Abstract
Comparisons of complete genome sequences allow the most objective and comprehensive descriptions possible of a lineage's evolution. This communication uses the completed genomes from four major euryarchaeal taxa to define a genomic signature for the Euryarchaeota and, by extension, the Archaea as a whole. The signature is defined in terms of the set of protein-encoding genes found in at least two diverse members of the euryarchaeal taxa that function uniquely within the Archaea; most signature proteins have no recognizable bacterial or eukaryal homologs. By this definition, 351 clusters of signature proteins have been identified. Functions of most proteins in this signature set are currently unknown. At least 70% of the clusters that contain proteins from all the euryarchaeal genomes also have crenarchaeal homologs. This conservative set, which appears refractory to horizontal gene transfer to the Bacteria or the Eukarya, would seem to reflect the significant innovations that were unique and fundamental to the archaeal “design fabric.” Genomic protein signature analysis methods may be extended to characterize the evolution of any phylogenetically defined lineage. The complete set of protein clusters for the archaeal genomic signature is presented as supplementary material (see the PNAS web site, www.pnas.org).
Keywords: taxonomic domains, Archaea, biological classification, protein clusters
Classification lies at the heart of biology, for it is the essential starting point to making sense of any complex system. At its most superficial, biological classification serves merely to group similar things. A second more difficult goal of taxonomy is to rank groups in a hierarchy such as the Linnaean binomial classification scheme. These groups and ranks were based on collections of characteristic properties, “signatures,” that codified relationships. Because the main purpose of this classification was pragmatic, to facilitate identification, there have been innumerable arguments over how to most usefully define these signatures. With Darwin, biological classification advanced significantly. Darwin criticized “our ignorance of what we are searching after in our natural classifications.” Thereafter, groups and ranks were to reflect evolutionary descent (1).
Bacterial taxonomy faced all of the problems of general biological classification plus new ones: bacterial cells seemed structurally homogenous (both internally and externally), they did not interbreed in any conventional manner, and they appeared to be geographically ubiquitous. As a result, bacterial taxonomy amounted to little more than a convenient filing system for the better part of this century (2). This taxonomic quagmire was resolved only by the introduction of techniques for macromolecular sequencing and comparison. The important lesson of molecular evolution was that a gene's history is recorded in its nucleotide sequence (3). Similarly, an organism's history is recorded in its complement of genes and their individual sequences. By the 1970s, small subunit ribosomal RNA sequences emerged as the basis on which a universal phylogenetic tree could be constructed (4). This tree reflected organismal evolution and, for the first time, an objective taxonomy based on Darwin's criterion of evolutionary descent became knowable. This phylogeny contradicted many classical groups and ranks, most notably the “prokaryotic” rank that joined the Archaea to the Bacteria (5). These two major groups of diverse microorganisms were no more related genetically to one another than either was to the Eukarya.
But this great advance came with a price, for the molecular approach based on the sequence of a single “representative” gene (whose function and distribution were universal) had made classification abstract. There were no physiological, ecological, or structural characteristics intrinsic to a group that was defined by single gene phylogeny. Therefore classical signatures were identified for the new groups and ranks. In the case of the Archaea, members share antibiotic resistances (6), characteristic modified nucleotides in tRNA (7), ether-linked isoprenoid lipids (8), proteinaceous cell walls (9), novel coenzymes (10), and unique structures of the DNA-dependent RNA polymerase (11). Although these results somewhat elaborate the history and nature of the group, they are not necessarily defining characteristics. Furthermore, some shared characteristics may be analogies rather than homologies (e.g., surface-layer proteins).
With the advent of complete genome sequences, phylogenetically derived groups can be described objectively and comprehensively by their shared gene complements. Herein we define an archaeal signature in terms of the set of genes that function uniquely within the archaeal lineage. Previous genetic and biochemical experiments have demonstrated that some of these genes function uniquely in the Archaea. Nevertheless, most signature genes, present in two or more complete archaeal genome sequences, have no known function and no homologs outside of the Archaea.
Previous works have compared full genome protein complements pairwise (12) or by summary signature by using heuristic confidence levels (13). By focusing our attention on clusters of proteins unique to Archaea and by incorporating published experimental results, we have collected the most definitive characters describing the archaeal lineage. This set was assembled through extensive manual editing of gene clusters computationally derived from protein sequence similarity data. The set described here is necessarily a conservative one, which we expect will grow with an increase in archaeal sequence data, in genetic/biochemical evidence, and in our understanding of the evolutionary process.
Genomic signatures describe taxa in intricate detail. Their breadth complements rather than replaces single molecular phylogenies, which are less expensive, better understood in function and structure, and directly comparable across huge evolutionary distances. Crucially, the signatures do not require a priori guesses as to which phenotypes best define the lineage. As stated above, the archaeal signature presented herein consists predominantly of uncharacterized genes, suggesting that we have much to learn about the workings of archaeal cells. Such signatures can be compiled for the other phylogenetic domains and groups as well. In the aggregate, signatures from all major lineages will be fundamental to understanding the evolution of modern cell types.
Materials and Methods
Identification of Open Reading Frames.
Assembled genomic DNA sequences from genome sequencing projects for Methanococcus jannaschii JAL-1 (14), Methanobacterium thermoautotrophicum ΔH (15), Archaeoglobus fulgidus VC-16 (16), Pyrococcus horikoshii OT3 (17), Pyrococcus furiosus (http://www.genome.utah.edu), Pyrococcus abyssi (http://www.genoscope.cns.fr/Pab/), Aeropyrum pernix K1 (18), Pyrobaculum aerophilum (http://genome.caltech.edu/pyrobaculum), and Sulfolobus solfataricus P2 (http://niji.imb.nrc.ca/sulfolobus) were analyzed for ORF protein coding regions (ORFs) by using the critica program (19). ORF assignments were edited and reconciled with previous annotation both automatically (using Perl scripts) and manually (on the basis of comparative analysis and gene overlap). Where these ORFs corresponded to published ORFs, the published identifier was used. Other ORFs derived from genome sequencing projects were identified by a local accession number. All ORF assignments are available on the WIT2 server (http://wit.mcs.anl.gov/WIT2).
Formation of Homologous Archaeal Protein Clusters.
Each ORF in the total set of archaeal proteins was compared pairwise against ORFs from other complete (and partial) genome sequences by using version 3 of the fasta program with the blosum50 matrix and default gap penalties (20). Matches with expectation values less than 1.0 × 10−20 were used to cluster additional ORFs to each query sequence. Significantly similar relatives of the newly introduced proteins were then added to the cluster and the recursive process continued until all significant relatives in the data set had been introduced into the cluster. From the full set of these transitive protein clusters, those clusters with only archaeal members (no bacterial or eukaryal homologs) were retained for further analysis. Genes from Pyrococcus OT3, P. furiosus, and P. abysii were considered interchangeable records of that lineage.
Confirmation and Editing of Clusters.
To verify the integrity of the automatically generated clusters, one or more members were compared against the GenBank nonredundant protein database at National Center for Biotechnology Information by using the blastp program (Ver. 2) with the blosum62 matrix and default gap penalties (21). Protein sequence alignments were produced for some clusters by using clustalw (22) to disambiguate and delimit membership in the clusters. Alignments were manually examined and improved by using the AE2 alignment editor [T. Macke, Ribosomal Database Project (http://www.cme.msu.edu/RDP)]. Clusters judged to contain similar bacterial or eukaryal homologs were removed from the set. Those clusters linked by substantial similarity over a single domain in member proteins were also discarded (23). Signature clusters containing short protein sequences missed by the automatic clustering system because of sequence length-dependent scoring (24) were introduced. Several proteins with substantial similarity to bacterial or eukaryal proteins, but known to function uniquely in Archaea, were added back into the clusters. Each cluster by definition contains proteins from at least two of the four euryarchaeal genomes. The final set of clusters is termed the “archaeal signature.” The complete set of data containing these signature clusters is available on the PNAS web site as supplementary material (www.pnas.org).
Functional Assignment of Clusters.
Where sufficient genetic, biochemical, or comparative evidence was presented in published literature to support attributing a function to members of the cluster, the signature clusters were so annotated. In the absence of such information, proteins were labeled “hypothetical.”
Results
The archaeal signature contains 351 clusters representing 1,149 archaeal protein-encoding genes. These signature genes account for a significant portion (9–15%) of genomic DNA in each organism (Table 1). For comparison, the ribosomal translation apparatus of M. jannaschii, including its two ribosomal RNA operons, is encoded by 3% of the genome.
Table 1.
Archaeal signature cluster statistics
| Statistic | M. jannaschii JAL-1 | M. thermoautotrophicum ΔH | A. fulgidus VC-16 | P. horikoshii OT3 | A. pernix K1 |
|---|---|---|---|---|---|
| Genome size, bp | 1,739,934 | 1,751,377 | 2,178,400 | 1,738,505 | 1,669,695 |
| DNA proportion encoding protein | 86% | 90% | 92% | 91% | 86% |
| DNA proportion encoding signature proteins | 15% | 12% | 9% | 9% | 3% |
| Total proteins predicted | 1,797 | 1,870 | 2,494 | 1,826 | 1,633 |
| Total proteins in archaeal signature | 19% (345) | 15% (289) | 11% (286) | 11% (201) | 3% (57) |
| Median molecular weight of all predicted proteins | 26,740 | 26,809 | 26,182 | 28,682 | 28,561 |
| Median molecular weight of all signature proteins | 25,837 | 25,174 | 22,096 | 25,147 | 22,053 |
| Signature clusters represented | 81% (290) | 70% (252) | 62% (222) | 57% (203) | 22% (76) |
Genome sizes include all extrachromosomal elements identified in the sequenced strain. Protein encoding regions do not include regulatory regions or terminator codons. Coding region proportions are relative to complete genome size. Percentages of signature clusters represented in each organism are calculated relative to the complete set identified here (351 clusters).
Most genes in the signature are classified in the “hypothetical” category and have no known function (Table 2). This is not necessarily surprising in that most archaeal genes have been identified by studies of homologous bacterial and eukaryal proteins. Signature proteins, by definition, have no counterparts in these more extensively studied systems. This “hypothetical” subset includes 12 clusters of ATP-/GTP-binding proteins (defined by a common P-loop motif). The next largest subset comprises the methanogenesis pathway in M. jannaschii and M. thermoautotrophicum and the related methyl oxidation pathway of A. fulgidus. Ten clusters of DNA or RNA processing enzymes are unique to the Archaea. By definition, the clusters include no universally conserved proteins; translation, transcription, central metabolism, cell division, amino acid, and nucleotide biosynthesis are largely absent.
Table 2.
Functions assigned to major groups of archaeal signature clusters
| Functional category | Number of clusters in archaeal signature | Proportion of signature, % |
|---|---|---|
| Hypothetical | 283 | 81 |
| C1 transfer enzymes and cofactor biosynthesis (methanogenesis) | 34 | 10 |
| DNA/RNA processing (replication, transcription, translation) | 10 | 3 |
| DNA binding (helix-turn-helix) | 8 | 2 |
| Flagellar biosynthesis | 7 | 2 |
| Other | 9 | 3 |
| Total | 351 |
Although most proteins in these signature clusters have no close homologs outside of the archaeal genomes, some signature proteins are functionally diverged from their homologs in bacterial or eukaryal genomes. For example, genes encoding the methyl reduction pathway of methanogenesis are homologous to those encoding oxidative tetrahydromethanopterin-/methanofuran-dependent proteins in a methylotrophic bacterial species, Methylobacterium extorquens AM1 (25). Despite these genes' presence in M. extorquens, they are included in the signature because their ancestral operation in a reductive methanogenic direction (in deeply branching euryarchaeal organisms) fits the criterion of unique function in the archaeal lineage. In contrast, subunits of the archaeal acetyl-CoA/carbon monoxide dehydrogenase enzymes are similar to clostridial subunits and operate in the same synthetic direction under autotrophic methanogenesis. These are excluded from the signature.
For several proteins, available tools for primary sequence comparison are too imprecise to decide functional relationships. The euryarchaeal histones might be described as uniquely archaeal because of their divergent primary amino acid sequences. Yet careful study has demonstrated functional equivalence and tertiary structure similarity to eukaryal histone subunits (26). Thus they are excluded from the archaeal signature.
The median molecular weight of predicted proteins in the archaeal signature is less than that expected from a random sampling of archaeal proteins from their corresponding genomes. Although this difference is small, it is significant. (Median molecular weights of bootstrap samples of a comparable set of proteins were greater than that of the signature set in 83–99% of the replicates for each genome.) The largest ORFs in each genome are not members of the signature: some of these excluded proteins are universal in distribution, some contain intein insertion elements, and some are hypervariable structural proteins such as paracrystalline surface layer proteins. Yet the absence of these extremely large proteins from the signature may not completely account for the size shift; the signature set may also be enriched for short proteins such as transcription factors or electron carriers.
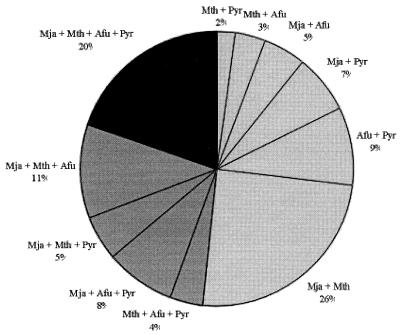
Each of the four euryarchaeal genomes is represented in at least 50% of the signature clusters (Table 1). Twenty percent of the clusters contain at least one ORF from each genome, whereas another 28% have members in three of the four genomes (Fig. 1). Among the set of clusters found in two archaeal genomes, the M. jannaschii + M. thermoautotrophicum relationship stands out, accounting for a quarter of the signature clusters (Fig. 1). In addition to the recognized contributions of methyl-coenzyme M reductase and methyltransferase subunits, this subset probably contains unrecognized proteins integral to the methanogenic physiology. Despite superficial similarities in their shared heterotrophic physiologies, A. fulgidus and Pyrococcus spp. share exclusively only one-third the number of signature clusters.
Figure 1.
Distribution of signature clusters in four euryarchaeal genomes. Mja, M. jannaschii; Mth, M. thermoautotrophicum; Afu, A. fulgidus; Pyr, Pyrococcus spp.
Our compilation of signature proteins treats all Pyrococcus spp. ORFs as representatives of the same lineage. Although there are differences among the three strains, the addition of signature proteins from P. furiosus introduces 23 clusters not represented by ORFs observed in Pyrococcus sp. OT3. Signature proteins from P. abyssi then add one more cluster. The predominant differences among Pyrococcus spp. are DNA translocations (27) and loss (or gain) of some biosynthetic genes (28).
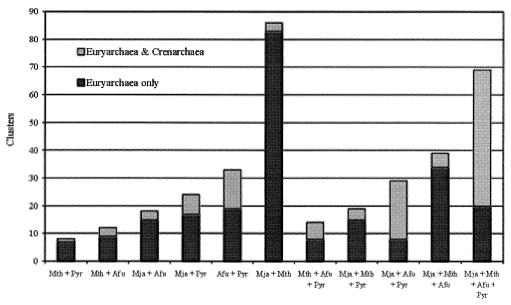
Partial and complete genome sequences from three Crenarchaea show that at least one-third of the euryarchaeal signature clusters have crenarchaeal homologs. The distribution of these homologs varies significantly across signature categories (Fig. 2). Approximately 70% of the clusters with proteins in all four Euryarchaea contain crenarchaeal homologs. Crenarchaeal sequences have a similar representation in the set of clusters found in M. jannaschii, A. fulgidus, and Pyrococcus spp. but not M. thermoautotrophicum. Only the group of clusters shared specifically by M. jannaschii and M. thermoautotrophicum has a very small crenarchaeal proportion (3%).
Figure 2.
Portions of each category of signature clusters (in Fig. 1) with crenarchaeal homologs. Organism abbreviations are as in Fig. 1.
Discussion
This set of archaeal signature genes upsets the classical view of what best distinguishes members of the Archaea from Bacteria or Eukarya. Antibiotic resistance factors, cell-wall proteins, and most of the information-processing system are missing from the signature. In their stead are key energetic systems, cofactor biosynthesis, and many uncharacterized genes. Almost 15% of the proteins encoded by each archaeal genome sequence are members of the signature and therefore unique to the Archaea. Such a high proportion of unique genes in the archaeal lineage is inconsistent with a random assortment mechanism, in which any gene could be transferred, lost, or replaced. Therefore, this signature supports the phylogenetic conclusion that the Archaea are an anciently diverged major lineage, containing a substantial proportion of unique genes.
These signature clusters necessarily emphasize the euryarchaeal component because only one crenarchaeal genome sequence has been completed to date. Nevertheless, preliminary comparisons between these euryarchaeal signature clusters and proteins from crenarchaeal genomes show that half of all signature clusters containing at least three euryarchaeal members also contain crenarchaeal members. We expect that complete genome sequences from several crenarchaea will introduce new signature clusters: some will be specific to the crenarchaea, whereas others will include single euryarchaeal genes.
Groups of clusters (Fig. 1) show the fundamental nature of the signature set. In the absence of strong selective pressures, we expect that related lineages would share progressively fewer unique proteins over time. This erosion is most apparent in those signature proteins found in only two archaea: M. thermoautotrophicum and M. jannaschii share a significant number of genes (presumably required by their methanogenic biochemistry), whereas M. thermoautotrophicum shares relatively few genes exclusively with Pyrococcus spp. or with A. fulgidus. Yet, a substantial core of euryarchaeal genes remains, and it is well represented in the Crenarchaea.
Synthesis.
Given a long evolutionary history, with ample opportunity for horizontal genetic exchange and erosion of the shared gene complement, why should so many genes be uniquely associated with a single lineage? The chromosome of Escherichia coli has been estimated to acquire 31 kbp of DNA every million years (29). Innumerable cloning experiments have shown that most DNA can be artificially propagated and expressed in E. coli. Nevertheless, with 15% of their proteins functioning uniquely in Archaea, the four Euryarchaea examined here support the existence of long-term constraints on gene assimilation or loss. The addition of new diverse sequence data to the molecular databases (e.g., Thermotoga maritima) has had relatively little impact on the archaeal core signature. Therefore we are left with the question of the relationship between these signature genes and the cell.
Cells may not incorporate new genes for a variety of reasons including: (i) the cells have intrinsic barriers to transfer (codon usage, restriction modification systems, and regulatory apparatus) that hinder gene acquisition and expression; (ii) they already have functional equivalents; (iii) they cannot use them (because of cofactor or substrate requirements or environmental conditions); or (iv) the new genes would be detrimental (futile cycles, antibiotic or bacteriophage sensitivity, etc.). These constraints are a reflection of the cell's “design fabric,” the product of all evolutionary commitments that the lineage has made. Although these constraints alone do not preclude a gene's horizontal transfer, the recipient must have a compatible design fabric or must recruit the genes to a new pathway if they are to become a useful part of that cell.
A previous paper, “The Universal Ancestor,” proposed a model of genomic evolution based on the successive “crystallization” of differentiated cellular subsystems (30). That model posited that organisms at one time were so simple, so uncomplicated in their overall design, and their components so simple and modular in their structures and functions that most, if not all, of an organism's genes could be exchanged with other organisms and were in effect shared communally. Through evolution, proteins and their complexes became more specific, catalytically efficient, and precisely defined. They also became less modular, and their genes became less interchangeable: they “crystallized out” of the horizontal gene exchange pool. The emerging constraints fostered congruent histories among many genes. Because this “crystallization” process was gradual, many genes could have become fixed in the genome only after the divergence of organisms into fundamentally distinct lineages. This hypothesis thus predicts the existence of signature genes—genes that are uniquely essential to or function only in the context of a cell's design fabric.
The Universal Ancestor model also predicts a continuum of gene specialization in an organism, ranging from genes integral to the design fabric to genes constrained by the fabric to genes peripheral to the fabric. Besides explaining the observed congruence among universally distributed genes involved in transcriptional and translational systems, this idea predicts that some genes may have become integral to the design fabric only during the evolution of the various modern cell types. Proteins in the archaeal signature support this hypothesis. For example, archaeal ribosomes contain proteins and RNA homologous to eukaryotic and bacterial versions. Ribosomal protein LX, however, is unique to archaeal ribosomes and may functionally differentiate the archaeal translational system from all others. Another protein acts on archaeal tRNAs to convert a specific guanosine nucleotide to archaeosine, a hypermodified derivative. Both ribosomal protein LX and the archaeosine insertion enzyme may function uniquely in the context of evolutionary constraints imposed by the design fabric within which they operate.
This strategy of identifying genes that function uniquely in a lineage can be applied to any phylogenetically related group of organisms. The comprehensive nature of genomic analysis brings an unprecedented objectivity to describing cell lineages: genomics raises taxonomy to a new level. Whereas earlier taxonomies identified and related organisms, the new taxonomy will elaborate those relationships, allowing the biologist to see the essential character of a group and (to some extent) the mode of that group's evolution.
Supplementary Material
Acknowledgments
We are grateful to Nikos Kyrides, Norman Pace, Roger Garrett, and William Whitman for their comments on and interest in this project. The project was supported by National Aeronautics and Space Administration Grant NAG5–8479 to C.R.W. and G.J.O. and by a University of Illinois Distinguished Fellowship and National Institutes of Health Cellular and Molecular Biology Training Grant to D.E.G.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050564797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050564797
References
- 1.Darwin C. Letter to George Robert Waterhouse. Cambridge, U.K.: Cambridge Univ. Press; 1843. [Google Scholar]
- 2.Woese C R. Microbiol Rev. 1994;58:1–9. doi: 10.1128/mr.58.1.1-9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuckerkandl E, Pauling L. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 4.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese C R, Fox G E. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böck A, Kandler O. The Bacteria. VIII. New York: Academic; 1985. pp. 525–544. [Google Scholar]
- 7.Gupta R, Woese C R. Curr Microbiol. 1980;4:245–249. [Google Scholar]
- 8.Langworthy T A. The Bacteria. VIII. New York: Academic; 1985. pp. 459–497. [Google Scholar]
- 9.Kandler O, König H. In: The Biochemistry of Archaea (Archaebacteria) Kates M, Kushner D J, Matheson A T, editors. Amsterdam: Elsevier; 1993. pp. 223–259. [Google Scholar]
- 10.DiMarco A A, Bobik T A, Wolfe R S. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 11.Zillig W, Stetter K O, Schnabel R, Thomm M. In: The Bacteria: A Treatise on Structure and Function. Gunsalus I C, editor. Vol. 8. New York: Academic; 1985. pp. 499–524. [Google Scholar]
- 12.Snel B, Bork P, Huynen M A. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 13.Gaasterland T, Ragan M A. Microbiol Comp Genomics. 1998;3:177–192. doi: 10.1089/omi.1.1998.3.177. [DOI] [PubMed] [Google Scholar]
- 14.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1017–1140. [Google Scholar]
- 15.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klenk H-P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 17.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 18.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. , 145–152. [DOI] [PubMed] [Google Scholar]
- 19.Badger J H, Olsen G J. Mol Biol Evol. 1998;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 20.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson W R. Protein Sci. 1995;4:1145–1160. doi: 10.1002/pro.5560040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins J F, Coulson A F, Lyall A. Comput Appl Bioscience. 1988;4:67–71. doi: 10.1093/bioinformatics/4.1.67. [DOI] [PubMed] [Google Scholar]
- 25.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 26.Pereira S L, Grayling R A, Lurz R, Reeve J N. Proc Natl Acad Sci USA. 1997;94:12633–12637. doi: 10.1073/pnas.94.23.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suckow J M, Suzuki M. Proc Jpn Acad. 1999;75(B):10–15. [Google Scholar]
- 28.Hoaki T, Nishijima M, Kato M, Adachi K, Mizobuchi S, Hanzawa N, Maruyama T. Appl Environ Microbiol. 1994;60:2898–2904. doi: 10.1128/aem.60.8.2898-2904.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence J G, Ochman H. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 30.Woese C. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.