23 template, creating #1 and Δ23#1 (Fig. 1C and Table 1). The symbol # indicates that the termination codon of the uORF is mutated and that the corresponding uORFs were connected to the GFP ORF. In the case of uORF1, this required the insertion of an A (at position 164). Both primers contained a restriction enzyme recognition site, so that PCR products could be cloned into the vectors. uORF2 and uORF3 were connected to the GFP ORF by destroying the corresponding termination codon as well as two in-frame downstream stop codons in a PCR with primers 5F1 and 5R5 (Table 2) and the WT template, creating the #23 mutant plasmid (Table 1). For all primer and template combinations, the same PCR programme was used. After the initial denaturation (5 min, 95°C), 20 cycles were performed for denaturation (15 s, 95°C), annealing (15 s, 60°C) and elongation (30 s, 72°C), followed by an extra elongation period of 10 min (72°C).
Table 2. Primers used for plasmid construction.
| Name | Sequence (5′–3′)a,b,c | Position | Remarks |
|---|---|---|---|
| 5F1 | CCGCCAAGCTTGCTCTCTTATCACACAGC | 1–18 | HindIII |
| 5F2 | GGCACATACGGAATTTATAAGGCTTTACCAGTGAAG | 23–58 | Mutagenesis uATG1 |
| 5F3 | CTACACACAATTTCTGGGCAWGCCAAWGCAGGTGGGGTGCC | 78–118 | Mutagenesis uATG2+3 |
| 5F5 | CCGCCAAGCTTGCTCTCTTATCACACAGCTCTTGGCACATACGGAATTTATATGGCGTTACCGGTAAAGATTGGCATTACAACTAGCTC | 1–78 | HindIII, mutagenesis ALPVd into rare codons |
| 5F6 | CCGCCAAGCTTGCTCTCTTATCACACAGCTCTTGGCACATACGGAATTTATATGGCTCTGCCAGTGAAGATTGGCATTACAACTAGCTC | 1–78 | HindIII, mutagenesis ALPVd into common codons |
| 5R1 | CAGTCTCCCATGGTGCTTTGATCTTAG | 185–159 | NcoI |
| 5R4 | TTGCTCACCATGGTGCTTTGATTCTTAGTATTCCCCCAGCAATAATCAAAGTAGATCTTCAAGCTATAGGCACCCCACC | 178–108 | NcoI, mutagenesis stop codon uORF1 |
| 5R5 | TTGCTCACCATGGTGCTTTGATCTTTGTATTCCCCCAGCAATAATCTAAGTAGATCTTCTAGCTTTAGGCACCCCACCTGCATTG | 178–101 | NcoI, mutagenesis stop codon uORF2+3 |
aRestriction sites are underlined.
bCx41 nucleotides are in bold.
cW = A or T.
dALPV = alanine, leucine, proline, valine, amino acid residues 2–5 of uORF1.
The codons of amino acid residues 2–5 of uORF1 were either made optimal (codon bias or CB+ variants: only common codons) or minimal (CB–: four rare codons) (Table 1). The CB variants were made in different vectors: WT, #1, Δ23 and Δ23#1, in a PCR containing the primers and templates as described in Table 1. After incubation for 5 min at 95°C, three cycles were performed for denaturation (20 s, 95°C), annealing (20 s, 55°C) and elongation (40 s, 72°C), followed by 27 additional cycles with an annealing temperature of 60°C. After the last cycle, an extra elongation period of 10 min (72°C) was included. All PCR fragments were sequenced.
For two constructs, Δ23 and Δ23#1, the GFP ORF was replaced by the chloramphenicol acetyl transferase (CAT) ORF, using the NcoI restriction site between the 5′ UTR and the ORF, and the PstI site in the vector (just downstream of the A30C30 tail). The CAT ORF was derived from a construct containing the IGFl1 5′ UTR, the CAT ORF, the globin 3′ UTR and an A30C30 tail (16).
Analysis of translation efficiency
Linearisation of DNA downstream of the A30C30 tail, in vitro transcription and in vitro translation assays were done as described (14). Theoretical capping efficiency is 80%; the uncapped mRNAs do not contribute to protein synthesis as they are quickly degraded as well as poorly translated (15). Transcript yield and integrity were determined after Sephadex G50 gel filtration by spectrophotometry (1 A260 U = 40 µg) and agarose gel electrophoresis (14,15). The concentration of mRNA during in vitro translation was 10 µg/ml or 26 nM. All constructs had the same molecular mass and were tested at equal molar concentration. Injection into one-cell stage X.laevis embryos and in vivo translation analyses by western and northern blot analysis were performed as described (14). All embryos were injected with 1 ng (2.6 fmol) of mRNA (in 10 nl). GFP antibody (Clontech) was used in a 100-fold dilution, CAT antibody (5′→3′) was 500-fold diluted, and the eIF4A antibody, a generous gift from Dr C. Kuhlemeier, was 6000-fold diluted. Quantification of the amount of protein and mRNA after injection was done as before (17). Statistical analysis of the data was done with a one-tailed t-test. The tRNA added in the in vitro translation assays was isolated from rat liver and deacetylated as described (18).
Immunoprecipitation
Protein A–Sepharose CL-4B (Amersham Pharmacia Biotech) was suspended in RIPA buffer [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% (v/v) Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 5 mM EDTA pH 8.0, 1 mM phenylmethylsulfonyl fluoride]. Per assay, 4 mg of protein A–Sepharose was incubated with the CAT antibody (5′→3′, 0.3 µl) in 60 µl of RIPA buffer for 1 h at room temperature. Unbound antibody was removed by washing with RIPA buffer, and the protein A–Sepharose was resuspended in 56.5 µl of RIPA buffer. After addition of 8.5 µl of in vitro synthesised 35S-labelled protein, the mixture was incubated for 2 h at room temperature. Protein A–Sepharose with bound antibody and CAT protein was precipitated by centrifugation. The pellet was washed twice in high-salt buffer [500 mM NaCl, 1% (v/v) Triton X-100, 20 mM Tris–HCl pH 7.4] and twice in low-salt buffer [150 mM NaCl, 1% (v/v) Triton X-100, 20 mM Tris–HCl pH 7.4], and resuspended in Laemmli sample buffer. The unbound proteins in the supernatant were precipitated with 5% trichloroacetic acid. The resulting pellet was resuspended in Laemmli sample buffer complemented with 0.2 M Tris. Bound and unbound CAT protein was analysed by SDS–PAGE.
RESULTS
Efficiency of uAUG recognition
Mutation of the three uAUGs in the 5′ UTR of Cx41 strongly increased translation of the GFP reporter (Fig. 1). The leaky scanning model suggests that the strong increase of GFP translation after removal of the uAUGs is due to efficient recognition of the uAUGs. To visualise initiation at uAUG1, the termination codon of uORF1 was mutated and the uORF was made in-frame with the GFP ORF (Table 1). Initiation at uAUG1 will result in the synthesis of an elongated GFP protein, designated #1 GFP.
Transcripts consisting of the (mutated) Cx41 5′ UTR, the GFP ORF, the globin 3′ UTR and an A30C30 tail were synthesised in vitro. In the Δ1 and Δ23 transcripts, uAUG1 or uAUG2 and uAUG3 were mutated to AAG codons. The #1 transcript lacks the termination codon of uORF1, and the uORF was connected to the GFP ORF. To analyse the upstream initiation efficiency at uAUG1 in the absence of uORF2 and uORF3, Δ23#1 (no uAUG2 and uAUG3, uORF1 linked to the GFP ORF) was constructed.
Transcripts lacking initiation and/or termination codons in the 5′ UTR were translated in a rabbit reticulocyte lysate for a qualitative analysis to determine whether the uAUGs were recognised in vitro and whether correctly sized proteins can be detected (Fig. 2). Translation of all transcripts resulted in the synthesis of correctly sized GFP. The nearly equal synthesis of GFP in all lanes was not unexpected, because earlier analysis in the in vitro reticulocyte system had shown that uAUG codons in the Cx41 5′ UTR hardly influenced GFP translation in this assay (14). Mutation of the termination codon resulted nonetheless in the synthesis of elongated GFP, showing the recognition of uAUG1 in vitro.
Figure 2.
In vitro analysis of the effect of mutation of the uORF1 termination codon. All transcripts contain the (mutated) Cx41 5′ UTR, the GFP ORF, the X.laevis β-globin 3′ UTR and a tail of 30 A and 30 C nucleotides. Transcripts (Fig. 1C and Table 1) were translated in a rabbit reticulocyte lysate in the presence of [35S]methionine, the resulting proteins were separated on a 15% 30:0.18 acrylamide/bisacrylamide gel, and visualised by autoradiography. Migration of GFP and elongated GFP (#1 GFP) is indicated on the right.
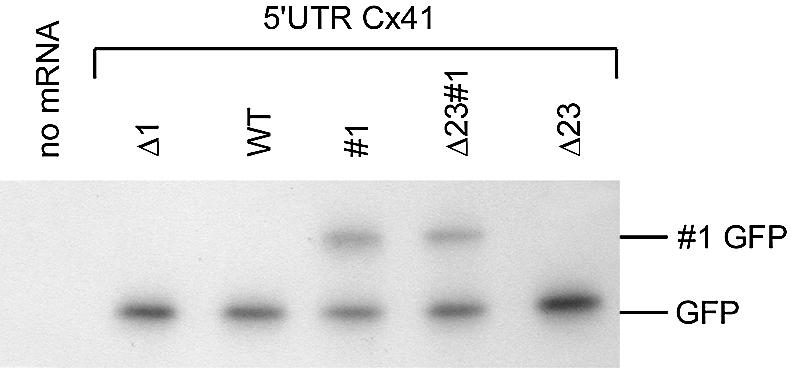
The same transcripts were analysed in vivo by injecting them into one cell-stage Xenopus embryos. After 24 h, pools of 10 embryos were made and proteins and RNA were isolated. Synthesised GFP protein was visualised by western blotting (Fig. 3A). The amount of eIF4A was used as a loading control (Fig. 3B). The amount of injected RNA was analysed by northern blotting, using a GFP probe, and found to be similar (Fig. 3C). After stripping, the blot was hybridised with a Xenopus histone 3 (XH3) probe to correct for the amount of RNA loaded (Fig. 3D). The translation efficiency of the different transcripts was calculated by dividing the amount of GFP protein by the amount of GFP mRNA, and both figures were corrected for the loading controls. The translation efficiency of the most efficiently translated transcript (Δ1) was set to 100; the other translation efficiencies were calculated relative to Δ1.
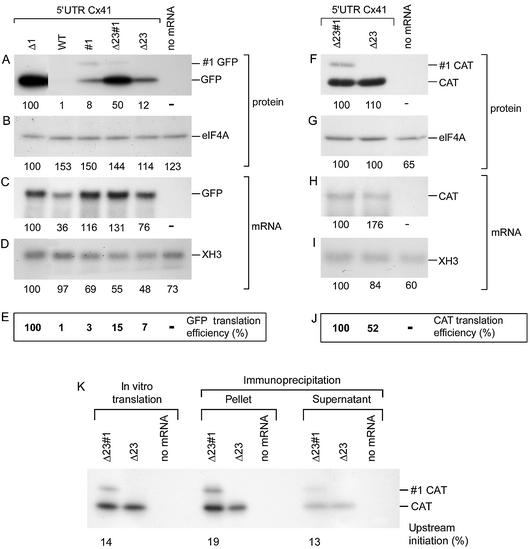
Figure 3.
The uAUG1 codon is barely recognised in vivo. (A–E) One cell-stage Xenopus embryos were injected with GFP encoding transcripts containing the (mutated) Cx41 5′ UTR, as indicated above each lane. Protein and mRNA were isolated after 24 h. Protein derived from two embryos was separated on a 15% 30:0.18 acrylamide/bisacrylamide gel. The blot was divided into two parts; the lower part was incubated with an antibody against GFP (A), whereas the upper part was incubated with an eIF4A antibody as loading control (B). All lanes were from the same experiment and blot. A 5 µg aliquot of mRNA was glyoxylated and analysed on a sodium phosphate-buffered agarose gel. The blot was subsequently analysed by incubation with a GFP (C) and a Xenopus histone 3 (D) probe. GFP translation efficiency was calculated by dividing the amount of GFP protein by the amount of injected GFP mRNA (E). Both figures were corrected for the loading controls. (F–J) For two transcripts, the GFP ORF was replaced by the CAT ORF. Translation analysis was the same as described for the GFP transcripts except that for the western blot the GFP antibody was replaced by a CAT antibody. (K) The affinity of the CAT antibody for the two CAT forms is similar. After in vitro translation of the Δ23#1-CAT and Δ23-CAT transcripts in the presence of 35S-labelled methionine, the resulting proteins were precipitated by the same CAT antibody used for the translation analysis. Proteins were separated on an acrylamide/bisacrylamide gel prior to and after immunoprecipitation.
As also shown previously (14), mutation of the first uAUG (Δ1) strongly increased the translation of the GFP ORF (100-fold, Fig. 3E). This suggests that the majority of ribosomes that enter the mRNA translate uORF1 and are released after the termination codon. However, mutation of the termination codon of uORF1 did not result in large amounts of elongated GFP (indicated as #1 GFP). The amount of #1 GFP was on average 8% (see also Fig. 7F). Normal GFP translation was slightly enhanced with #1 mRNA compared with WT mRNA (3-fold after corrections, Fig. 3E), suggesting that prevention of termination induced 3-fold more 40S subunits to scan the entire 5′ UTR and reach the GFP ORF. Similar results were obtained with the Cx41 variant in which uAUG2 and uAUG3 were mutated to AAG. The inactivation of these uAUGs led to 7-fold more GFP, in agreement with earlier data (14). The removal of the termination codon in Δ23#1 led to higher GFP synthesis, as also observed with the #1 variant, suggesting that termination at uORF1 is a translational barrier.
Figure 7.
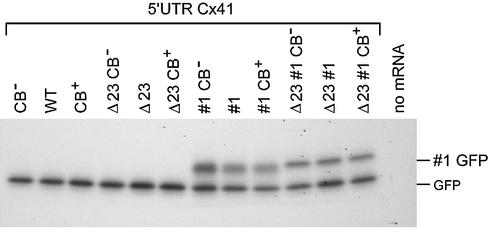
A rare leucine codon in uORF1 inhibits GFP translation. Cx41–GFP transcripts with a modified codon usage in uORF1 were injected into Xenopus embryos. Initiation at uAUG1 (#1 GFP) and at the GFP AUG was visualised by western blotting using a GFP antibody (A). The amount of injected mRNA was determined by northern blotting with a GFP probe (C). eIF4A antibody (B) and a XH3 probe (D) were used as loading controls. Translation efficiency was calculated by dividing the amount of GFP protein by the amount of GFP mRNA, after correction for the loading controls (E). The data were normalised towards the data for the variants with the wild-type codon usage. The percentage of upstream initiation was calculated by dividing the amount of #1 GFP by the total amount of GFP (F). The relative translation efficiency (E) and the percentage upstream initiation (F) are presented as mean values of five analyses. The SEM is indicated.
The small amount of #1 GFP contrasts with the large amount of GFP in the Δ1 mutant. This might be due to a low affinity of the GFP antibody for the elongated #1 GFP form, which may therefore result in an underestimation of the initiation frequency at uAUG1. To exclude this effect, and to exclude the possibility that AUG recognition was dependent on the reporter sequence, the GFP ORF was replaced by the CAT ORF. The activity of two mutants, Δ23#1 and Δ23, was analysed by western and northern blotting, as described for the GFP constructs. The translation efficiency directed by the Δ23#1 and Δ23 transcripts was determined (Fig. 3F–J). The termination codon-mutated variant (Δ23#1) was twice as active as the Δ23 variant, just as has been found for the GFP constructs (Fig. 3E and J). Importantly, the upstream initiation efficiency measured as #1 GFP and #1 CAT was also similar (Fig. 3A and F).
Theoretically, the protein stability of the #1 GFP might be lower than that of GFP, resulting in an underestimation of the initiation frequency at uAUG1. This explanation is considered unlikely since the ratio of the elongated proteins to the normal forms of CAT and GFP is similar for both forms (Fig. 3A and F). Moreover, we will show later (see Fig. 7) that the amount of GFP and #1 GFP can be manipulated by changing the mRNA sequence, but keeping the amino acid sequence, and thus the protein stability, constant.
The amount of GFP and #1 GFP could not be directly assayed by immunoprecipitation with GFP antibodies (not shown). In contrast, the affinity of the CAT antibody for both forms could be measured in an immunoprecipitation assay. Analysis of the CAT products after in vitro translation of Δ23#1-CAT and Δ23-CAT transcripts in the presence of 35S-labelled methionine showed that the ratio of CAT isoforms in immunoprecipitated CAT was similar to the amount directly analysed. These controls indicate that the GFP and CAT antibodies were equally active towards the normal and the elongated forms, and thus that the initiation frequency at uAUG1 is quite low, in contrast to the expectation after analysis of the Δ1 transcript. In spite of the strong stimulation of GFP translation in the Δ1 variant, upstream initiation is apparently a rather rare event as measured by the amount of #1 GFP in the Δ23#1 variant. Removal of uAUG2 and uAUG3 induced even less ribosome initiation at uAUG1 in the #1 variant (Fig. 3A), suggesting that initiation at uAUG2 and uAUG3 influenced upstream initiation at uAUG1. This result resembles the increased initiation when a hairpin is positioned just downstream of an initiation codon (19).
Codon usage
The translation of uORF1 in the Cx41 5′ UTR, albeit rather rare, apparently has an enormous inhibitory effect on translation of the main ORF. This inhibitory effect was not caused by efficient initiation at uAUG1 as only a minor portion of the ribosomes initiate at this uAUG. Therefore, the initiation, the elongation or the termination process at uORF1 might block scanning ribosomal subunits. The data in Figure 3 indicate that the termination codon of uORF1 indeed caused some delay for ribosomes heading towards the AUG of GFP, and thus that termination at uORF1 caused fewer ribosomes to reach the AUG of GFP. This effect (3-fold) is relatively minor compared with the 100-fold increase when uAUG1 is mutated. The total amount of initiations at the #1 transcript (visualised as GFP and #1 GFP together) is very small compared with the initiations at the Δ1 transcript. This suggests that the initiation at uORF1 is not a major rate-limiting step in the translation of uORF1, although it will contribute. Therefore, we looked at other features in uORF1 that may cause the strong hindrance of scanning 40S subunits trying to reach the initiation codon of GFP. One striking feature is a rare leucine codon, the third codon of uORF1 (Fig. 4A). Most amino acids are encoded by more than one codon. The percentages presented under the codons of uORF1 (Fig. 4A) represent the relative frequency with which the codon is used to encode the corresponding amino acid. When the codon for a particular amino acid is unique (as for tryptophan), this percentage is 100.
Figure 4.
Analysis of codon usage in uORF1. (A) Sequence of the peptide encoded by uORF1. The numbers under the codons represent the frequency with which the codons are used to encode the corresponding amino acid in X.laevis ORFs (27). The rare leucine codon is in bold. (B) Codon usage of amino acids 2–5 in the codon bias (CB) mutants. The codons were made either minimal (CB–, four rare codons) or optimal (CB+, the rare leucine codon was replaced by a common leucine codon). Mutated nucleotides are underlined. (C) Translation efficiency of RNAs after injection into one cell-stage Xenopus embryos. For explanation of the different mutants, see Figure 1C. The relative translation efficiencies are calculated based on the results presented in Figures 3 and 7.
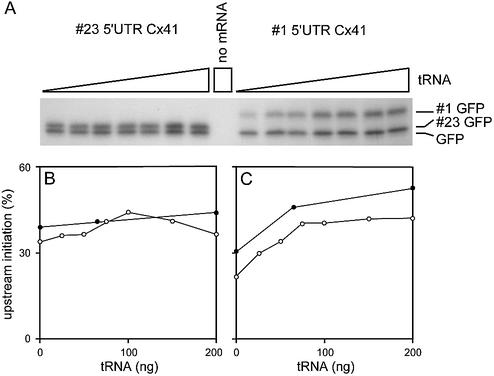
Examples have been described in which codon bias can regulate translational efficiency (20). In eukaryotes, examples of this type of translational control are still rare. To test whether codon usage could indeed be responsible for the translational control by uORF1, we first used the rabbit reticulocyte assay. The reticulocyte lysate contains an amount of tRNA sufficient to drive translation to a level of 60%, compared with the normalised 100% level obtained after addition of sufficient exogenous tRNA (unpublished data). We tested whether addition of tRNA to reticulocyte lysate would improve translation of the elongated #1 GFP more than GFP itself. As a control, a construct was used that synthesised three GFP forms, initiated at uAUG2, uAUG3 and the start AUG of GFP. The GFP forms initiated at uAUG2 or uAUG3 could not be separated and were thus quantified together (Fig. 5). For both mRNAs, total protein synthesis was stimulated 30–40% after addition of tRNA (compare the first and last lanes in the autoradiogram of Fig. 5). The percentage of upstream initiation was calculated by dividing the amount of elongated GFP (#1 GFP or #23 GFP) by the total amount of GFP. At the lower tRNA concentrations, initiation at uAUG1 was inhibited (Fig. 5C), whereas initiation at uAUG2 and uAUG3 was comparable with initiation at the main AUG (Fig. 5B). This result suggested indeed that sequences unique for uORF1 are responsible for the relatively inefficient translation, and that tRNA addition can reverse this inefficiency.
Figure 5.
Effect of tRNA addition on uAUG recognition in vitro. Transcripts encoding different GFP forms, indicated above each panel, were translated in rabbit reticulocyte lysate complemented with 0–200 ng of tRNA per 5 µl assay. The resulting proteins were separated on a 15% 30:0.18 acrylamide/bisacrylamide gel. The gel was dried, and incorporated [35S]methionine was visualised by exposure to film (A). The bands were quantified. Percentage upstream initiation was calculated by dividing the amount of elongated GFP (#1 GFP or #23 GFP) by the total amount of GFP (B and C). The data were corrected for the number of methionines present in the different GFP forms. The open and closed circles represent two independent experiments.
This encouraged us to construct a new set of mutants (Fig. 4B and C). In the first series, CB+, the rare leucine codon was mutated to the common CUG leucine codon that is three times more frequent in codon regions of X.laevis (http://www.kazusa.or.jp/codon). In the second series, CB–, the rare leucine codon was retained, and the surrounding codons for alanine, proline and valine were also mutated into rare codons. The codons downstream of the valine codon could not be mutated into rare codons, because all codons for these amino acids were used with similar frequency. The codon bias mutants were made in four different 5′ UTRs, the wild-type Cx41 5′ UTR, the Δ23 form, which is more efficient and allows better detection of changes in translation efficiency, the #1 form, allowing direct analysis of the codon bias on upstream initiation, and the Δ23#1 form, combining both purposes.
These CB– and CB+ mutants were transcribed and translated in vitro (Fig. 6). All variants produced similar amounts of GFP and #1 GFP. This result shows that the transcripts were of similar quality, allowing their use in vivo. The migration of #1 GFP synthesised from the Δ23#1 transcript is slightly changed compared with #1 GFP encoded by the #1 transcript due to the mutation of uAUG2 and uAUG3, which causes two amino acid changes in uORF1: C20S and C22S.
Figure 6.
In vitro translation of codon bias transcripts. The indicated Cx41–GFP transcripts were translated in a rabbit reticulocyte lysate in the presence of [35S]methionine. The resulting proteins were separated on a 15% 30:0.18 acrylamide/bisacrylamide gel. Deletion of uAUG2 and uAUG3 slightly changed the migration of #1 GFP due to two amino acid changes.
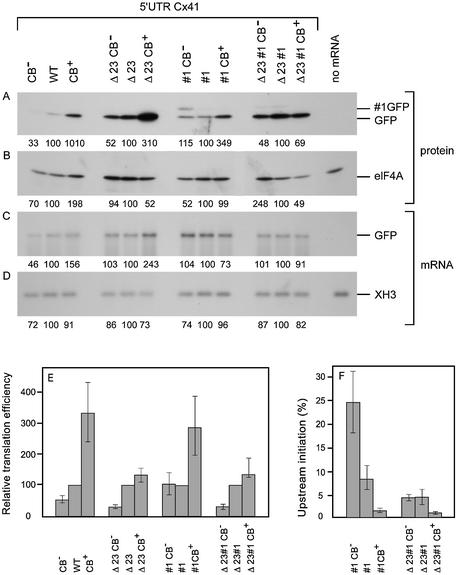
Transcripts were injected into Xenopus embryos, and protein and mRNA were isolated after 24 h to analyse the amount of produced GFP protein (Fig. 7A) relative to the amount of injected GFP mRNA present at the time of harvesting (Fig. 7C). Both figures were corrected for the loading controls (Fig. 7B and D). Since it is technically impossible to inject 12 transcripts in one experiment before the embryos start to divide, the transcripts were divided into four groups. Translation efficiency in each group was calculated relative to the transcript with the wild-type codon usage. The translation efficiencies are presented as the mean of five analyses (Figs 4C and 7E).
Replacement of the rare wild-type leucine codon with a common leucine codon in the wild-type Cx41 5′ UTR increased GFP translation 3.4-fold, indicating that this codon indeed delays progress of translation of uORF1. Addition of three rare codons next to the leucine codon reduced GFP translation below the wild-type level. The effect of codon usage was also analysed in the absence of uAUG2 and uAUG3. The stimulating effect of the removal of the rare leucine codon was less prominent, but the addition of the three rare codons repressed GFP translation strongly. A similar trend in GFP expression was also measured with the #1 constructs: removal of the rare UUA leucine codon increased GFP synthesis. Upstream initiation was visualised by the appearance of the #1 GFP protein after translation of the #1 transcripts. Exchange of the UUA for the CUG leucine codon effectively annihilated upstream initiation, an unexpected result. Apparently, the presence of the rare leucine codon improves translation initiation on the AUG codon just upstream, similar to the initiation-enhancing effect of a hairpin structure just downstream of an initiation codon (12).
DISCUSSION
uORF1 in the Cx41 5′ UTR strongly inhibited translation of the GFP reporter, as mutation of uAUG1 induced a 100-fold increase in GFP translation. This suggested a very efficient initiation at uAUG1, and consequently a very low number of ribosomes capable of initiation at the GFP AUG. The frequency of upstream initiation was measured by using a construct in which uORF1 was linked to the GFP ORF, resulting in an N-terminal elongated GFP form after initiation at uAUG1. In contrast to the large impact of uAUG1 on GFP translation, this codon was barely used for initiation. Therefore, translation of uORF1 must be a relatively slow event, preventing leaky scanning of the Cx41 5′ UTR by ribosomal subunits. The result implies that scanning ribosomes transverse the mRNA much faster than translating ribosomes.
Although initiation in general is the rate-limiting step in the translation of an ORF, some examples are described in which elongation or termination is the restrictive process. Translation of AdoMetDC is strongly dependent on the sequence of a peptide encoded by a small uORF. Elevated polyamines inhibit the termination of translation of the uORF by stabilising the complete nascent peptide, linked to the tRNA that decodes the final codon. This peptidyl-tRNA molecule interacts with a component of the translational apparatus. The resulting complex blocks upstream scanning 40S subunits and therefore inhibits translation (21). Termination at the second uORF of gpUL4 is a slow process because the peptidyl-tRNA complex is not released and this complex causes queuing of upstream scanning ribosomal subunits (22,23). Translation of Arg2 is dependent on a conserved uORF encoding the arginine attenuator peptide, AAP. The synthesis of this peptide also causes stalling, independently of the termination codon (24). Introduction of a rare leucine codon directly downstream of the removed termination codon resulted in an increased toe-print signal compared with the introduction of a common leucine codon. Remarkably, this extra stalling did not have any effect on downstream translation efficiency (24).
Mutation of the termination codon of uORF1 in the Cx41 5′ UTR caused a 3-fold increase in GFP translation, showing that the termination indeed delayed the arrival of ribosomes at the main ORF. Replacement of the rare leucine codon, the third codon of uORF1, by a common leucine codon also resulted in an increased GFP translation. The impact of mutation of uAUG1 is much stronger than the effect of the rare leucine codon and the termination codon. Therefore, additional delaying elements in uORF1 are expected. The leucine codon was the only rare codon in uORF1. Other candidates for a role in the slow translation of uORF1 are two tryptophan and two cysteine codons. Tryptophan is encoded by only one codon, whereas cysteine is encoded by two equally used codons. Both tryptophan and cysteine are infrequently used amino acids. The presence of two tryptophan codons and two identical cysteine codons in a uORF encoding a 28 amino acid peptide is remarkable.
Stalling of ribosomes can be shown by toe-printing: reverse transcription of polysomal RNA will terminate when 40S or 80S ribosomes block reverse transcription. Unfortunately, the reticulocyte data do not reflect the Xenopus data, and toe-printing proved impossible in Xenopus lysates: the lysates were only marginally active in translation, and the majority of ribosomes were not bound to mRNA.
The elimination of the rare leucine codon caused a less frequent initiation at uAUG1. Pausing of the elongating ribosome at the leucine codon apparently stimulated uAUG1 recognition. The block formed by the stalled ribosome might function comparably with a hairpin just downstream of an AUG codon (19). The optimal stimulatory effect of the hairpin was achieved when the distance between the AUG and the hairpin was 14 nt (19), corresponding to half the size of a 40S ribosomal subunit. The explanation of the stimulatory effect of a stalled ribosome at codon 3 of uORF1 is more complex. Only one codon separates the uAUG1 initiation codon from the rare leucine codon. The size of the elongating ribosome seems to exclude stalling of the scanning 40S subunit with the anticodon of Met-tRNAi placed opposite the uAUG codon. The distance between the 5′ cap structure and the rare leucine codon is 46 nt. A stalling ribosome might therefore prevent landing of a new 40S subunit next to the cap due to steric hindrance. Consequently, a new 40S subunit can land only when a rare leucine tRNA becomes bound to the ribosomal A-site and when the ribosome can continue elongation. A scanning 40S subunit, queued behind the elongating ribosome, will scan the 5′ UTR as fast as the elongating ribosome can incorporate the amino acids. This will result in slow scanning of the 5′ UTR. Apparently, this resulted in a more careful scanning and a better recognition of uAUG1. The rare leucine codon counterbalances the inefficient recognition of uAUG1 by delaying the translation of uORF1 and by improving the initiation frequency at uAUG1. Initiation at uAUG1 was stimulated by uAUG2 and uAUG3. Translation of uORF2 and uORF3 therefore might slow down upstream scanning ribosomal subunits in a way comparable with ribosomes translating uORF1. Probably, any event downstream of uAUG1 that slows down the scanning 40S subunits upstream of uAUG1 results in a more careful scanning process and a better recognition of the initiation codon. A comparable mechanism was described for Fli-1 (13). Fli-1 translation can be initiated at two in-frame AUG codons, resulting in the synthesis of two protein isoforms of 51 and 48 kDa. The ratio between these isoforms was determined by two uORFs, both overlapping with the 51 kDa Fli-1 initiation codon. Translation of the uORFs stimulated initiation at both Fli-1 AUG codons. Synthesis of the 48 kDa isoform was dependent on reinitiation after translation of a uORF, whereas synthesis of the 51 kDa form was stimulated by scanning 40S subunits queued upstream of terminating ribosomes (13).
It can be argued that the removal of the termination codon of uORF1 caused faster scanning, thereby decreasing the efficiency of uAUG1 recognition, resulting in an underestimation of the initiation frequency at uAUG1 in the #1 transcript. If removal of the uORF1 termination codon indeed caused a poorer recognition of uAUG1, a substantial increase in GFP translation would be expected. This was not found: GFP translation only increased 3-fold. This cannot account for the 100-fold difference in GFP translation efficiency between the WT and Δ1 Cx41 5′ UTR (Fig. 3).
Inefficient translation due to selective codon usage was shown for the HIV-1 env (envelope glycoprotein) mRNA (20). Conversion of the codon usage of Thy-1, an abundant cell-surface protein of rodent thymocytes, by the HIV-1 env codon usage severely attenuated translation. In contrast, the replacement of the endogenous codons of GFP, a jellyfish protein relatively poorly translated in mammalian cells, with those of highly expressed mammalian proteins caused an increase in translation. Therefore, codon bias can play a role in the efficiency of translation of mammalian genes (20).
Analyses of codon usage in Caenorhabditis elegans, Drosophila melanogaster and Arabidopsis thaliana showed a strong correlation between codons frequently used to encode a specific amino acid and codons mainly present in abundant transcripts (25) and proteins (26). This correlation is stronger in short than in long proteins (25), which implies that codon usage of short uORFs can play a distinct role in the control of translation efficiency.
Results of the reticulocyte assays did not reflect the results obtained with Xenopus embryos. As shown previously (14), mutation of the uAUGs in the Cx41 5′ UTR had no consistent effect on GFP translation in reticulocyte lysate. This suggested that the uAUGs were inefficiently recognised in vitro. Nevertheless, in vitro translation of the #1 and Δ23#1 transcripts showed an initiation efficiency at uAUG1 almost identical to the initiation at the GFP AUG. The lack of competition for translation initiation factors in the reticulocyte lysate might induce a better upstream initiation than measured in vivo. For the initiation at uAUG1, but not for uAUG2 and uAUG3, the in vivo circumstances were approached by omitting extra tRNA. With extra tRNA, initiation at uAUG1 and GFP AUG was almost identical, 48% of the ribosomes starting at uAUG1, and this was reduced to 26% without addition of tRNA (Fig. 6). This percentage approaches the in vivo initiation efficiency at uAUG1 (8%, Fig. 7). Further depletion of the reticulocyte lysate from tRNA might further decrease the difference between in vitro and in vivo translation analyses. This suggests that the ratio between mRNA and tRNA or eIFs is important for the translational control of Cx41.
In conclusion, the first uAUG in the Cx41 5′ UTR was recognised with low efficiency, but the presence of this uAUG strongly reduced GFP synthesis. Translation of uORF1 itself was inhibitory, but the effect was stronger due to the rare leucine codon and the termination codon. Remarkably, uAUG2 and uAUG3, the rare leucine codon and the termination codon all induced a more frequent initiation at uAUG1, most probably because the slower scanning induced a better recognition of AUG codons. This suggests that any translational event will cause better scanning of upstream sequences and thus that once a uORF is translated, the queuing 40S subunits will have a higher efficiency of initiation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank P. A. Krieg (University of Texas, USA) for the pT7TS clone, C. Kuhlemeier (Berne University, Switzerland) for the eIF4A antibody, the Hubrecht Laboratory (Utrecht, The Netherlands) for providing X.laevis embryos, H. A. Wagemaker for technical assistance, the Department of Image Processing and Design for help in artwork, M. van Hattum for typing the manuscript, and W. J. A. G. Dictus and H. O. Voorma for their interest in our work and for critical reading of the manuscript (all Utrecht University, The Netherlands).
REFERENCES
- 1.Hershey J.W.B. and Merrick,W.C. (2000) Pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- 2.Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5 and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- 4.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pesole G., Gissi,C., Grillo,G., Licciulli,F., Liuni,S. and Saccone,C. (2000) Analysis of oligonucleotide AUG start codon context in eukaryotic mRNAs. Gene, 261, 85–91. [DOI] [PubMed] [Google Scholar]
- 6.Meijer H.A. and Thomas,A.A.M. (2002) Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J., 367, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene, 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak M. (1978) How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell, 15, 1109–1123. [DOI] [PubMed] [Google Scholar]
- 9.Harding H.P., Novoa,I., Zhang,Y., Zeng,H., Wek,R., Schapira,M. and Ron,D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell, 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch A.G. (1996) Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 199–239.
- 11.Pooggin M.M., Hohn,T. and Fütterer,J. (2000) Role of a short open reading frame in ribosome shunt on the cauliflower mosaic virus RNA leader. J. Biol. Chem., 275, 17288–17296. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds K., Zimmer,A.M. and Zimmer,A. (1996) Regulation of RARβ2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J. Cell Biol., 134, 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarrazin S., Starck,J., Gonnet,C., Doubeikovski,A., Melet,F. and Morle,F. (2000) Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol. Cell. Biol., 20, 2959–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meijer H.A., Dictus,W.J.A.G., Keuning,E.D. and Thomas,A.A.M. (2000) Translational control of the Xenopus laevis Connexin-41 5′-untranslated region by three upstream open reading frames. J. Biol. Chem., 275, 30787–30793. [DOI] [PubMed] [Google Scholar]
- 15.van der Velden A.W., Voorma,H.O. and Thomas,A.A.M. (2001) Vector design for optimal protein expression. Biotechniques, 31, 572–580. [DOI] [PubMed] [Google Scholar]
- 16.van der Velden A.W., Destrée,O.H.J., Voorma,H.O. and Thomas,A.A.M. (2000) Controlled translation initiation on insulin-like growth factor 2-leader 1 during Xenopus laevis embryogenesis. Int. J. Dev. Biol., 44, 843–850. [PubMed] [Google Scholar]
- 17.Meijer H.A., Dictus,W.J.A.G. and Thomas,A.A.M. (2000) Cloning and analysis of the untranslated regions of the Xenopus laevis Connexin30 mRNA. Gene, 258, 71–76. [DOI] [PubMed] [Google Scholar]
- 18.Schreier M.H., Erni,B. and Staehelin,T. (1977) Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J. Mol. Biol., 116, 727–753. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. (1990) Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA, 87, 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas J., Park,E.-C. and Seed,B. (1996) Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol., 6, 315–324. [DOI] [PubMed] [Google Scholar]
- 21.Raney A., Law,G.L., Mize,G.J. and Morris,D.R. (2002) Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem., 277, 5988–5994. [DOI] [PubMed] [Google Scholar]
- 22.Cao J and Geballe,A.P. (1996) Inhibition of nascent-peptide release at translation termination. Mol. Cell. Biol., 16, 7109–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alderete J.P., Jarrahian,S. and Geballe,A.P. (1999) Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J. Virol., 73, 8330–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang P., Wang,Z. and Sachs,M.S. (2000) Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem., 275, 26710–26719. [DOI] [PubMed] [Google Scholar]
- 25.Duret L. and Mouchiroud,D. (1999) Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila and Arabidopsis. Proc. Natl Acad. Sci. USA, 96, 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morton B.R. (1998) Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol., 46, 449–459. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y., Gojobori,T. and Ikemura,T. (2000) Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res., 28, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]