Abstract
Escherichia coli MutY is an adenine and a weak guanine DNA glycosylase involved in reducing mutagenic effects of 7,8-dihydro-8-oxo-guanine (8-oxoG). The C-terminal domain of MutY is required for 8-oxoG recognition and is critical for mutation avoidance of oxidative damage. To determine which residues of this domain are involved in 8-oxoG recognition, we constructed four MutY mutants based on similarities to MutT, which hydrolyzes specifically 8-oxo-dGTP to 8-oxo-dGMP. F294A-MutY has a slightly reduced binding affinity to A/G mismatch but has a severe defect in A/8-oxoG binding at 20°C. The catalytic activity of F294A-MutY is much weaker than that of the wild-type MutY. The DNA binding activity of R249A-MutY is comparable to that of the wild-type enzyme but the catalytic activity is reduced with both A/G and A/8-oxoG mismatches. The biochemical activities of F261A-MutY are nearly similar to those of the wild-type enzyme. The solubility of P262A-MutY was improved as a fusion protein containing streptococcal protein G (GB1 domain) at its N-terminus. The binding of GB1-P262A-MutY with both A/G and A/8-oxoG mismatches are slightly weaker than those of the wild-type protein. The catalytic activity of GB1-P262A-MutY is weaker than that of the wild-type enzyme at lower enzyme concentrations. Importantly, all four mutants can complement mutY mutants in vivo when expressed at high levels; however, F294A, R249A and P262A, but not F261A, are partially defective in vivo when they are expressed at low levels. These results strongly support that the C-terminal domain of MutY is involved not only in 8-oxoG recognition, but also affects the binding and catalytic activities toward A/G mismatches.
INTRODUCTION
Oxidative damage induced by reactive oxygen species (ROS) is a major source of mutation load in living organisms and is believed to play a causative role in degenerative diseases such as aging and cancer (1,2). ROS are produced during cellular metabolism as well as exogenous stimuli such as ionizing radiation and various chemical oxidants (3). 7,8-Dihydro-8-oxo-guanine (8-oxoG or GO) is one of the most stable products of oxidative DNA damage and has the most deleterious effects because it can mispair with adenine (4,5). Several repair pathways are involved in repair of DNA lesions caused by oxidative stress (6). In Escherichia coli, MutT, MutS, MutM and MutY are involved in defending against the mutagenic effects of 8-oxoG lesions. The MutT protein eliminates 8-oxo-dGTP from the nucleotide pool with its pyrophosphohydrolase activity (7), whereas the MutM glycosylase (Fpg protein) removes both mutagenic GO adducts and ring-opened purine lesions (8). When C/GO is not repaired by MutM, adenines are frequently incorporated opposite GO bases during DNA replication (9,10). MutM cannot efficiently remove GO from A/GO mismatch. A second round of replication through an A/GO mismatch subsequently leads to a G·C to T·A transversion (10–12). MutS and MutY increase replication fidelity by removing the adenines misincorporated opposite GO or G (4,13–15). The MutS-dependent mismatch repair removes mismatched A on the daughter DNA strands (16). MutY is an adenine and a weak guanine DNA glcosylase active on A/G, A/GO, A/C or G/GO (14,17–20). A/GO and G/GO mismatches are particularly important biological substrates of MutY glycosylase. Escherichia coli mutT mutants have a very high mutation rate of T·A to G·C transversions (21). Cells with a single mutation in mutY and mutM genes are moderate mutators; however, cells with double mutations in mutY and mutM genes have mutation rates of C·G to A·T transversions three orders of magnitude higher than the wild-type cells (14). The mutY mutants also have increased G·C to C·G transversions, suggesting that G/GO mispairs are formed during DNA replication (20).
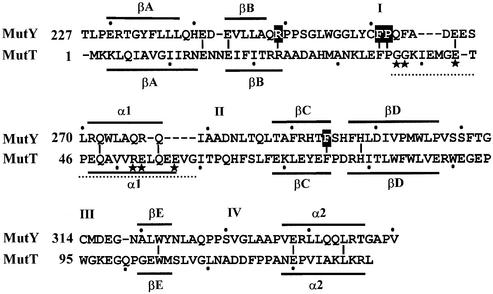
The E.coli 39 kDa MutY glycosylase contains a [4Fe–4S] iron–sulfur cluster (22,23). The N-terminal domain of the MutY protein retains the catalytic activity (18,24–27) and shares similar structure with AlkA, endonuclease III and 8-oxoG glycosylase (OGG1) (28–30). The X-ray crystal structure of the catalytic domain of D138N-MutY with bound adenine shows that adenine is buried in the active site of the catalytic domain, suggesting that the mismatched adenine must flip out of the DNA helix for glycosylase action (29). The protein–DNA co-crystal structures of AlkA and hOGG1 show that both enzymes distort their bound DNA substrates which are bent about 66° and the substrate bases are flipped out of the helix (28,30). The C-terminal domain of MutY has similarities to MutT (27,31) (Fig. 1) but not to other members of the helix–hairpin–helix (HhH) family. Preliminary NMR data indicate that the C-terminal domain of MutY contains five β-strands and two α-helices (31,32) which are very similar to those of MutT (33,34) (Figs 1 and 2). The C-terminal domain of MutY has been shown to play an important role in the recognition of GO lesions (18,24,27). The binding affinity and reaction rate of a truncated MutY on A/GO-containing DNA are reduced by >20-fold as compared with those of the intact MutY (18,27). Moreover, deletion of the C-terminal domain of MutY confers a mutator phenotype in vivo (18).
Figure 1.
Structure-based sequence alignment of the C-terminal domain of MutY and MutT. Boxed residues were mutated to alanines. Lines above or underneath of the sequences represent the α-helixes (α1 and α2) or β-sheets (βA–βE). The loops connecting α-helixes or β-sheets are marked with I–IV. The region G37–G59 (marked by a dotted line) of MutT is the pyrophosphohydrolase module with essential residues marked with stars. Identical residues between the C-terminal domain of MutY and MutT are marked with short vertical lines.
Figure 2.
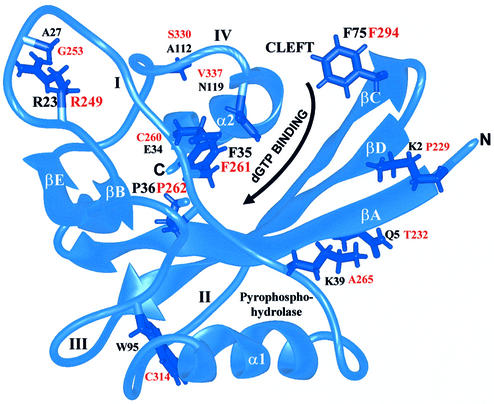
MutT structure (34) showing residues (labeled in black) corresponding to MutY residues (labeled in red). The ribbon diagram was produced with the graphic program MIDAS (58). Four conserved residues of MutY (R249, F261, P262 and F294) were mutated to Ala. The α-helixes (α1 and α2), β-strands (βA–βE) and loops (I–IV) connecting the α-helix or β-sheet are marked. The nucleotide binding site is located in a cleft defined by β-strands A, C and D on one side, and loop I, the end of loop IV and the beginning of helix 2 on the other side. The reaction center of pyrophosphohydrolase consists of residues from loop I, helix 1 and loop III.
To further investigate the substrate recognition of MutY on A/GO-containing DNA and to identify which residues are involved in defining the GO specificity, we have constructed four MutY mutants at the C-terminal domain based on its sequence and structural similarities to MutT through site-directed mutagenesis. F294A, R249A and P262A MutY mutants, but not F261A-MutY, have partially defective in vivo activities when they are expressed at low levels. The in vivo activities of these mutants are reflected by their in vitro biochemical activities. Mutagenesis studies of MutY described here support that a region corresponding to the cleft of MutT is involved in substrate recognition. Surprisingly, F294A, R249A and P262A MutY mutants have reduced binding and catalytic activities not only with A/GO but also with A/G mismatches. These findings suggest that the C-terminal domain of MutY may modulate the N-terminal domain of MutY in DNA binding and catalytic activities.
MATERIALS AND METHODS
Bacteria
Escherichia coli strains PR8 (Su- lacZ X74 galU galK Smr) and mutY-mutant strain PR70 (similar to PR8 but micA68:: Tn10kan) were obtained from M. S. Fox. The strain CC104 containing a lacZ mutation at residue 461 of β-galactosidase and its derivative CC104 mutM::mini-kan mutY::mini-Tn10 were generous gifts from J. H. Miller. DE3 lysogenic strains were constructed according to the procedures described by Invitrogen.
Plasmids
Plasmid pMYW-1 containing the entire mutY gene in the pET11a (Novagen) expression vector has been described (35). The expression vector pGEV1 (obtained from M. Clore) (36) was used to link the streptococcal protein G (GB1 domain) to the N-terminus of the desired proteins.
Cloning of mutY mutants
Mutant mutY genes encoding proteins containing F294A, R249A, F261A or P262A mutations were constructed by the PCR splicing overlap extension method (37). Plasmid pMYW-1 was used as template. Oligonucleotide pairs designed as mutagenesis PCR primers for the desired amino acid substitutions are listed in Table 1. A restriction enzyme site was incorporated within each primer set for easy screening. Primers Chang222 and Chang90 (see Table 1) were used as N-terminal region and C-terminal region primers, respectively. To create a site-directed mutation, primer Chang222 and a bottom strand mutagenesis primer were used to amplify the N-terminal region of the mutY gene. Primer Chang90 and an upper strand mutagenesis primer were used to amplify the C-terminal region of the mutY gene. Both purified PCR products were then mixed in a 1:1 molar ratio and used as templates for another PCR containing Chang222 and Chang90 flanking primers. The PCR product was digested with NdeI and BamHI and ligated into the NdeI–BamHI-digested pET11a vector. pET-F294A, pET-R249A, pET-F261A and pET-P262A mutant clones were first screened for the generation of a NheI, ApalI, FspI or NarI site, respectively, and then confirmed by DNA sequencing.
Table 1. Site-directed mutagenesis primers.
Both the wild-type and P262A mutant mutY genes were transferred to pGEV1 (obtained from M. Clore) to express MutY as fusion proteins containing streptococcal protein G (GB1 domain) at their N-termini. PCR reactions with pMYW-1 and pET-P262A as templates and Chang 348 (5′ GCGACGGCTAGCATGCAAGCGTCGCAATTTTC 3′) and Chang 349 (5′ GCCGGACTCGAGCTAAACCGGCGCG CCAGTGC 3′) as primers were performed. The PCR products were digested with NheI and XhoI and ligated into the NheI–XhoI-digested pGEV1 vector (36), a derivative from pET21a (Novagen). The sequences of pGEV-MutY (containing the wild-type mutY gene) and pGEV-P262A (containing the P262A mutant mutY gene) were confirmed by DNA sequencing. The mutY gene and its derivatives in the plasmid pET11a and pGEV1 were under the control of the T7 promoter.
Western blot analysis
Escherichia coli cells were grown overnight in Luria–Bertani (LB) media with appropriate antibiotics. Cell paste from 1 ml of the culture was resuspended in cracking dye containing 60 mM Tris–HCl (pH 6.8), 10% glycerol, 1% SDS, 1% β-mercaptoethanol and 0.01% bromophenol blue (0.1 ml per OD unit) and boiled for 10 min. Cell lysates and purified MutY were fractionated by 10% SDS–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (38). The membrane was subjected to the enhanced chemiluminescence analysis system from Amersham Pharmacia International according to the manufacturer’s protocol. The anti-MutY antibodies were affinity purified by reaction with the membrane-bound MutY protein (39).
Measurement of mutation frequency
Independent overnight cultures (0.1 ml) of each strain were plated onto LB agar plates containing 0.1 mg/ml rifampicin. The cell titer of each culture was determined by plating 0.1 ml of a 10–6 dilution onto LB agar plates. For each measurement, four independent cultures were plated, and the experiments were repeated three times. The ratio of RifR cells to total cells was the mutation frequency.
Protein expression and purification
Escherichia coli strains PR70/DE3 harboring expression plasmids pET-F294A, pET-R249A, pET-F261A, pGEV-MutY or pGEV-P262A were grown in LB broth containing 50 µg/ml ampicillin at 37°C. The expression of proteins was induced at an OD600 of 0.6 by adding isopropyl β-D-thiogalactoside to a final concentration of 0.2 mM to the culture at 20°C. The cells were harvested 16 h after induction.
The MutY mutant proteins were purified from E.coli PR70/DE3 cells harboring the over-production plasmids pET-F294A, pET-R249A, pET-F261A, pGEV-MutY or pGEV-P262A similar to the method used with the wild-type MutY enzyme (23). All column chromatography steps were conducted in a Waters 650E FPLC system at 4°C; all buffer solutions were flushed with helium gas; and centrifugation was done at 15 000 g for 30 min. Cells (12–32 g of cell paste) were resuspended in 50–125 ml of buffer T [50 mM Tris–HCl (pH 7.6), 0.1 mM EDTA, 1 mM dithiothreitol, 0.1 mM phenylmethanesulfonyl fluoride] and disrupted with a bead beater (Biospec Products, Bartlesville, OK) using 0.1 mm glass beads. Cell debris was removed by centrifugation and the supernatant was then treated with 5% streptomycin sulfate. After stirring for 45 min, the solution was centrifuged, and the supernatant was collected as Fraction I. Ammonium sulfate was added to Fraction I to a final concentration of 65%. The solution was stirred for 45 min and the protein was pre cipitated overnight. After centrifugation, the protein pellets were resuspended in 15 ml of buffer T and dialyzed against two changes of 1 l of the same buffer for 1.5 h each. The dialyzed protein sample was centrifuged, and the supernatant was diluted to 150 ml (Fraction II) with buffer A [20 mM potassium phosphate (pH 7.4), 0.1 mM EDTA, 10% glycerol, 0.5 mM dithiothreitol and 0.1 mM phenylmethanesulfonyl fluoride] containing 25 mM KCl. Fraction II was loaded onto a 30 ml phosphocellulose column, which had been equilibrated with buffer A containing 25 mM KCl. After washing with 75 ml of equilibration buffer, proteins were eluted with a 300 ml linear gradient of KCl (0.025–0.55 M) in buffer A. Fractions eluted at ∼0.25 mM KCl were pooled (Fraction III). Fraction III was loaded onto an 18 ml hydroxylapatite column equilibrated with buffer B [10 mM potassium phosphate (pH 7.4), 10 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethanesulfonyl fluoride]. The flow-through and early elution fractions were pooled and diluted with TEG buffer [50 mM Tris–HCl (pH 7.6), 0.1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethanesulfonyl fluoride] containing 25 mM KCl (Fraction IV). Fraction IV was loaded onto a 6 ml heparin column equilibrated with TEG buffer containing 25 mM KCl. Upon washing with 12 ml of equilibration buffer, the column was eluted with a 60 ml linear gradient of KCl (0.025–0.65 M) in TEG buffer. Fractions containing the MutY protein were pooled and diluted with TEG buffer containing 25 mM KCl to yield Fraction V. Fraction V was then applied to a 1 ml MonoS column (Amersham Pharmacia Biotech) that had been equilibrated in TEG buffer containing 25 mM KCl. After washing with 10 ml of equilibration buffer, the protein(s) was eluted with a 20 ml linear gradient of KCl (0.025–0.5 M) in TEG buffer. Fractions containing the MutY protein were pooled (Fraction VI), divided into small aliquots, and stored at –80°C. Protein concentration was determined by the Bradford method (40).
Oligonucleotide substrates
The nucleotide sequences of the mismatch-containing heteroduplexes used in this study were as follows: 19mer
5′ CCGAGGAATTAGCCTTCTG 3′
3′ GCTCCTTAAXCGGAAGACG 5′,
(X = G or GO).
The first strand is the mismatched adenine-containing strand (A-strand) and the second strand is the mismatched G- or GO-containing strand (G- or GO-strand). The 19mers were labeled at the 3′ end with [α-32P]dCTP on the A-strand and were converted to 20mers after the sticky ends were filled in with the Klenow fragment of DNA polymerase I as described by Lu et al. (13).
MutY binding, trapping and glycosylase assays
The MutY activity assays with labeled oligonucleotide substrates were performed as described by Lu (41) with some modifications. The MutY enzyme was diluted with diluent containing 20 mM potassium phosphate (pH 7.4), 50 mM KCl, 1.5 mM dithiothreitol, 0.1 mM EDTA, 200 µg/ml bovine serum albumin and 10% glycerol before use. The MutY binding reaction mixture contained 20 mM Tris–HCl (pH 7.6), 80 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 2.9% glycerol, 20 ng of poly(dI-dC) and 1.8 fmol of labeled DNA in a total volume of 20 µl. After incubation at 37 or 20°C for 30 min, the mixtures were supplemented with 3 µl of 50% glycerol and analyzed on 8% polyacrylamide gels in 50 mM Tris–borate (pH 8.3) and 1 mM EDTA. To determine the apparent dissociation constant (Kd) values, nine different MutY enzyme concentrations were used to bind DNA substrates (1.8 fmol) and experiments were repeated at least three times. Bands corresponding to enzyme-bound and free DNA were quantified from PhosphorImager images and Kd values were obtained from analyses by a computer-fitted curve generated by the Enzfitter program (42).
Covalent complexes of MutY with 20mer DNA substrates (trapping assays) were formed in a 10 µl reaction containing 20 mM Tris–HCl (pH 7.6), 1 mM dithiothreitol, 1 mM EDTA, 2.9% glycerol and 0.1 M NaBH4. An NaBH4 (1 M) stock solution was freshly prepared and was added immediately after the enzyme was added. After incubation at 37°C for 30 min, 5-fold concentrated dye buffer containing 25% glycerol, 5% SDS, 155 mM Tris–HCl (pH 6.8), 5% β-mercaptoethanol and 0.5 mg/ml bromophenol blue was added to the samples, which were heated at 90°C for 2 min and separated on a 12% polyacrylamide gel in the presence of SDS according to Laemmli (43).
The glycosylase assays were carried out similarly to the trapping assay except 50 µg/ml of bovine serum albumin was added and NaBH4 was omitted. After incubation at different temperatures for various times, the reaction mixtures were lyophilized, resuspended in 3 µl of formamide dye (90% formamide, 10 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue), heated at 90°C for 2 min, and loaded onto 14% 7 M urea sequencing gels. For time-course studies, a 75 µl reaction containing enzyme in 20 mM Tris–HCl (pH 7.6), 1 mM dithiothreitol, 1 mM EDTA, 2.9% glycerol and 50 µg/ml of bovine serum albumin was performed, and 10 µl of the reaction mixtures was withdrawn at different time points. Samples (10 µl) were immediately frozen at –70°C, followed by heating at 90°C for 30 min in the presence of 0.1 M NaOH, supplemented with 5 µl of formamide dye and heated at 90°C for 2 min, and 5 µl of the mixture was loaded onto 14% 7 M urea sequencing gels.
RESULTS
Choice of MutY mutants
The C-terminal domain of MutY has been shown to play an important role in the recognition of GO lesions (18,24,27). In order to study the function of this domain, we analyzed MutY mutants at this domain. Although this domain shares some sequence and structural similarities to MutT (27,31) (Fig. 1), they have several different structural and functional features. First, β-strands A and B, α-helix 1, loops I and II are shorter in the C-terminal domain of MutY than in MutT (see Fig. 1). Second, MutT, but not MutY, has pyrophosphohydrolase activity (see Fig. 1). Five out of six essential residues for the pyrophosphohydrolase activity (44) are not present in MutY. Third, MutT but not MutY binds to metals. MutT requires two divalent cations for activity at the reaction center (34). It has been suggested that MutT interacts with dGTP and dGTP in a different manner because K39 has more crucial role for dGTPase than for 8-oxo-dGTPase (44). In spite of these differences, the similarities between the C-terminal domain of MutY and MutT were used as a guide for our site-directed mutagenesis. Upon inspection of the MutT structure, the binding site of dGTP is located at a cleft defined by three β-strands A, C, and D on one side, and loop I, the end of loop IV and the beginning of helix II on the other side (45) (Fig. 2). These cleft walls contain several amino acid residues conserved between MutT and MutY. F294 is located at the edge of the cleft and points into the cleft. Residues R249, F261 and P262 of loop I are located on the wall or base of the cleft. These four residues were selected for analysis by mutation to alanines.
Expression and purification of mutant MutY proteins
MutY proteins of F294A, R249A and F261A mutants were purified from PR70/DE3 containing the pET11a expression vectors with high yields: 33.6 mg of F294A-MutY protein from 32 g cell paste, 54.5 mg of R249A-MutY protein from 12 g cell paste, and 52.0 mg of F261A-MutY protein from 14 g cell paste. The mutant P262A-MutY expressed in the pET11a vector has poor solubility and cannot be purified to homogeneity. To improve the yield of this protein, we expressed the P262A mutant protein as a fusion protein. We employed the strategy described by Huth et al. (36) to express the mutant MutY as a fusion with the GB1 domain at its N-terminus. As expected, the solubility of the GB1-P262A-MutY protein was substantially improved. We were able to purify 0.7 mg of the mutant protein from 12 g of PR70/DE3/pGEV-P262A cell paste. In contrast, 125 mg of the GB1-tagged wild-type MutY was purified from 24 g of cell paste. As judged on a 10% SDS–polyacrylamide gel, all the MutY and GB1-MutY proteins were purified to >98% homogeneity (data not shown).
Glycosylase activities of mutant MutY proteins
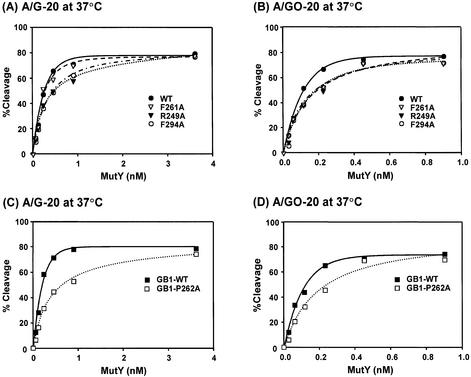
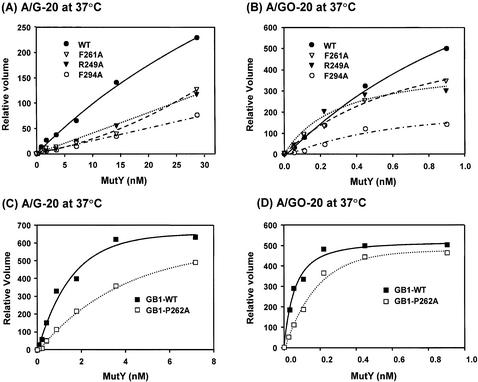
With purified mutant MutY proteins, we compared their catalytic activities with those of the wild-type enzyme. In the first assay, the adenine glycosylase activity of MutY was measured with different concentrations of MutY proteins at 37°C. At lower protein concentrations, F294A-MutY and R249A-MutY proteins had slightly lower glycosylase activities toward both A/G and A/GO mismatches than the wild-type enzyme (Fig. 3A and B). However, at concentrations >3.6 and 0.9 nM, both enzymes had similar activities with A/G and A/GO mismatches, respectively. The F261A-MutY protein had a slightly lower glycosylase activity towards A/GO mismatches than the wild-type enzyme (Fig. 3B), but had a similar glycosylase activity towards A/G mismatch DNA as compared to the wild-type MutY (Fig. 3A). The adenine glycosylase activities of GB1-tagged MutY with A/GO and A/G mismatches are shown in Figure 3C and D. The GB1-P262A-MutY protein had lower glycosylase activities toward both A/G and A/GO mismatches than the wild-type enzyme at lower protein concentrations.
Figure 3.
Glycosylase activities of the wild-type and mutant MutY proteins at different enzyme concentrations. (A) and (C), A/G-containing DNA (1.8 fmol) was incubated with MutY proteins at concentrations of 0.056, 0.1125, 0.225, 0.45, 0.9 and 3.6 nM in a 10 µl reaction at 37°C for 30 min. (B) and (D), A/GO-containing DNA (1.8 fmol) was incubated with MutY proteins at concentrations of 0.028, 0.056, 0.1125, 0.225, 0.45 and 0.9 nM at 37°C for 30 min. After reaction, the products were dried, resuspended in formamide dye, heated at 90°C for 2 min, and analyzed on a 14% denaturing sequencing gel. Data were from PhosphorImager quantitative analyses of gel images over three experiments. Percentages of DNA cleaved were plotted versus protein concentrations. Wild-type MutY (WT, solid circle), F261A (open triangle), R249A-MutY (solid triangle), F294A-MutY (open circle), GB-tagged MutY, (GB1-WT, solid rectangle), and GB-F261A-MutY (open rectangle).
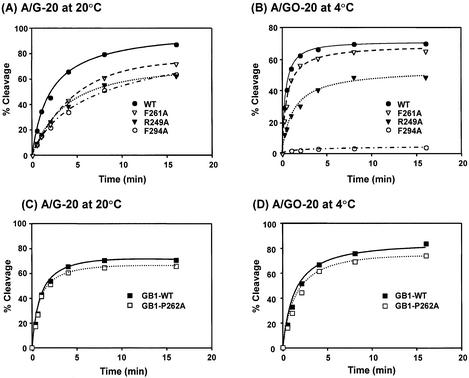
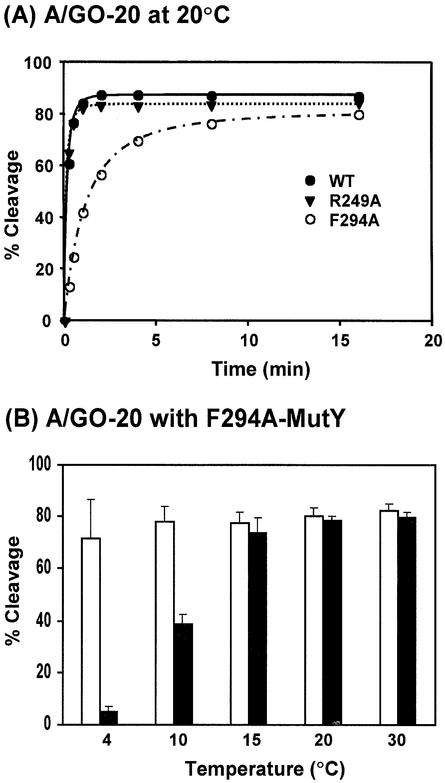
Because of the slow turn-over rate of MutY (46,47), the steady-state kinetics of the MutY reaction, as measured at 37°C for 30 min, may not reflect the true reactivity. Thus, we used single-turnover glycosylase kinetics to compare the activities of wild-type and mutant MutY proteins. Because the cleavage rates on A/GO-containing DNA are too fast for manual measurements, the time-course reactions of glycosylase with A/G and A/GO mismatches were performed at 20 and 4°C, respectively. As shown in Fig. 4, the rates of cleavage on both A/G- and A/GO-containing DNA by F261A-MutY and GB1-P262A-MutY were only slightly lower than those of the wild-type MutY proteins. However, the rates of cleavage of both A/G- and A/GO-containing DNA were lower for F294A-MutY and R249A-MutY than those of the wild-type MutY (Fig. 4A and B). The rate of cleavage on A/GO-containing DNA by F294A-MutY was barely detected at 4°C. Therefore, the F249A mutation affects the glycosylase activity with A/GO mismatch more substantially than with A/G mismatches. Because the rates of glycosylase activity of F294A-MutY and F249A-MutY were much slower at 4°C, we then performed the time-course studies of these proteins on A/GO mismatches at 20°C in comparison with the wild-type MutY. As shown in Figure 5A, the cleavage rates of the wild-type and R249A MutY proteins were very fast (the reactions plateau at 1 min). At 20°C, F294A-MutY was more active than at 4°C; however, it was still less active than the wild-type MutY (Fig. 5A).
Figure 4.
Time course studies of glycosylase activities of the wild-type and mutant MutY proteins. (A) and (C), A/G-containing DNA (1.8 fmol) was incubated with 3.6 nM proteins in a 10 µl reaction at 20°C. (B) and (D), A/GO-containing DNA (1.8 fmol) was incubated with 0.45 nM proteins in a 10 µl reaction at 4°C. After reaction, the products were treated with 0.1 M NaOH as described in Materials and Methods and analyzed on a 14% denaturing sequencing gel. Data were from PhosphorImager quantitative analyses of gel images over three experiments. Percentages of DNA cleaved were plotted as a function of time. Wild-type MutY (WT, solid circle), F261A (open triangle), R249A-MutY (solid triangle), F294A-MutY (open circle), GB-tagged MutY, (GB1-WT, solid rectangle), and GB-F261A-MutY (open rectangle).
Figure 5.
(A) Time course studies of glycosylase activities of the wild-type and mutant MutY proteins with A/GO-containing DNA at 20°C. Reactions were similar to Figure 4B with 0.45 nM proteins but were performed at 20°C. Wild-type MutY (WT, solid circle), R249A-MutY (solid triangle) and F294A-MutY (open circle). (B) The temperature effect on glycosylase activities of the wild-type MutY and F294A-MutY with A/GO-containing DNA. Reactions were performed at 4, 10, 15, 20 and 30°C for 30 min with 0.45 nM proteins.
To investigate the temperature effect on the glycosylase activity of the F294A-MutY protein with A/GO-containing DNA, reactions were performed at temperatures of 4, 10, 15, 20 and 30°C for 30 min. As shown in Fig. 5B, the glycosylase activity with A/GO mismatches of the wild-type MutY was almost not affected by temperature. In contrast, the activity of F294A-MutY was ∼7 and 50% at 4 and 10°C, respectively, of those at 15–30°C. Thus, the glycosylase activity of F294A-MutY toward A/GO-containing DNA is significantly reduced at lower temperatures.
Trapping activities of mutant MutY proteins
The MutY protein can form a covalent Schiff base intermediate with DNA through Lys142 residue (35,48–50), thus we performed the trapping assay in the presence of sodium borohydride. At all protein concentrations tested, all four mutant MutY proteins produced about 2-fold less of the covalent protein–DNA complex on A/G-containing DNA than the wild-type MutY (Fig. 6A and C). Among them, F294A mutant is the weakest. With A/GO mismatches, F249A-MutY also had much reduced trapping activity as compared to the wild-type enzyme (Fig. 6B). In contrast, the R249A, F261A and GB1-P262A mutant proteins had slightly weaker trapping activities than the wild-type enzymes (Fig. 6B and D). These results indicate that mutations at the C-terminal domain of MutY can alter the Schiff base formation between K142 and DNA and can modulate the MutY catalytic activity. Specifically, the F294A mutant has a severe defect in glycosylase and trapping activities with A/GO mismatches.
Figure 6.
Trapping activities of the wild-type and mutant MutY proteins. A/G- (A and C) and A/GO- (B and D) containing DNA (1.8 fmol) was incubated with various amounts of proteins in the presence of NaBH4 in a 10 µl reaction at 37°C for 30 min. The samples were heated at 90°C for 2 min and separated on a 12% polyacrylamide gel in the presence of SDS. Data were from PhosphorImager quantitative analyses of gel images over three experiments. Volumes of covalent complexes were plotted versus protein concentrations. The percentages of covalent complexes could not be calculated because the free DNA and free [α-32P]dCTP were not separated far enough in the gel. Wild-type MutY (WT, solid circle), F261A (open triangle), R249A-MutY (solid triangle), F294A-MutY (open circle), GB-tagged MutY (GB1-WT, solid rectangle), and GB-F261A-MutY (open rectangle).
DNA binding affinities of mutant MutY proteins
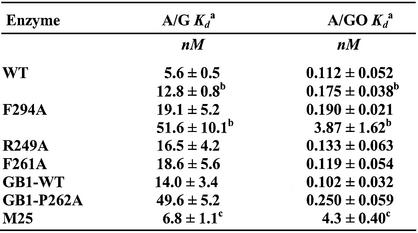
Using gel retardation assays, we compared the apparent dissociation constant (Kd) values of the wild-type and mutant MutY proteins with A/G and A/GO-containing DNA substrates. Binding assays were performed with fixed DNA concentrations and various MutY concentrations. The Kd values of the MutY mutants with both A/G and A/GO mismatches are presented in Table 2. For binding reaction at 37°C for 30 min, the Kd values of F294A, R249A and F261A mutants with A/G mismatches were ∼3-fold higher than that of the wild-type protein whereas the Kd values of these mutants with A/GO were comparable with that of the wild-type protein. The apparent Kd values of GB1-tagged wild-type MutY with 20mer DNA containing A/G and A/GO mismatches were 14.0 and 0.102 nM, respectively. On the other hand, the apparent Kd values of GB1-P262A-MutY with A/G- and A/GO-containing 20mer DNA were 49.6 and 0.250 nM, respectively. Compared with untagged MutY, the GB1-tagged wild-type MutY had a slightly higher Kd value with A/G mismatch but a similar Kd value with A/GO-containing DNA. GB1-P262A-MutY had slightly higher Kd values with A/G and A/GO mismatches than those of the wild-type GB1-MutY protein. Thus, to our surprise, the binding affinities with A/GO mismatches are not significantly reduced in all four MutY mutants.
Table 2. Apparent dissociation constants (Kd) of MutY mutants for mismatch-containing DNA.
aKd values were determined from three experiments using nine protein concentrations with 20mer oligonucleotides.
bBinding reactions are performed at 20°C for 30 min and others are performed at 37°C for 30 min.
cDissociation constants for a truncated MutY (M25) are derived from Li et al. (18).
As shown above, F294A-MutY had a defect in glycosylase activity at lower temperatures. Thus, we measured the apparent Kd values of the wild-type and F294A MutY proteins with A/G and A/GO-containing DNA at 20°C. When binding reactions were performed at 20°C, the apparent Kd values of the wild-type MutY with A/G and A/GO mismatches were 12.8 and 0.175 nM (Table 2, values with b), respectively, which are 2.3- and 1.6-folds of those measured at 37°C. The apparent Kd values for F294A-MutY with A/G and A/GO mismatches at 20°C were 51.6 and 3.87 nM, respectively, which are 2.7- and 20.4-fold of those measured at 37°C. At 20°C, F294A-MutY had Kd values with A/G and A/GO mismatches, respectively, 4- and 22-fold higher than those of the wild-type protein. These results indicate that the F294A mutation causes a severe defect in A/GO binding at 20°C. This is consistent with the glycosylase activity with A/GO-containing DNA results at lower temperatures (Fig. 5). Therefore, at lower temperatures, both DNA binding and glycosylase activities of the F294A mutant with A/GO mismatches are substantially weaker than those of the wild-type enzyme.
In vivo complementation activity of MutY mutants
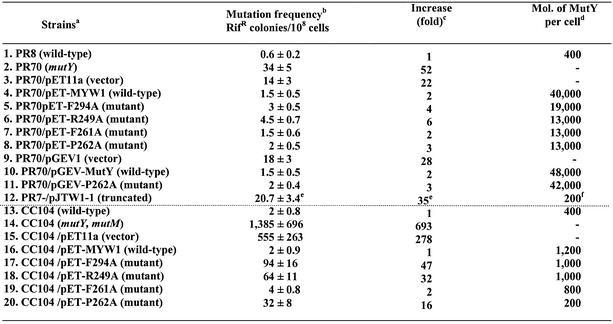
The E.coli mutY mutant is a mutator because it fails to correct replication errors. It has been shown that mutY mutants have higher mutation frequencies than wild-type cells (51) as measured by rifampicin resistance and Lac+ reversion (52,53). Rifampicin-resistant cells contain mutations in the gene of RNA polymerase. In the absence of functional MutY, a high level of mutations in the rifampicin binding site of RNA polymerase renders the cell resistant to rifampicin. To study the in vivo complementation activity of MutY mutants, recombinant plasmids of pET-F294A, pET-F249A, pET-F261A and pET-P262A were transformed into hosts of PR70(mutY)/DE3 and CC104/mutYmutM/DE3 cells. As shown in Table 3 (lines 1 and 2; columns 2 and 3), PR70 (mutY) had a 52-fold higher mutation frequency than wild-type PR8 cells. All four MutY mutants expressed in PR70/mutY/DE3 had mutation frequencies (lines 5–8; columns 2 and 3) comparable to the expressed wild-type MutY (line 4; columns 2 and 3). The PR70(mutY)/DE3 cells expressing the GB1-tagged wild-type and P262A MutY also had similar mutation frequencies (Table 3, lines 10 and 11; columns 2 and 3). Thus, all four mutant proteins expressed from overproducing vectors are functional in the PR70/DE3 strain. Even some of the mutant proteins have reduced binding and catalytic activities (see above). To test the possibility that over-expression of the mutant proteins, even without induction, is sufficiently high to overcome their weak biochemical activities, we then performed western blot analysis to determine their protein expression levels. By comparison with a known amount of purified MutY, the MutY molecules per cell were estimated. As shown in Table 3, the total protein expression levels of these untagged MutY mutants from plasmids in PR70/DE3 were lower than the levels of wild-type MutY protein expressed from this same vector (lines 4–8, last column); but the levels were at least 30-fold higher than MutY protein when it is expressed from the chromosome in strain PR8 (line 1, last column). Although the GB1-tagged P262A had a higher expression level and solubility than untagged protein, they both functioned similarly in vivo.
Table 3. Mutation frequencies of E.coli mutY and mutYmutM mutants expressing mutant MutY proteins.
aE.coli strains PR8 and CC104 contain wild-type mutY gene, PR70 has a mutated mutY gene, and CC104/mutYmutM is a double mutant of mutY and mutM. All strains contain λDE3 lysogen but the lysogen in CC104 derivatives is not properly established.
bThe in vivo activity of MutY is measured by the frequency of rifampicin-resistant colonies by averaging at least three separate experiments. The errors reported are the standard deviations of the averages.
cFold increase compared with the respective wild-type (WT) strains.
dMutY protein expression was detected by western blotting.
eThe mutation rate of PR70/pJTW1-1 is derived from Li et al. (18).
fThe expression level the truncated MutY in PR70/pJTW1-1 is estimated from a western blot presented by Li et al. (18).
The CC104/mutYmutM cells with double mutations at mutY and mutM genes have a very high mutation frequency (14) (Table 3, line 14; columns 2 and 3). Surprisingly, when the same plasmids containing the wild-type and mutant mutY genes were transformed into the CC104/mutYmutM/DE3 cells, different results were observed from those plasmids in the PR70/DE3 strain. While the mutation frequency of F261A was similar to that of the wild type; F294A, R249A and P262 mutants could not reduce the mutation frequency of CC104/mutYmutM to the wild-type level (Table 3, lines 16–20; columns 2 and 3). Among these mutants, the F294A is the most defective. Another surprising finding was that these proteins were not well expressed in the CC104/mutYmutM/DE3 strain. All four mutant and wild-type proteins were not visible by Coomassie Blue stain and could only be detected by western blotting (data not shown). Because CC104/mutYmutM is lacI–, the T7 RNA polymerase is constitutively expressed from the λDE3 lysogen and the gene under the control of the T7 promoter is expected to be constitutively expressed. Further investigation revealed that the λDE lysogen was not properly established in the CC104/mutYmutM strain. The tester T7 phage with a defective T7 RNA polymerase could only form very tiny plaques on the cells. Repeated isolation of a better lysogen was unsuccessful. Because the gene expression from pET11a is controlled by the T7 promoter, improper λDE lysogen resulted in poor expression of the proteins. By comparison to a known amount of purified MutY, wild type as well as F294, R249 and F261 MutY mutants were expressed as ∼1000 molecules per CC104/mutYmutM cell (Table 3, lines 16–19, last column), which were only 2–3 fold higher than the MutY protein expressed from its own chromosomal promoter in stain CC104 (Table 3, line 13, last column). The expression level of P262A-MutY was even 2-fold lower than that of wild-type CC104 cell. Therefore, the in vivo activities of the four mutants (Table 3, lines 17–20; columns 2 and 3) are consistent with their biochemical activities as described above only at low protein expression levels.
DISCUSSION
Because the C-terminal domain of MutY has been shown to play an important role in the recognition of GO lesions (18,24,27) and its structure has not been solved from limited NMR information (31,32,54), we have compared four MutY mutants at the C-terminal domain with the wild-type enzyme for catalytic activity and substrate recognition on A/G- and A/GO-containing DNA. F294A-MutY and R249A-MutY have weaker glycosylase and trapping activities than the wild-type MutY protein. The catalytic activity of F294A-MutY is almost abolished at 4°C. In addition, F294A-MutY has a severe defect in A/8-oxoG binding but has only slightly reduced binding to A/G mismatch at 20°C. The DNA glycosylase activity of GB1-P262A-MutY is weaker at low enzyme concentrations but is comparable to that of the wild-type enzyme at high enzyme concentrations. The biochemical activities of F261A-MutY are nearly similar to those of the wild-type enzyme. All four mutants expressed at high levels can complement mutY mutants in vivo; however, F294A, R249A and P262A, but not F261A, have partially defective in vivo activities when they are expressed at low levels. Among the four MutY mutants, F294A has the most defective in vivo activity in CC104/mutYmutM cells and has the most defective catalytic and binding activities with A/GO-containing DNA.
A very small amount of a truncated 249-residue polypeptide is expressed in the PR70 strain which contains a transposon insertion at the mutY gene on the micA68 allele (18). The expression level of the truncated 249-residue polypeptide from plasmid pJTW1-1, which contains the micA68 allele of the mutY gene with its own promoter in pBR322 (55), is estimated to be about 200 molecules/cell from a western blot in Figure 5 presented by Li et al. (18). The truncated 249-residue polypeptide has a lower binding affinity to A/GO mismatches than the intact MutY. Li et al. (18) showed that PR70 containing pJTW1-1 has a 35-fold higher mutation frequency than the wild-type PR8 cell (Table 3, line 12; columns 2 and 3). Their data are in agreement with the results presented here that the in vivo activities of the C-terminal MutY mutants can only be reflected by their in vitro biochemical activities when they are expressed at the levels comparable to that of wild-type protein expressed from the chromosome. The F294A, R249A and P262 mutations in the chromosomal mutY gene will probably lead to a mutator phenotype.
The sequence and structure of the C-terminal domain of MutY are similar to those of MutT (27,31) (Fig. 1). The dGTP binding site of MutT is located at a cleft defined by three β-strands A, C and D on one side, and loop I, the end of loop IV and the beginning of helix II on the other side (Fig. 2). F294 of MutY is analogous to F75 of MutT in β-strand C (Figs 1 and 2). F75 of MutT is exposed to water, points to the edge of dGTP binding cleft, and is involved in MutT substrate recognition (45). Our data indicate that F294 of MutY, corresponding to F75 of MutT, is also involved in GO recognition by hydrophobic interactions. Another hydrophobic residue, Tyr73 of MutT, is also implicated in binding to the nucleotide (45). The corresponding residue of MutY is His292. This region may undergo conformation change because a F294A mutation causes a severe defect in binding and catalysis at lower temperatures. R249A-MutY mutant, although not as severe as F294A, also has a similar phenotype as F294A-MutY. R249 of MutY is homologous to R23 of MutT in loop I and is on one side of the cleft wall (Fig. 2). F261 and P262 residues in loop I are located at the base of the cleft at the proximity to the corresponding reaction center of MutT. While F261A functions as well as the wild-type enzyme, P262A has reduced in vivo activity. This deficiency of P262A-MutY in vivo activity may be due to its poor solubility and slightly weaker binding and catalytic activities. The P262A mutation may increase the flexibility of loop I because the proline residue assumes only two fixed angles in the polypeptide. Our mutagenesis studies of MutY described here support the model that a region at the C-terminal domain of MutY corresponding to the cleft of MutT is involved in substrate recognition. However, because there are structural and functional differences between the C-terminal domain of MutY and MutT, these two proteins may bind the nucleotide differently. For example, MutY with a short loop I may have a more shallow cleft as compared to MutT. In addition, the C-terminal domain of MutY has no pyrophosphohydrolase activity. Because MutY does not cleave the phosphodiester bond on the G- or GO-strand, the domain of the MutT reaction center must be different in MutY.
It is interesting to note that mutants in the C-terminal domain of MutY and truncated MutY polypeptides bind to and react with the A/G mismatch differently. F294A, R249A and P262A MutY mutants have reduced binding and catalytic activities not only with A/GO but also with A/G mismatches. However, the truncated MutY without the C-terminal domain binds A/G mismatches and excises adenines from A/G mismatches in the same manner as the intact MutY (18,24,27). Li et al. (18) found that the truncated MutY (M25) has more than 30-fold lower binding affinity with an A/8-oxoG mismatch than the intact MutY but has a similar Kd value with an A/G mismatch as the intact MutY (18) (see last line in Table 2). In addition, deletion of the C-terminal domain of MutY does not affect the glycosylase activity with A/G-containing DNA although its catalytic activity on A/GO-containing DNA is much reduced (18,27). Thus, the C-terminal domain of MutY may have an additional function in A/G recognition. Our findings suggest that the C-terminal domain may modulate the N-terminal domain of MutY for DNA binding and catalytic activities besides GO recognition. The possible role(s) of the C-terminal domain may be (i) direct binding to GO, (ii) promoting base flipping of mismatched GO, (iii) facilitating conformation change of the catalytic domain, or/and (iv) promoting base flipping of mismatched A.
DNA glycosylases recognize damaged bases within a vast excess of normal DNA and then cleave the target nucleotides. How a DNA glycosylase searches for base damage remains unclear. Based on the structures of several glycosylases complexed to DNA or inhibitors, a pinch-push-pull mechanism has been proposed for DNA damage detection (56). The glycosylase scans the DNA substrate by compressing the DNA intra-strand phosphate distance and binds to the lesion site in a specific way that causes the DNA to kink. The enzyme then forces the target base to flip out the distorted DNA into a pocket on the enzyme surface. Base flipping makes the glycosyl bond accessible for cleavage. The specificity of the glycosylase on the target base is determined by base-flipping specificity of the push and by the chemical specificity of the pull by the binding pocket. In addition, it has been suggested that MutY uses its unique C-terminal domain to recognize the base opposite to the excised base and increase the overall specificity and catalysis (18,24,27). A double flip mechanism, with both bases of an A/GO mismatch flipped into the A and GO binding pockets, has been proposed for the MutY protein (27,31). Our results do not support a double flip mechanism. First, F294 at the edge of the cleft has strong interaction with the GO base; however, F261 and P262 at the base of the cleft have minimal contact with the G or GO base. If the orphan GO is flipped out into the cleft, F261 and P262 are expected to have contacts with the nucleotide. Second, there may be conformation changes in both the N- and C-terminal domains of MutY with bound DNA because the glycosylase activity of F294A with A/GO mismatch is more temperature-sensitive than that of the wild-type enzyme. Third, if MutY flips out the mismatched adenine and bends the DNA about 66° similarly to AlkA and hOGG1 (28), the GO base in its helical conformation in the wider minor groove may be easily accessed by the C-terminal domain of MutY (30). Finally, an interaction between the N- and C-terminal domains of MutY may exist. Some portions of the C-terminal domain of MutY may penetrate and interact with the N-terminal domain. This is consistent with a proposed clamp model (57) in which the DNA is embedded between the N- and C-terminal domains of MutY. Li et al. (18) showed that the C-terminal domain of MutY contacts five phosphates (one on the 3′ side and four on the 5′ side of the mismatched GO) and several purines on the GO-strand, and the N3 position of the mismatched adenine. They suggested that the C-terminal domain of MutY is positioned on the opposite side of the HhH motif, which binds to two phosphate groups on the 3′ end of the mismatched A. This clamp model is different from that of Volk et al. (31) regarding the orientation of the C-terminal domain. A detailed picture of how MutY specifically recognizes DNA substrates and then cleaves adenine awaits structural information.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ms Ruiqing Yang of our department for generating the figure of MutT structure, Mr Patrick Wright for critical reading of the manuscript, and Dr Marius Clore at NIH for the expression plasmid pGEV1. We also thank Drs Marice Fox and Jeffery H. Miller for providing E.coli strains. This work is supported by Grant GM 35132 from the National Institutes of General Medical Science, National Institutes of Health.
REFERENCES
- 1.Ames B.N. and Shigenaga,M.K. (1993) Oxidants are a major contributor to cancer and aging. In Halliwell,B. and Aruoma,O. (eds), DNA and Free Radicals. Ellis Horwood, New York, NY, pp. 1–18.
- 2.Wiseman H. and Halliwell,B. (1996) Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J., 313, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halliwell B. and Gutteridge,J.M.C. (1989) Free radicals in Biology and Medicine. Oxford University Press, New York.
- 4.Michaels M.L. and Miller,J.H. (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxo-guanine). J. Bacteriol., 174, 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tchou J. and Grollman,A.P. (1993) Repair of DNA containing the oxidatively-damaged base 8-hydroxyguanine. Mutat. Res., 299, 277–287. [DOI] [PubMed] [Google Scholar]
- 6.Lu A.-L., Li,X., Gu,Y., Wright,P.M. and Chang,D.-Y. (2001) Repair of oxidative DNA damage. Cell Biochem. Biophys., 35, 141–170. [DOI] [PubMed] [Google Scholar]
- 7.Maki H. and Sekiguchi,M. (1992) MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature, 355, 273–275. [DOI] [PubMed] [Google Scholar]
- 8.Tchou J., Kasai,H., Shibutani,S., Chung,M.-H., Grollman,A.P. and Nishimura,S. (1991) 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl Acad. Sci. USA, 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 10.Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J.M. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 29, 7024–7032. [DOI] [PubMed] [Google Scholar]
- 11.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 12.Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G·C to T·A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA, 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu A.-L., Tsai-Wu,J.-J. and Cillo,J. (1995) DNA determinants and substrate specificities of Escherichia coli MutY. J. Biol. Chem., 270, 23582–23588. [DOI] [PubMed] [Google Scholar]
- 14.Michaels M.L., Cruz,C., Grollman,A.P. and Miller,J.H. (1992) Evidence that MutM and MutY combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl Acad. Sci. USA, 89, 7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J. and Winkler,M.E. (2000) Reduction of GC → TA transversion mutation by overexpression of MutS in Escherichia coli K-12. J. Bacteriol., 182, 5025–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modrich P. and Lahue,R.S. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 17.Au K.G., Cabrera,M., Miller,J.H. and Modrich,P. (1988) Escherichia coli mutY gene product is required for specific A/G to C:G mismatch correction. Proc. Natl Acad. Sci. USA, 85, 9163–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Wright,P.M. and Lu,A.-L. (2000) The C-terminal domain of MutY glycosylase determines the 7,8-dihydro-8-oxo-guanine specificity and is crucial for mutation avoidance. J. Biol. Chem., 275, 8448–8455. [DOI] [PubMed] [Google Scholar]
- 19.Lu A.-L. and Chang,D.-Y. (1988) A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell, 54, 805–812. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q.M., Ishikawa,N., Nakahara,T. and Yonei,S. (1998) Esherichia coli MutY protein has a guanine-DNA glycosylase that acts on 7,8-dihydro-8-oxoguanine:guanine mispair to prevent spontaneous G:C to C:G transversions. Nucleic Acids Res., 26, 4669–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajiri T., Maki,H. and Sekiguchi,M. (1995) Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res., 336, 257–267. [DOI] [PubMed] [Google Scholar]
- 22.Michaels M.L., Pham,L., Nghiem,Y., Cruz,C. and Miller,J.H. (1990) MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res., 18, 3841–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai-Wu J.-J., Liu,H.-F. and Lu,A.-L. (1992) Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A·C and A·G mispairs. Proc. Natl Acad. Sci. USA, 89, 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gogos A., Cillo,J., Clarke,N.D. and Lu,A.-L. (1996) Specific recognition of A/G and A/8-oxoG mismatches by Escherichia coli MutY:removal of the C-terminal domain preferentially affects A/8-oxoG recognition. Biochemistry, 35, 16665–16671. [DOI] [PubMed] [Google Scholar]
- 25.Manuel R.C., Czerwinski,E.W. and Lloyd,R.S. (1996) Identification of the structural and functional domains of MutY, an Escherichia coli DNA mismatch repair enzyme. J. Biol. Chem., 271, 16218–16226. [DOI] [PubMed] [Google Scholar]
- 26.Manuel R.C. and Lloyd,R.S. (1997) Cloning, overexpression, and biochemical characterization of the catalytic domain of MutY. Biochemistry, 36, 11140–11152. [DOI] [PubMed] [Google Scholar]
- 27.Noll D.M., Gogos,A., Granek,J.A. and Clarke,N.D. (1999) The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine·adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry, 38, 6374–6579. [DOI] [PubMed] [Google Scholar]
- 28.Bruner S.D., Norman,D.P. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 29.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 30.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volk D.E., House,P.G., Thiviyanathan,V., Luxon,B.A., Zhang,S., Lloyd,R.S. and Gorenstein,D.G. (2000) Structural similarities between MutT and the C-terminal domain of MutY. Biochemistry, 39, 7331–7336. [DOI] [PubMed] [Google Scholar]
- 32.Volk D.E., Thiviyanathan,V., House,P.G., Lloyd,R.S. and Gorenstein,D.G. (1999) 1H, 13C and 15N resonance assignments of the C-terminal domain of MutY: an adenine glycosylase active on G:A mismatches. J. Biomol. NMR, 14, 385–386. [DOI] [PubMed] [Google Scholar]
- 33.Abeygunawardana C., Weber,D.J., Gittis,A.G., Frick,D.N., Lin,J., Miller,A.F., Bessman,M.J. and Mildvan,A.S. (1995) Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry, 34, 14997–15005. [DOI] [PubMed] [Google Scholar]
- 34.Lin J., Abeygunawardana,C., Frick,D.N., Bessman,M.J. and Mildvan,A.S. (1997) Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action. Biochemistry, 36, 1199–1211. [DOI] [PubMed] [Google Scholar]
- 35.Wright P.M., Yu,J., Cillo,J. and Lu,A.-L. (1999) The active site of the Escherichia coli MutY DNA adenine glycosylase. J. Biol. Chem., 274, 29011–29018. [DOI] [PubMed] [Google Scholar]
- 36.Huth J.R., Bewley,C.A., Jackson,B.M., Hinnebusch,A.G., Clore,G.M. and Gronenborn,A.M. (1997) Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci., 6, 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 38.Towbin H.T., Staehlin,T. and Gordon,J. (1979) Eletrophoretic transfer of proteins from polyacrylamide gel to nitrocellulose sheets procedure. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGoldrick J.P., Yeh,Y.-C., Solomon,M., Essigmann,J.M. and Lu,A.-L. (1995) Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol. Cell. Biol., 15, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradford M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 41.Lu A.L. (2000) Repair of A/G and A/8-oxoG mismatches by MutY adenine DNA glycosylase. In Vaughan,P. (ed.), DNA Repair Protocols: Prokaryotic Systems. Humana Press Inc., Totowa, NJ, pp. 3–16.
- 42.Leatherbarrow R.J. (1987) Enzfitter: A Non-linear Regression Analysis Program for IBM PC. Elsevier Science Publisher BV, Amsterdam.
- 43.Laemmli U.K. (1970) Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 44.Shimokawa H., Fujii,Y., Furuichi,M., Sekiguchi,M. and Nakabeppu,Y. (2000) Functional significance of conserved residues in the phosphohydrolase module of Escherichia coli MutT protein. Nucleic Acids Res., 28, 3240–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J., Abeygunawardana,C., Frick,D.N., Bessman,M.J. and Mildvan,A.S. (1996) The role of Glu 57 in the mechanism of the Escherichia coli MutT enzyme by mutagenesis and heteronuclear NMR. Biochemistry, 35, 6715–6726. [DOI] [PubMed] [Google Scholar]
- 46.David S.S. and Williams,S.D. (1998) Chemistry of glycosylase and endonuclease involved in base-excision repair. Chem. Rev., 98, 1221–1261. [DOI] [PubMed] [Google Scholar]
- 47.Porello S.L., Leyes,A.E. and David,S.S. (1998) Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry, 37, 14756–14764. [DOI] [PubMed] [Google Scholar]
- 48.Williams S.D. and David,S.S. (1999) Formation of a Schiff base intermediate is not required for the adenine glycosylase activity of Escherichia coli MutY. Biochemistry, 38, 15417–15424. [DOI] [PubMed] [Google Scholar]
- 49.Zharkov D.O. and Grollman,A.P. (1998) MutY DNA glycosylase: base release and intermediate complex formation. Biochemistry, 37, 12384–12394. [DOI] [PubMed] [Google Scholar]
- 50.Zharkov D.O., Gilboa,R., Yagil,I., Kycia,J.H., Gerchman,S.E., Shoham,G. and Grollman,A.P. (2000) Role for lysine 142 in the excision of adenine from A:G mispairs by MutY DNA glycosylase of Escherichia coli. Biochemistry, 39, 14768–14778. [DOI] [PubMed] [Google Scholar]
- 51.Lu A.-L. and Fawcett,W.P. (1998) Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from Schizosacchromyces pombe. J. Biol. Chem., 273, 25098–25105. [DOI] [PubMed] [Google Scholar]
- 52.Michaels M.L., Tchou,J., Grollman,A.P. and Miller,J.H. (1992) A repair system for 8-oxo-7,8-dihydrodeoxyguanine (8-hydroxyguanine). Biochemistry, 31, 10964–10968. [DOI] [PubMed] [Google Scholar]
- 53.Radicella J.P., Clark,E.A. and Fox,M.S. (1988) Some mismatch repair activities in Escherichia coli. Proc. Natl Acad. Sci. USA, 85, 9674–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.House P.G., Volk,D.E., Thiviyanathan,V., Manuel,R.C., Luxon,B.A., Gorenstein,D.G. and Lloyd,R.S. (2001) Potential double-flipping mechanism by E. coli MutY. Prog. Nucleic Acid Res. Mol. Biol., 68, 349–364. [DOI] [PubMed] [Google Scholar]
- 55.Tsai-Wu J.-J., Radicella,J.P. and Lu,A.-L. (1991) Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of MicA and MutY. J. Bacteriol., 173, 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mol C.D., Parikh,S.S., Putnam,C.D., Lo,T.P. and Tainer,J.A. (1999) DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys Biomol. Struct., 28, 101–128. [DOI] [PubMed] [Google Scholar]
- 57.Li X. and Lu,A.L. (2000) Intact MutY and its catalytic domain differentially contact with A/8-oxoG-containing DNA. Nucleic Acids Res., 28, 4593–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrin T.E., Huang,C.C., Jarvis,L.E. and Langridge,R. (1988) The MIDAS display system. J. Mol. Graphics, 6, 13–27. [Google Scholar]