Abstract
mRNAs containing premature translation termination codons (nonsense mRNAs) are targeted for deadenylation-independent degradation in a mechanism that depends on Upf1p, Upf2p and Upf3p. This decay pathway is often called nonsense- mediated mRNA decay (NMD). Nonsense mRNAs are decapped by Dcp1p and then degraded 5′ to 3′ by Xrn1p. In the yeast Saccharomyces cerevisiae, a significant number of wild-type mRNAs accumulate in upf mutants. Wild-type PPR1 mRNA is one of these mRNAs. Here we show that PPR1 mRNA degradation depends on the Upf proteins, Dcp1p, Xrn1p and Hrp1p. We have mapped an Upf1p-dependent destabilizing element to a region located within the 5′-UTR and the first 92 bases of the PPR1 ORF. This element targets PPR1 mRNA for Upf-dependent decay by a novel mechanism.
INTRODUCTION
The conserved nonsense-mediated mRNA decay (NMD) pathway targets specific mRNAs for rapid decay (reviewed in 1,2). In yeast, three trans-acting factors are required for NMD; Upf1p, Upf2p and Upf3p (3–7). Deletion of any one of these three genes selectively stabilizes mRNAs that are degraded by the NMD pathway without affecting other mRNAs (3–7). Humans, mice and Caenorhabditis elegans all have homologs of Upf1p, Upf2p and Upf3p (8–14). The Upf proteins detect nonsense mRNAs. The nonsense mRNAs are then decapped by Dcp1p and degraded 5′ to 3′ by Xrn1p (15–17).
The NMD pathway degrades mRNAs containing premature stop codons. In yeast, these mRNAs result from nonsense mutations, frameshift mutations and intron-containing transcripts (3,4,18). NMD of representative premature stop codon-containing mRNAs have been characterized (19–21). In addition to the three trans-acting factors, these mRNAs require cis-acting sequences located 3′ to the premature stop codon. The cis-acting sequence element is referred to as the downstream sequence element (DSE). Deletion of the DSE from mRNAs containing a premature stop codon renders these mRNAs insensitive to NMD. PGK1, ADE3 and HIS4 mRNAs all have a functional DSE (19–22). Seventy-five percent of the yeast genes are predicted to contain at least one imperfect match to the DSE (21). Hrp1p specifically binds to the PGK1 DSE and interacts with Upf1p to target the nonsense-containing mRNA for NMD (23).
Potential wild-type NMD targets in yeast were identified by using microarrays to compare mRNA levels in wild-type and upf mutant yeast strains (24). The abundance of 225 of ∼6000 potential yeast mRNAs are increased or marginally increased in an NMD-dependent manner. The accumulation of the majority of mRNAs is unchanged, and ∼40 mRNAs are decreased in upf mutant yeast strains (24). The proteins encoded by mRNAs with Upf-dependent accumulation have a broad range of functions. Some of these mRNAs are co-regulated (24). One subset of mRNAs whose abundance increases in upf– yeast strains consists of PPR1, URA1, URA3 and URA4 mRNAs. Ppr1p is a transcription activator of URA1, URA3 and URA4 (25). PPR1 mRNA half-life increases 3-fold with the loss of Upf1p (26), while URA1, URA3 and URA4 mRNA accumulation increases in upf– yeast strains due to an increase in transcription (3,24,26).
Here, we confirm the results reported by Peltz and Jacobson (26) in a review that PPR1 mRNA decay is dependent on Upf1p. We extend that observation by showing that PPR1 mRNA degradation is also dependent on Upf2p, Upf3p, Dcp1p, Xrn1p and Hrp1p. We map a region of PPR1 mRNA that contains a targeting element for this pathway. We call this element an Upf1p-dependent destabilizing element (UDE). This element targets PPR1 mRNA for Upf1p-dependent decay by a novel mechanism.
MATERIALS AND METHODS
Yeast and bacterial strains
Escherichia coli DH5α (Life Technologies Inc., Rockville, MD) was used for plasmid DNA preparation. Escherichia coli was grown, maintained and transformed using standard techniques (27). Plasmid DNAs were prepared from E.coli using the QIAprep spin plasmid miniprep kit (Qiagen Inc., Chatsworth, CA).
The Saccharomyces cerevisiae strains used in this study and their genotypes are listed in Table 1. Yeast strains were constructed, grown and maintained using standard techniques (27). Yeast transformations were done by the lithium acetate method (28) or electroporation (29).
Table 1. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| W303a | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | (50) |
| PLY167 | MATahis4-713 ura3-52 trp1-Δ1 upf1-Δ2a leu2-3,112 lys2-20 rpb1-1 | (3) |
| BY4741 ppr1Δ | MATa his3D1 leu2D0 met15D0 ura3D0 ppr1Δ | Research Genetics |
| AAY320 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 upf1-Δ2a | (35) |
| AAY334 | MATa ura3- his3-11,15 trp1-1 leu2-3,112 rpb1-1 | (35) |
| AAY335 | MATa ura3- his3-11,15 trp1-1 leu2-3,112 rpb1-1 upf1-Δ2a | (35) |
| HFY1200 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 | (34) |
| HFY870 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::HIS3 NMD2 UPF3 | (34) |
| HFY1300 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 nmd2::HIS3 UPF3 | (34) |
| HFY861 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 upf3::HIS3 | (34) |
| HFY1067 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 dcp1::URA3 | (51) |
| HFY1081 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 UPF1 NMD2 UPF3 xrn1::ADE2 | (51) |
| Y137 | MATa ura3-1 leu2-3 his3-11 trp1-1 ade2-1 | (23) |
| Y191 | MATa ura3-1 leu2-3 his3-11 trp1-1 ade2-1 hrp1-3 | (23) |
aupf1-Δ2 is a UPF1 deletion allele in which the entire ORF has been replaced with URA3 (4).
DNA methods
The plasmids used in this study are listed in Table 2. Standard DNA cloning methods were used for plasmid DNA construction (27). Cloning-free PCR was used to create precise fusions between 5′-UTR and ORF sequences (30). All DNAs produced by PCR for sub-cloning and fusion junctions were sequenced. Probe DNA templates were produced by PCR.
Table 2. Plasmids used in this study.
| Plasmids | Description | Source |
|---|---|---|
| pRS315 | LEU2, CEN6, ARSH4, AmpR, LacZ | (52) |
| pAA79 | LEU2, CEN6, ARSH4, UPF1 | (36) |
| pAA319 | ACT1-PPR1, LEU2, CEN6, ARSH4, AmpR, LacZ | This study |
| pAA330 | Mini-PPR1, LEU2, 2µ, AmpR, LacZ | This study |
| PAA381 | PPR1-ACT1, LEU2, 2µ, AmpR, LacZ | This study |
| pAA401 | ACT1 5′-UTR mini-PPR1, LEU2, 2µ, AmpR, LacZ | This study |
| pAA416 | PPR1 5′-UTR ACT1-PPR1, LEU2, 2µ, AmpR, LacZ | This study |
RNA methods
Yeast total RNA used for mRNA steady-state levels was extracted by a hot phenol method from yeast cells harvested at mid-log phase (4). Total RNA, used to measure mRNA decay rates, was harvested as described in Herrick et al. (31) with some modifications. The cells were shifted to 39°C by the addition of an equal amount of media preheated to 60°C. The cells were placed in a water bath at 39°C and then harvested at various time points. rpb1-1 mutant cells are usually transferred to the non-permissive temperature of 37°C for half-life measurement. However, we found that at 37°C transcription did not arrest until ∼3 min. We used 39°C because at that temperature, the arrest of transcription was essentially immediate.
For both steady-state levels and mRNA decay northerns, equal amounts of RNA (15 µg) were resolved on 1.0% agarose–formaldehyde gels. mRNA steady-state northern blots were transferred to GeneScreen Plus® (NEN™ Life Science Products, Inc., Boston, MA) using the capillary blot transfer protocol recommended by the manufacturer. The NorthernMax™ Complete Northern Blotting Kit (Ambion, Inc., Austin, TX) was used for northern transfer to GeneScreen Plus® and hybridization of mRNA decay northern blots. The northern blots were probed with oligolabeled DNA probes. The probes were labeled with 32P using an Oligolabelling Kit (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) as described in Atkin et al. (32). Northern blots were PhosphorImaged™ using a Storm PhosphorImager (Amersham Pharmacia Biotech, Inc.). mRNA abundances were normalized using ScR1 RNA, a RNA polymerase III transcript that is insensitive to NMD (33,34). mRNA decay rates were determined by graphing log10 of the percent mRNA remaining versus time using SigmaPlot 2000, Version 6.10 (SPSS Science, Chicago, IL). Steady-state mRNA levels and mRNA half-lives were averaged from a minimum of three independent experiments.
RESULTS
Decay of PPR1 mRNA is Upf1p-dependent
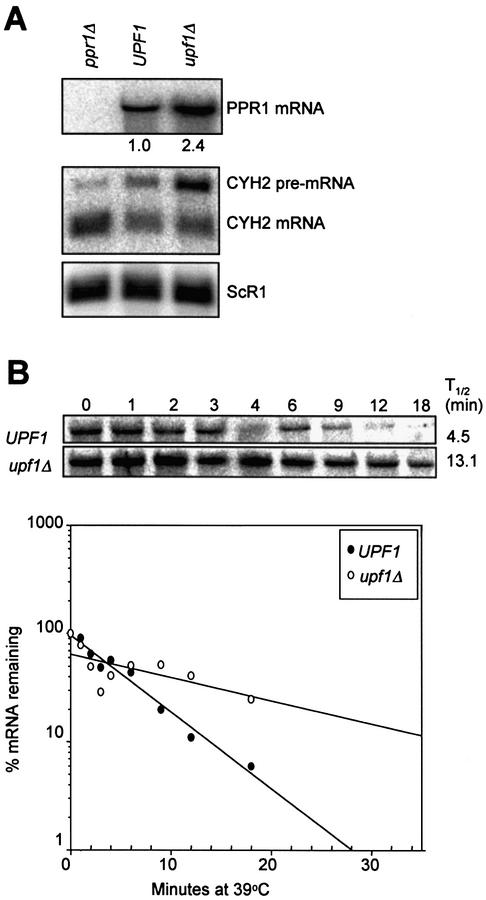
Earlier studies showed that PPR1 mRNA is degraded more rapidly in UPF1 yeast strains than upf1– yeast strains [presented in a review by Peltz and Jacobson (26)]. We have confirmed these results. The steady-state level of the PPR1 mRNA was 2.4 (±0.2) fold higher in the upf1Δ cells than in UPF1 cells (Fig. 1A), and the half-life of PPR1 mRNA was 4.3 (±1.1) min in UPF1 cells and 13.1 (±1.2) min in upf1Δ cells (Fig. 1B). Thus, the results of Peltz and Jacobson (26) and those presented here demonstrate that wild-type PPR1 mRNA is degraded more rapidly in yeast cells with Upf1p.
Figure 1.
The decay of wild-type PPR1 mRNA is Upf1p-dependent. (A) PPR1 mRNA accumulation in BY4741 (ppr1Δ) grown in YPD, and PLY167 [pAA79] (UPF1) and PLY167 [pRS315] (upf1Δ) grown in CM-leucine. Northern blots were prepared with 15 µg of total RNA and hybridized with radiolabeled PPR1, CYH2 and ScR1 DNA. CYH2 pre-mRNA is a target for NMD used to confirm the NMD phenotype of our yeast cells (18). ScR1 mRNA is transcribed by RNA polymerase III and is not degraded by NMD (33,34). It is used as a loading control. The relative PPR1 steady-state levels are indicated under the northern blot hybridized with radiolabeled PPR1 DNA. (B) PPR1 mRNA half-lives were measured in isogenic yeast strains AAY334 (UPF1, upper image) and AAY335 (upf1Δ, lower image) both grown in YAPD. The PhosphorImages are of representative northern blots hybridized with radiolabeled PPR1 DNA. The northern blots were prepared with 15 µg total RNA harvested at different time points following inhibition of RNA polymerase II. The percent mRNA remaining at each time point following inhibition of transcription was calculated by dividing the amount of probe hybridized to a particular band (corrected for loading with ScR1) by the amount of probe hybridized to the band at time point zero. The percent mRNA remaining versus time after transcriptional inhibition was plotted using Sigmaplot™. The PPR1 mRNA half-lives was determined and is shown to the right of the PhosphorImages.
PPR1 mRNA half-lives were reported as 3 min in upf1 yeast cells and 1 min in UPF1 yeast cells by Peltz and Jacobson (26). These reported PPR1 mRNA half-lives are much shorter than the PPR1 mRNA half-lives determined by us. We measured the PPR1 mRNA half-lives in a new pair of yeast strains derived from a W303 genetic background (35). This strain pair has an enhanced CYH2 pre-mRNA accumulation phenotype resulting from a larger difference in CYH2 pre-mRNA half-lives in wild-type and NMD mutants than in other yeast strains used to measure mRNA half-lives. This is due to genetic differences between W303 and other yeast strains that have been used to measure mRNA half-lives. Thus, genetic background also affects PPR1 mRNA half-life.
The accumulation of PPR1 mRNA is also dependent on Upf2p and Upf3p
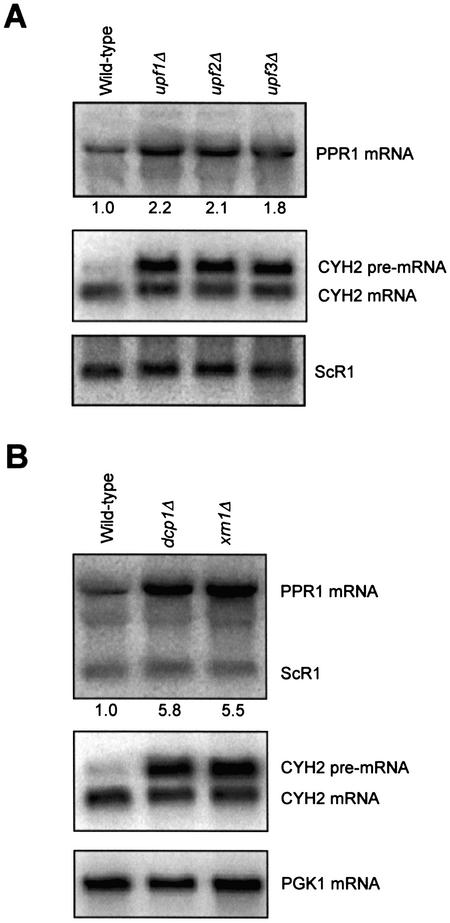
Upf1p functions with Upf2p and Upf3p to degrade nonsense mRNAs. Single, double and triple disruptions of the UPF genes have essentially identical effects on nonsense mRNA accumulation (36,37). We tested whether PPR1 mRNA decay is also dependent on Upf2p and Upf3p by examining PPR1 mRNA steady-state levels in isogenic wild-type, upf1Δ, upf2Δ and upf3Δ cells (Fig. 2A). The steady-state level of the PPR1 mRNA was 2.1 (±0.2) fold higher in the upf2Δ cells and 1.8 (±0.1) fold higher in the upf3Δ cells than in wild-type cells. The increase in PPR1 mRNA accumulation in upf2Δ and upf3Δ yeast cells is similar to the increase in steady-state levels measured in upf1Δ cells (2.2 ± 0.4). Therefore, PPR1 mRNA accumulation is dependent on Upf2p and Upf3p in addition to Upf1p.
Figure 2.
Wild-type PPR1 mRNA also accumulates in upf2Δ, upf3Δ, dcp1Δ and xrn1Δ cells. (A) PPR1 mRNA accumulation in HFY1200 (wild-type), HFY870 (upf1Δ), HFY1300 (upf2Δ) and HFY861 (upf3Δ) yeast cells grown in YAPD. (B) PPR1 mRNA accumulation in yeast cells HFY1200 (wild-type), HFY1067 (dcp1Δ) and HFY1081 (xrn1Δ) grown in YAPD. The northern blots were prepared using 15 µg of total RNA and hybridized with radiolabeled PPR1, CYH2 and ScR1 DNA. The relative PPR1 steady-state levels are indicated under the northern blots hybridized with radiolabeled PPR1 DNA.
PPR1 mRNA accumulates in dcp1Δ and xrn1Δ mutants
Nonsense mRNAs are degraded by deadenylation-independent decapping followed by 5′ to 3′ decay (17). Dcp1p and Xrn1p are responsible for decapping and 5′ to 3′ decay, respectively (15–17). We determined steady-state levels for PPR1 mRNA in wild-type, dcp1Δ and xrn1Δ cells (Fig. 2B). The steady-state level of PPR1 mRNA was 5.8 (±2.8) fold higher in dcp1Δ cells and 5.5 (±1.8) fold higher in xrn1Δ cells than in wild-type cells. Therefore, PPR1 mRNA is degraded by a mechanism that is dependent on the presence of Dcp1p and Xrn1p.
PPR1 mRNA accumulation depends on Hrp1p
Hrp1p binds the PGK1 DSE, interacts with Upf1p and is required for the nonsense-mediated decay of nonsense mRNAs containing the PGK1 DSE (23). The ability of Hrp1p to bind the DSE and interact with Upf1p is probably important for determining whether translation termination occurs before or after the DSE. Mutations in HRP1 stabilize nonsense-containing mini-PGK1 and GCN4-PGK1 mRNAs but not wild-type PGK1, GCN4, MFA4, HTB1 or OLE1 mRNAs (23). We used the temperature sensitive hrp1-3 to ask whether Hrp1p is also important for the accumulation of PPR1 mRNA (Fig. 3). PPR1 mRNA accumulates to higher levels in hrp1-3 cells at 37°C than hrp1-3 cells maintained at room temperature. The relative PPR1 mRNA accumulation was 2.6 (±0.5) at 37°C and 1.0 (±0.03) at room temperature in hrp1-3 cells. The higher PPR1 mRNA levels observed in the hrp1-3 cells shifted to 37°C is specific for the loss of Hrp1p because PPR1 mRNA levels do not show a corresponding increase when isogenic HRP1 cells are shifted to 37°C. The relative PPR1 mRNA accumulations were 1.0 (±0.2) and 1.3 (±0.1) in HRP1 cells at room temperature and 37°C, respectively.
Figure 3.
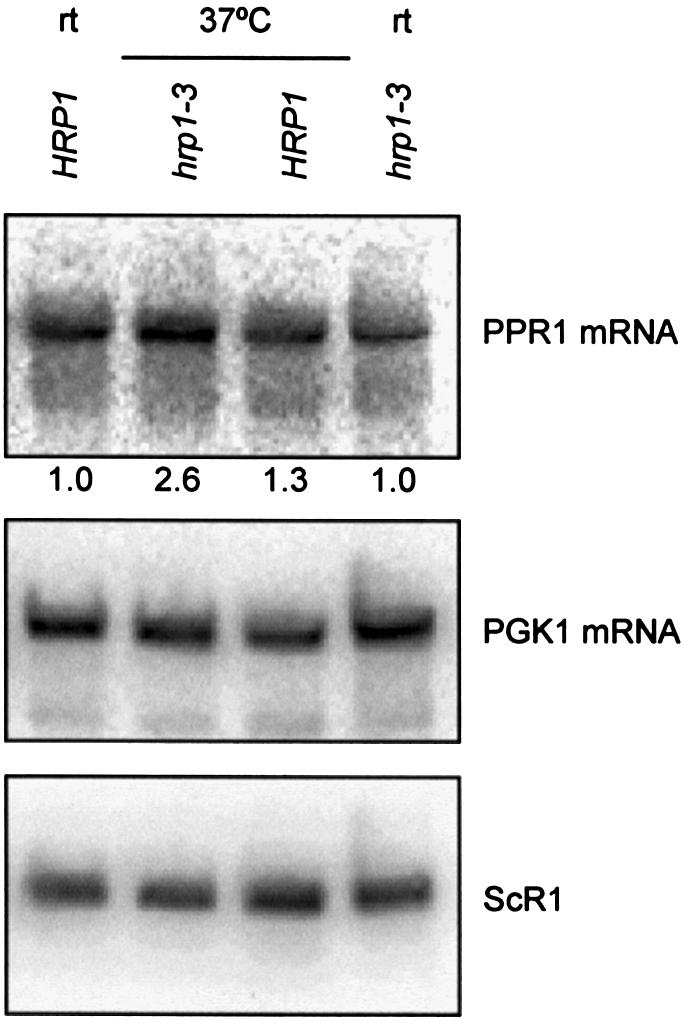
Wild-type PPR1 mRNA accumulates in hrp1-3 yeast cells at the non-permissive temperature. PPR1 mRNA accumulation in Y137 (wild-type) and Y191 (hrp1-3) at room temperature (rt) and after 1 h at 37°C. Y137 and Y191 were grown in YAPD. Northern blots were prepared using 15 µg of total RNA and hybridized with radiolabeled PPR1, PGK1 and ScR1 DNA. The relative PPR1 steady-state levels are indicated under the northern blot hybridized with radiolabeled PPR1 DNA.
PGK1 mRNA is a wild-type mRNA degraded by the major mRNA decay pathway (38). PGK1 mRNA did not accumulate to higher levels in the hrp1-3 mutant at 37°C (Fig. 3). This result is consistent with a previous study showing that PGK1 mRNA half-life is the same in wild-type and hrp1-3 cells following a temperature shift to 37°C for 1 h (23).
Thus, the decay of PPR1 mRNA is Hrp1p-dependent. PPR1 mRNA is one of only three mRNAs whose accumulation is known to be affected by mutations in HRP1.
A region of the PPR1 mRNA that maps from the 5′ end to +92 contains an UDE
mRNAs subject to leaky scanning, containing upstream ORFs (uORFs) and with extended 3′-UTRs are substrates for Upf-dependent deadenylation-independent decay (39–42). The PPR1 translation start codon is in an ideal context suggesting that this mRNA is probably not subject to leaky scanning. The 5′ and 3′ ends of the PPR1 mRNAs have been mapped and are fairly typical for yeast mRNAs (43). Transcription starts at four sites leading to mRNAs with 50, 45, 36, or 20 nt 5′-UTRs. All PPR1 mRNAs contain a short uORF; however, changes in PPR1 that eliminate the uORF do not affect PPR1 mRNA half-life (44). Thus, the uORF does not target PPR1 mRNA for Upf1p-dependent decay. The PPR1 mRNA 3′-UTR is ∼300 nt long (43). Most yeast mRNA 3′-UTRs are between 20 and 350 nt (45). Thus, the PPR1 mRNA 3′-UTR is within the size range of most yeast 3′-UTRs and considerably shorter than the aberrant 3′-UTRs (720– 3970 nt long) targeting CYC1 mRNA for NMD (42). Because the PPR1 mRNA is probably not targeted for Upf-dependent, deadenylation-independent decay by a previously described mechanism, we used fusion constructs to map a region of PPR1 mRNA containing an UDE.
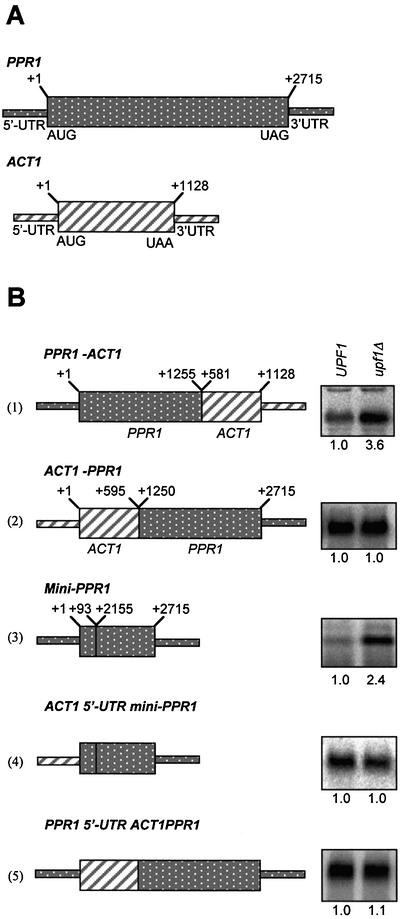
The region within PPR1 mRNA containing the UDE was determined by constructing fusions of PPR1 to ACT1 sequences, and a mini-PPR1 gene (Fig. 4). ACT1 was chosen for the fusions because it encodes a stable mRNA (t1/2 = 30 min), its decay is not Upf1p-dependent and it has been used previously in fusion constructs to map instability elements (3,46). The mRNAs encoded by the fusion genes and mini-PPR1 gene was evaluated by measuring mRNA steady state levels. Half-lives were then determined for mRNAs that accumulated to higher levels in an upf1Δ strain relative to the isogenic wild-type strain.
Figure 4.
Schematic diagrams of the mRNAs used to map the PPR1 mRNA UDE, and representative northern blots of the mRNA encoded by each construct in W303a (UPF1) and AAY320 (upf1Δ) yeast strains. The location of the 5′-UTR, 3′-UTR and ORF of PPR1 and ACT1 mRNAs are indicated. The first base of the ORF is +1. The fusion junction locations are indicated in nucleotides relative to the first base of the ORF. Construct 1, PPR1-ACT1, is an in-frame fusion of the PPR1 5′-UTR and the first 1255 nt of the PPR1 ORF fused to nucleotides +581 to +1128 of the ACT1 ORF and ACT1 3′-UTR. Construct 2, ACT1-PPR1, includes the 5′-UTR and nucleotides +1 to +595 of ACT1 fused in frame to nucleotides +1250 to +2715 of the PPR1 ORF and the PPR1 3′-UTR. Construct 3, mini-PPR1 gene, was constructed by deleting +93 to +2156 of the PPR1 ORF. Construct 4, ACT1 5′-UTR mini-PPR1 gene, is identical to construct 3 except the 5′-UTR of PPR1 has been replaced with the 5′-UTR of ACT1. Construct 5, PPR1 5′-UTR ACT1-PPR1, is identical to construct 2 except the ACT1 5′-UTR has been replaced with the 5′-UTR of PPR1. The abundance of the construct mRNAs in isogenic UPF1 and upf1Δ yeast strains grown in CM- leucine was determined by northern blotting. The relative construct mRNA steady-state levels are indicated under the northern blots. The northern blots were prepared using 15 µg of total RNA and hybridized with radiolabeled PPR1, ACT1, CYH2 (data not shown) and ScR1 (data not shown) DNA.
PPR1-ACT1 mRNA contains the 5′-UTR and the first 1255 nt of the PPR1 ORF fused in frame to nucleotides +581 to +1128 of the ACT1 ORF and 3′-UTR. Steady-state levels of PPR1-ACT1 mRNA are 3.6 (±0.7) fold higher in upf1Δ yeast cells compared with UPF1 yeast cells (Fig. 4B, construct 1). The PPR1-ACT1 mRNA half-life is 8.8 min (±2.3) in UPF1 yeast cells and 16.3 min (±1.5) in upf1Δ yeast cells (Fig. 5A). The PPR1-ACT1 mRNA contains an UDE.
Figure 5.
mRNAs encoded by the PPR1-ACT1 and the mini-PPR1 gene constructs are degraded more rapidly in AAY334, a UPF1 yeast strain, than the isogenic upf1Δ yeast strain, AAY335. mRNA half-lives for PPR1-ACT1 (A) and PPR1 mini-mRNA (B) were determined from northern blots of total RNA harvested from UPF1 and upf1Δ yeast cells grown in CM-leucine at time points following the arrest of transcription as described in the legend of Figure 1B. The time points at which the cells were harvested after inhibition of transcription are shown above the PhosphorImages and are different for the mRNAs encoded by the PPR1-ACT1 construct and the mini-PPR1 gene construct. The upper band in the PhosphorImages of (A) is the endogenous PPR1 mRNA.
ACT1-PPR1 mRNA includes the 5′-UTR and first 595 nt of the ACT1 ORF fused in frame to nucleotides 1250–2715 of the PPR1 ORF and 3′-UTR. Steady-state levels for the ACT1-PPR1 mRNA are identical in upf1Δ and UPF1 yeast cells (1.0 ± 0.03, Fig. 4B, construct 2). Because the change in steady-state levels between upf1Δ yeast cells and UPF1 yeast cells was not significant, nucleotides 1250–2715 of the PPR1 ORF and 3′-UTR do not contain a functional UDE.
A mini-PPR1 gene was constructed by an in-frame deletion of +93 to +2156 from the PPR1 ORF. Steady-state levels of mini-PPR1 mRNA were 2.4 (±0.4) fold higher in upf1Δ yeast cells compared with UPF1 yeast cells (Fig. 4B, construct 3). Mini-PPR1 mRNA half-life was 6.3 min (±0.4) in UPF1 yeast cells and 14.8 min (±2.5) in upf1Δ yeast cells (Fig. 5B). The mini-PPR1 gene mRNA contains an UDE.
Analysis of the PPR1-ACT1 mRNA, ACT1-PPR1 mRNA and the mini-PPR1 gene mRNA narrowed the location of the UDE to the 5′-UTR and the first 92 nt of the PPR1 ORF. To determine whether the UDE was located entirely within the 5′-UTR or the first 92 nt of the PPR1 ORF, ACT1 5′-UTR mini-PPR1 gene and PPR1 5′-UTR ACT1-PPR1 were constructed.
The ACT1 5′-UTR mini-PPR1 gene is identical to the mini-PPR1 gene except the 5′-UTR of PPR1 was exactly replaced with the 5′-UTR of ACT1 (Fig. 4B, construct 4). The steady-state levels of the ACT1 5′-UTR mini-PPR1 mRNA were essentially identical in UPF1 and upf1Δ yeast cells (1.0 ± 0.2; Fig. 4B, construct 4). Because the change in the mRNA steady state levels in upf1Δ yeast cells compared with UPF1 yeast cells was not significant, the first 92 nt of the PPR1 ORF are not sufficient to target the PPR1 mRNA for Upf1p-dependent decay.
PPR1 5′-UTR ACT1-PPR1 is identical to ACT1-PPR1 except the ACT1 5′-UTR was exactly replaced with the PPR1 5′-UTR (Fig. 4B, construct 5). Steady-state levels of the PPR1 5′-UTR ACT1-PPR1 mRNA was 1.1 (±0.3) fold higher in the upf1Δ yeast cells compared with the UPF1 yeast cells (Fig. 4B, construct 5). The change in steady-state levels in the upf1Δ yeast cells compared to the UPF1 yeast cells was not significant. Thus, the PPR1 5′-UTR is not sufficient to target the PPR1 mRNA for Upf1p-dependent decay.
The PPR1 5′-UTR ACT1-PPR1 has a uORF that overlaps the ACT1 ORF. The uORF begins in the PPR1 5′-UTR and extends to +197 in the ACT1 ORF. Because the steady-state levels of this mRNA are similar in UPF1 and upf1Δ yeast cells, the uORF itself is insufficient to trigger Upf1p-dependent decay. These data are consistent with the earlier work of Pierrat et al. (44). The PPR1 uORF does not target PPR1 mRNA for rapid decay.
Analyses with the five constructs used in this study are summarized in Figure 6. The data allowed us to make the following conclusions: a PPR1 mRNA Upf1p-UDE maps between the 5′ end and +92 nt of the PPR1 mRNA. The PPR1 5′-UTR or the first 92 nt of the PPR1 ORF alone are not sufficient to target the PPR1 mRNA for Upf1p-dependent decay. The UDE must span both the PPR1 5′-UTR and the 5′ end of the ORF. Nucleotides +93 to +2712 of the PPR1 ORF and the PPR1 3′-UTR do not contain an UDE. Nucleotides +93 to +2156 are dispensable for UDE function.
Figure 6.

Summary of construct analysis. The UDE maps to a region within the 5′ end to nucleotide +92 of the PPR1 mRNA. Nucleotides +93 to +2715 of the PPR1 ORF and the PPR1 3′-UTR do not contain an UDE. The schematic diagrams represent PPR1 mRNA (top) and the relevant portions of PPR1 mRNA in the five constructs used to map the location of the UDE. The diagrams are numbered relative to the PPR1 transcription start point at +1.
DISCUSSION
We find that the PPR1 mRNA is degraded 3-fold slower in upf1Δ yeast cells compared with UPF1 yeast cells (Fig. 1) and that PPR1 mRNA steady-state levels are increased in upf2Δ, upf3Δ, dcp1Δ, xrn1Δ and hrp1-3 yeast cells (Figs 2 and 3). Thus, the factors required for the degradation of nonsense mRNAs by NMD are also required for the degradation of PPR1 mRNA. The PPR1 mRNA cis-acting sequence, called the UDE, maps to a region from the 5′-UTR to +92 in PPR1 mRNA (Figs 4–6). This sequence targets PPR1 mRNA for accelerated decay by a novel mechanism (see below).
PPR1 mRNA is an unstable mRNA. Pierrat et al. (44) measured the half-lives of mRNAs encoded by chimeric genes constructed by swapping regions of PPR1 and URA3 in wild-type cells to identify what parts of PPR1 are responsible for the rapid decay of PPR1 mRNA. They showed that the PPR1 5′-UTR contains a general instability element because it is sufficient to destabilize the URA3 mRNA. In contrast, UDE function requires sequences from both the 5′-UTR and the first 92 nt of the ORF. Thus, the general instability element may be distinct from the UDE. Alternatively, the UDE may have two parts, a general instability region and a region targeting the mRNA for Upf1p-dependent decay. The later possibility predicts that the general instability region functions independently, while the Upf1p-targeting region requires the presence of the general instability region to target PPR1 mRNA for Upf1p-dependent decay. We plan to test these possibilities by more precisely mapping the location of the UDE, and functional characterization and mutational analysis of the UDE.
Six wild-type mRNAs, PPR1, SPT10, UBP7, STE50 and CPA1 mRNAs, are degraded more rapidly in UPF1 yeast strains than upf mutants (26,39,47 and this study). Wild-type mRNAs could potentially be subject to Upf-dependent decay either because cis-elements trigger a premature translation termination event in an otherwise wild-type mRNA or by a novel mechanism. Premature translation termination can be triggered in an otherwise wild-type mRNA by uORF, extended 3′-UTRs, leaky scanning and potentially a ribosome frameshift.
mRNAs with artificially created uORFs are degraded by NMD (40,41). However, natural uORFs do not necessarily target mRNAs for NMD. GCN4 and YAP1 mRNAs contain uORFs but are not subject to NMD-dependent decay (48,49). The PPR1 and SPT10 uORFs do not target these mRNAs for Upf-dependent decay (39,44). CPA1 mRNA also contains a uORF, but its role in the NMD-dependent decay of this mRNA has not been tested (47).
mRNAs with abnormally long 3′-UTRs are also degraded by NMD (42). A mutation in the CYC1 gene, deleting sequences required for proper 3′ end formation, results in the synthesis of mRNAs with abnormally extended 3′-UTRs ranging between 720 and 3970 nt. PPR1 mRNA does not appear to be targeted for Upf1p-dependent decay by an extended 3′-UTR for two reasons. First, the PPR1 mRNA 3′-UTR is within the normal range and considerably shorter than the 3′-UTRs resulting from deletion of the CYC1 3′ end processing signals. In yeast, most 3′-UTRs are ∼100 nt in length and they can range in size from ∼20 to ∼350 nt (45). The 3′-UTR of PPR1 is ∼300 nt (43). Secondly, the ACT1-PPR1 and PPR1 5′-UTR ACT1-PPR1 fusion constructs contain the PPR1 3′-UTR sequence and 3′ end processing signals (Fig. 4B, constructs 2 and 5). If an extended 3′-UTR targeted PPR1 mRNA for Upf1p-dependent decay, we would expect these constructs to encode mRNAs that are stabilized by inactivation of the NMD pathway. Because these mRNAs are not stabilized by inactivation of the NMD pathway, PPR1 mRNA is not targeted for Upf1p-dependent decay by an extended 3′-UTR (Fig. 4B, constructs 2 and 5).
Leaky scanning followed by translation initiation at an internal ORF triggers the NMD-dependent decay of SPT10 mRNA and probably UBP7, REV7 and STE50 (39). These mRNAs share two key features: (i) a start codon in a sub-optimal context; (ii) an out-of-frame downstream AUG in an optimal context for translation initiation within 90 nt of the ORF start codon followed by an in-frame translation termination codon. The S.cerevisiae optimal start codon context is ANNAUGPuPuPu were N is any base and Pu is an A or a G. The PPR1 start codon context is ATCAUGAAG, the optimal context. Further, there are no AUG codons within the following 90 nt. Therefore, the Upf1p-dependent decay of PPR1 mRNA is probably not due to leaky scanning.
The UDE may target PPR1 for Upf-dependent, deadenylation-independent decay either by triggering a ribosome frameshift or a novel mechanism. The PPR1 UDE could be a binding site for trans-acting factor(s) that target PPR1 for decay by interacting directly with the surveillance complex containing Upf1p, Upf2p and Upf3p. Alternatively, these trans-acting factors could induce the translation machinery to shift to a new reading frame. This would result in premature termination of translation and trigger NMD of PPR1 mRNA.
The UDE could be unique to PPR1 mRNA or alternatively it could be part of a family of similar sequences targeting wild-type mRNAs for NMD. We BLAST searched the S.cerevisiae DNA sequence datasets for similarities to the PPR1 UDE-region. No matches were found to sequences spanning both the 5′-UTR and the ORF (data not shown). This suggests that UDE is unique to PPR1 or similar elements are difficult to identify using this strategy because they have dispersed sequence elements or a degenerate sequence. Identification and analysis of the UDE sequence will be necessary to distinguish between these possibilities.
The PPR1 UDE could function alone as a destabilizing element or in conjunction with a DSE. Nonsense mRNAs require both the premature termination codon and a DSE to be recognized as a substrate for NMD. PGK1, ADE3 and HIS4 mRNAs all have DSEs with a loosely conserved motif (5′-TGYYGATGYYYYY-3′) (19–22). There are four loose matches in PPR1 mRNA to the DSE consensus sequence. The potential matches (5′-TGCATGTAAACGATGT-3′, 5′-TGGCTGTCATGATGC-3′, 5′-TACGCCGATGT-3′ and 5′-TGCAAATAAATGATGC-3′) are located at +96, +251, +599 and +2249 in the PPR1 ORF. The first three of these potential DSEs are not essential for the Upf1p-dependent decay of PPR1 because when deleted, the resultant mRNA is still subject to Upf1p-dependent decay (See mini-PPR1 gene, Figs 4B construct 3 and 5B).
The accumulation of PPR1 mRNA in a hrp1-3 mutant at the non-permissive temperature provides indirect evidence that the putative DSE elements in PPR1 mRNA may be functional. Hrp1p specifically binds the PGK1 DSE and mutations in HRP1 stabilize nonsense mRNAs but not wild-type mRNAs (23). The stabilization of PPR1 mRNA in a hrp1-3 mutant at the non-permissive temperature suggests Hrp1p also binds PPR1 mRNA, probably at one or more of the putative DSE elements. This supports the model in which the UDE functions in conjunction with a DSE. Characterization of the potential PPR1 DSE would add to our understanding of a class of Hrp1p-binding DSEs.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Alan Jacobson, Stuart Peltz and Susan Wente for yeast strains used in this study. We are grateful to Lynne Maquat and all of the members of the Atkin laboratory for critical reading of this manuscript, helpful comments and discussions. This work was supported by a grant from the National Science Foundation (MCB-9874516). Any opinions, findings and conclusions, or recommendations expressed in this manuscript are those of the authors and do not necessarily reflect the views of the National Science Foundation.
REFERENCES
- 1.Maquat L.E. (2000) Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 849–869.
- 2.Gonzalez C.I., Bhattacharya,A., Wang,W. and Peltz,S.W. (2001) Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene, 274, 15–25. [DOI] [PubMed] [Google Scholar]
- 3.Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- 4.Leeds P., Wood,J.M., Lee,B.S. and Culbertson,M.R. (1992) Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Y., Hagan,K.W., Zhang,S. and Peltz,S.W. (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev., 9, 423–436. [DOI] [PubMed] [Google Scholar]
- 6.He F. and Jacobson,A. (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev., 9, 437–454. [DOI] [PubMed] [Google Scholar]
- 7.Lee B.S. and Culbertson,M.R. (1995) Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl Acad. Sci. USA, 92, 10354–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlick H.A., Medghalchi,S.M., Spencer,F.A., Kendzior,R.J.,Jr and Dietz,H.C. (1996) Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc. Natl Acad. Sci. USA, 93, 10928–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Applequist S.E., Selg,M., Raman,C. and Jack,H.M. (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res., 25, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page M.F., Carr,B., Anders,K.R., Grimson,A. and Anderson,P. (1999) SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol., 19, 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cali B.M., Kuchma,S.L., Latham,J. and Anderson,P. (1999) smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics, 151, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell J.T., Medghalchi,S.M., Lake,R.G., Noensie,E.N. and Dietz,H.C. (2000) Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol., 20, 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- 14.Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- 16.Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- 18.He F., Peltz,S.W., Donahue,J.L., Rosbash,M. and Jacobson,A. (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1– mutant. Proc. Natl Acad. Sci. USA, 90, 7034–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peltz S.W., Brown,A.H. and Jacobson,A. (1993) mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev., 7, 1737–1754. [DOI] [PubMed] [Google Scholar]
- 20.Hagan K.W., Ruiz-Echevarria,M.J., Quan,Y. and Peltz,S.W. (1995) Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol., 15, 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S., Ruiz-Echevarria,M.J., Quan,Y. and Peltz,S.W. (1995) Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol., 15, 2231–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Echevarria M.J. and Peltz,S.W. (1996) Utilizing the GCN4 leader region to investigate the role of the sequence determinants in nonsense-mediated mRNA decay. EMBO J., 15, 2810–2819. [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez C.I., Ruiz-Echevarria,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- 24.Lelivelt M.J. and Culbertson,M.R. (1999) Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol., 19, 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy A., Exinger,F. and Losson,R. (1990) Cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol. Cell. Biol., 10, 5257–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peltz S.W. and Jacobson,A. (1993) mRNA turnover in Saccharomyces cerevisiae. In Brawerman,G. and Belasco,J. (eds), Control of Messenger RNA Stability. Academic Press Inc., San Diego, CA, pp. 291–328.
- 27.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1998) Current Protocols in Molecular Biology. John Wiley and Sons, Inc.
- 28.Gietz D.R. and Woods,R.A. (1998) Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. In Brown,A.J.P. and Mick,F.T., (eds), Yeast Gene Analysis. Academic Press Inc., San Diego, CA.
- 29.Grey M. and Brendel,M. (1992) A ten-minute protocol for transforming Saccharomyces cerevisiae by electroporation. Curr. Genet., 22, 335–336. [DOI] [PubMed] [Google Scholar]
- 30.Erdeniz N., Mortensen,U.H. and Rothstein,R. (1997) Cloning-free PCR-based allele replacement methods. Genome Res., 7, 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrick D., Parker,R. and Jacobson,A. (1990) Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkin A.L., Altamura,N., Leeds,P. and Culbertson,M.R. (1995) The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell, 6, 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felici F., Cesareni,G. and Hughes,J.M.X. (1989) The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol., 9, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maderazo A.B., He,F., Mangus,D.A. and Jacobson,A. (2000) Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol., 20, 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kebaara B., Nazarenus,T., Taylor,R. and Atkin,A.L. (2003) Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr. Genet., DOI 10.1007/s00294-003-0386-3. [DOI] [PubMed] [Google Scholar]
- 36.Atkin A.L., Schenkman,L.R., Eastham,M., Dahlseid,J.N., Lelivelt,M.J. and Culbertson,M.R. (1997) Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem., 272, 22163–22172. [DOI] [PubMed] [Google Scholar]
- 37.He F., Brown,A.H. and Jacobson,A. (1997) Upf1p, Nmd2p and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- 39.Welch E.M. and Jacobson,A. (1999) An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J., 18, 6134–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun D.F. and Sherman,F. (1995) Initiation of translation can occur only in a restricted region of the CYC1 mRNA of Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira C.C. and McCarthy,J.E. (1995) The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 270, 8936–8943. [DOI] [PubMed] [Google Scholar]
- 42.Muhlrad D. and Parker,R. (1999) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA, 5, 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kammerer B., Guyonvarch,A. and Hubert,J.C. (1984) Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J. Mol. Biol., 180, 239–250. [DOI] [PubMed] [Google Scholar]
- 44.Pierrat B., Lacroute,F. and Losson,R. (1993) The 5′ untranslated region of the PPR1 regulatory gene dictates rapid mRNA decay in yeast. Gene, 131, 43–51. [DOI] [PubMed] [Google Scholar]
- 45.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker R. and Jacobson,A. (1990) Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MATα1 gene are involved in promoting rapid mRNA decay in yeast. Proc. Natl Acad. Sci. USA, 87, 2780–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz-Echevarria M.J. and Peltz,S.W. (2000) The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell, 101, 741–751. [DOI] [PubMed] [Google Scholar]
- 48.Vilela C., Linz,B., Rodrigues-Pousada,C. and McCarthy,J.E. (1998) The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res., 26, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruiz-Echevarria M.J., Gonzalez,C.I. and Peltz,S.W. (1998) Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J., 17, 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wente S.R., Rout,M.P. and Blobel,G. (1992) A new family of yeast nuclear pore complex proteins. J. Cell Biol., 119, 705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He F. and Jacobson,A. (2001) Upf1p, Nmd2p and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol., 21, 1515–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]