Abstract
Expression analysis using microarray technology implies a complex experimental procedure with a large number of parameters affecting the final result. We have demonstrated that optimization of such a complex protocol can be far better handled using design of experiments (DOE) than by working on a single parameter at a time. Based on the results of a screening design, we developed a spotting buffer composed of formamide, betaine and nitrocellulose. This buffer provides a 2-fold increase in signal-to-background ratio compared to 3× SSC. Comparison to seven other buffers tested on 10 different substrates revealed it had the highest sensitivity. DNA dissolved in this buffer can be spotted on epoxysilane-coated microscope slides at a density of up to 70 000 spots per slide. A second DOE approach characterized the RNA labeling process with regard to the concentration of fluorescent dyes, dNTPs and reverse transcriptase. Adjust ments of the concentrations of dNTPs, as well as reverse transcriptase, towards the optimum, produced an improvement in the performance of the labeling procedure by a factor of 3 (Cy3) and 10 (Cy5). These results demonstrate that the process of establishing a stable expression profiling protocol and its further optimization can be significantly shortened and improved by DOE.
INTRODUCTION
Microarray technology has become a widespread tool in functional genomics. cDNA arrays allow a comprehensive view of biological systems by measuring the expression of thousands of genes simultaneously (1). Using this approach, the workload for the selection of interesting genes from a large pool is greatly reduced. The increasing interest in microarrays in recent years has led to a number of studies establishing the necessary protocols (2,3).
One of the major recurrent problems is the overall complexity of the experimental approach. All parts of the protocol, e.g. probe amplification, array production, target labeling and hybridization, include a multitude of parameters which can be and need to be optimized to reach stable experimental results (2). Although the increasing number of commercial products generally helps to improve the situation by standardizing conditions, a laboratory establishing expression profiling based on cDNA arrays still needs to invest a substantial amount of time and resources.
The performance of such a complex protocol will be governed by the settings of all factors having an effect on this system. These ‘settings’ could be a concentration, e.g. of a spotting buffer component that has an influence on the intensity and sensitivity of a cDNA spot. Often not only a factor by itself will exert influence on the system, but the interactions between two or more factors may play an important role in the final response as well. The combination of two spotting buffer components can have a beneficial effect on the intensity even though the single components would not increase or could even decrease this value and thus the sensitivity of a spot.
An initial problem when optimizing a system with a large quantity of parameters influencing the result is the choice of factors that are considered important. This choice is often based on beliefs rather than on actual facts, largely due to the still incomplete knowledge of the physical aspects of microarrays (4,5). Choosing more than two or three factors to work on tremendously increases the number of necessary experiments, especially if factors are considered in a sequential fashion (6). This sequential approach will also completely ignore all interactions between factors and is biased towards parameters that are considered first. Another difficulty arises when searching for optimal concentration ranges for the significant parameters of an already established protocol. Often one parameter at a time is varied even though this approach is known to neither yield the maximum amount of possible information nor to provide an efficient way of finding an optimal setting (6).
These considerations strongly suggest a more sophisticated approach. Design of experiments (DOE) is especially suited for situations where parameter selection and optimization is the goal (7). So far, these methods have only sparsely been applied to protocol optimization in the microarray community (8), even though DOE is a common approach to comparable problems in other scientific fields (9–11). Similar mathematical methods have indeed been employed for microarray data analysis in the past (12), and recently the method is more and more often suggested as a tool to efficiently organize larger hybridization series (13).
DOE provides experimental schemes that are mathematically ‘optimal’ in a sense that they are not biased towards any of the factors and directly link the number of experiments to the amount of information that will be gained. The number of experiments can be reduced to the degree where only the direct (primary) effect of each factor can be determined or they can be increased to a level where all other interaction effects are determinable, too. Thus, it is easy to decide between a ‘screening’ experiment that will isolate factors with strong influence and experiments that will cover the system to a greater detail, like a ‘response surface’ design.
An important influence on the type of DOE scheme and the number of experiments is the expected relationship between factors and results. Assuming a linear relationship allows the study of each factor at only two different levels, since two points are sufficient to determine a straight line. To discover the influence of a factor assumed to have a quadratic relationship, one needs to characterize each factor at additional levels, thus necessitating a greater number of experiments. On the other hand, this will allow a correct prediction of the influence of this factor over a wider range of settings, whereas a linear relationship might easily fail outside of the investigated range (7). If one assumes that all factors do have a linear influence on the result, the number of parameters to be determined based on the generated data is greatly reduced. Though this is mostly a rough estimation and the relationship between variables and result might be as well quadratic or even more complex, this approximation will often be sufficient.
Here, we demonstrate that the use of DOE for the screening of microarray spotting buffer components and the optimization of sample labeling leads to a significantly improved experimental protocol.
MATERIALS AND METHODS
DOE schemes
The schemes provided by DOE use a generic encoding for the factor levels that are studied to make it applicable in many situations. To be independent of specific concentrations, these levels refer to a standard concentration and a specific slope just as in any linear equation. The standard concentration is encoded as level 0, while level 1 represents the standard concentration plus the slope. Level –1, for example, would represent a concentration of 4 U/µl if it were used in the context of a standard concentration of 5 U/µl and a slope of 1 U/µl.
cDNA libraries and probe preparation
For the screening of spotting buffer components four clones containing vectors with inserts specific for the human genes GCK, CDKN1B, TNFRSF10B and a human EST (IMAGp998G11343) were obtained from the German Resource Center for Genome Research (RZPD, Berlin, Germany). Amplification of the inserts was performed by an 80 µl PCR using modified M13 primers (MWG Biotech, Ebersberg, Germany). The PCR products were purified by isopropanol precipitation and dissolved in 30 µl of spotting buffer.
To screen for favorable components of this buffer, eight commonly used spotting buffer components were selected (Table 1). Out of these eight substances, 64 different mixtures were generated according to a factorial screening design [plan 6A.16 from Cochran and Cox (7)]. For each mixture, each of the eight substances was set to one of two possible levels: either the component was present in the concentration as given in Table 1 (level 1) or not (level –1). The PCR product of the four genes mentioned above was dissolved in each buffer and each gene/buffer combination was spotted in triplicate onto poly-l-lysine coated slides, as previously described (14).
Table 1. Spotting buffer components.
| Component | Reference | Concentration in Reference | Concentration in screening | P-value | Summed estimate |
|---|---|---|---|---|---|
| DMSO | Hedge et al. (2) | 50% (v/v) | 50% (v/v) | <1E–16*** | 377.9 |
| Tris–HCl | Hedge et al. (2) | 20 mM (pH 8.0) | 20 mM (pH 8.0) | 0.0055** | 257.6 |
| KCl | Hedge et al. (2) | 50 mM | 100 mM | 0.4797 | –0.1 |
| SDS | Lampel,S., Toldo,S. and Lichter,P. (unpublished results) | 0.1% (w/v) | 0.1% (w/v) | 0.0366* | –430.3 |
| SSC | Schena et al. (23) | 3× SSC | 3× SSC | 0.0426* | 98.2 |
| Betaine | Diehl et al.(22) | 1.5 M | 0.5 M | 1.09E–11*** | 867.4 |
| Formamide | – | – | 25% (v/v) | 1.53E–05*** | 249.7 |
| Nitrocellulose | Pinkel et al. (24) | 0.4 µg/µl in DMSO | 0.5 µg/µl | 0.0044** | 145.1 |
The solutions were tested as spotting buffer components in the screening design. The corresponding references and the concentrations at which they were employed are given as well. The final columns show the experimental results. P-values are given and coded according to their values for a better overview. The last column shows the summed effect estimates over all eight result vectors. The whole model had a P-value <1E–16.
Significance codes: ***, 0 < P < 0.001; **, 0.001 < P < 0.01; *, 0.01 < P < 0.05.
The performance of the buffer resulting from this screening was tested with 95 murine cDNA species picked from a commercially available 20K library (arrayTAG; LION bioscience AG, Heidelberg, Germany). Amplification was performed as given above. Here, the spotting buffer generated in the screening experiment was compared against seven other spotting buffers, as summarized in Table 2, on 10 different commercially available microarray substrates (Table 3) representing all common surface chemistries (aldehyde, epoxysilane, aminosilane, poly-l-lysine).
Table 2. Substrates used for microarray printing.
| Substrate | Supplier | Coating |
|---|---|---|
| SigmaScreen coated slides | Sigma | Aminopropyltriethoxysilane |
| Silane-Prep slides | Sigma | Aminoalkylsilane |
| Poly-Prep slides | Sigma | Poly-l-lysine |
| CreativeChip PCR slides | Eppendorf | Epoxysilane |
| SuperAldehyde substrates | Telechem | Organoaldehyde |
| SuperAmine substrates | Telechem | Organoamine |
| QMT Epoxy slides | Quantifoil Micro Tools | Epoxysilane |
| QMT aldehyde slides | Quantifoil Micro Tools | Organoaldehyde |
| GAPSII coated slides | Corning | Gamma-aminopropylsilane |
| SAL-1 slides | Asper Biotech | 3-Aminopropyltrimethoxysilane + 1,4-phenylenediisothiocyanate |
Table 3. Spotting buffers and their constituents.
| Name | Supplier | Composition | Reference |
|---|---|---|---|
| 3× SSC | Saline sodium citrate | (16,23) | |
| 3× SSC + 1.5 M betaine | Saline sodium citrate, betaine | (22) | |
| Micro Spotting Solution | Telechem | ND | http://arrayit.com/ |
| Micro Spotting Solution Plus | Telechem | ND | http://arrayit.com/ |
| QMT Spotting Solution I | Quantifoil Micro Tools | ND | http://www.quantifoil.com/ |
| GSS Amine Spotting Solution | Genetix Ltd | ND | http://www.genpakdna.com/ |
| GSSA Aldehyde Spotting Solution | Genetix Ltd | ND | http://www.genpakdna.com/ |
| FBNC buffer | Formamide 25% (v/v), nitrocellulose (0.5 µg/µl), betaine (0.5 M), pH 6.0 | This work |
ND, no data available.
For the response surface design to optimize RNA labeling as well as in the corresponding validation experiment, microarrays of 4500 human cDNA clones were used (a complete list of these clones is available at http://www.dkfz-heidelberg.de/kompl_genome/other/doe1.txt). The array production was performed according to the conclusions drawn from the first experiments. Formamide, betaine and nitrocellulose (FBNC) was chosen as spotting buffer and epoxysilane-coated slides as spotting substrate. FBNC is obtained by mixing 2.5 ml of formamide (Merck Eurolab, Darmstadt, Germany) with 250 µl of nitrocellulose (Sigma, München, Germany) dissolved at 20 µg/ml in DMSO (Merck). Finally, 2 ml of 2.5 M betainehydrochloride (pH 6.0, 23°C; Sigma) solution and 5.25 ml of H2O were added.
DNA spotting was performed using an OmniGrid Microarrayer (GeneMachines, San Carlos, USA) equipped with Stealth SMP3 Micro Spotting Pins (Telechem, Sunnyvale, CA, USA).
Post-processing of microarrays
DNA adhesion to the glass surface was usually achieved by 3 h of incubation at 80°C, followed by irradiation with UV light at 254 nm with an energy output of 2 × 120 mJ/cm2 in a Stratalinker Model 2400 UV illuminator (Stratagene). Prior to hybridization, slides were washed for 2 min in 0.2% SDS (w/v), followed by 2 min in ddH2O at room temperature and 2 min in heated ddH2O (95°C) (15).
Poly-l-lysine slides were instead treated according to the procedures given by DeRisi et al. (16). QMT Epoxy slides (Quantifoil Microtools, Jena, Germany), SAL substrates (Asper Biotech, Tartu, Estonia) and slides with aldehyde surfaces were treated according to the manufacturers’ instructions.
RNA extraction
RNA was extracted from the human cell line HL60 and the murine cell lines 3T3 and F9 (17–19), as well as from TPA-differentiated HL60 cells (20), 4 days after induction. Growth conditions were set according to the references.
Total RNA was isolated according to a protocol that combines both Trizol reagent (Invitrogen) and RNeasy Midi spin columns (Qiagen, Hilden, Germany). Poly-A RNA was extracted from total RNA using Oligotex resin purification (Qiagen).
Sample preparation
Samples for hybridization were processed by preparing mRNA from total RNA and labeled by reverse transcription. The labeling reaction was performed in a volume of 20 µl, containing 1× Omniscript buffer (Qiagen), 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.2 mM dTTP, 25 ng/µl oligo-dT12–18 (Amersham), 6 U Omniscript reverse transcriptase (Qiagen) and 2 U RNase inhibitor (Stratagene). Cy3- or Cy5-labeled dUTP (Perkin Elmer) was used at a concentration of 0.1 mM to incorporate a fluorescent marker in the resulting cDNA.
After labeling, both cDNA samples were combined, purified and concentrated to 10 µl using a Microcon YM-30 PCR filter unit (Millipore, Eschborn, Germany), as previously described (15).
DOE was employed to improve this initial protocol using a response surface design. Prior to this design, several reverse transcriptases [Omniscript (Qiagen), Superscript (Invitrogen), Superscript II (Invitrogen) and Fluoroscript (Invitrogen)] were compared to isolate the best enzyme among these. Based on that experiment, the DOE experiment investigated RNA labeling on the basis of the Fluoroscript cDNA Labeling System. Some modifications of the manufacturer’s standard protocol had to be introduced for the optimization of RNA labeling. Three components (concentrations of Fluoroscript, dNTPs and Cy-dUTP) that were assumed to have a major influence on the result of the labeling reaction were set variable. The initial concentration at level 0 (0.75 U/µl for Fluoroscript, 1/20 of stock solution for dNTPs and 0.1 mM for Cy-dUTP) was increased by 50, 30 and 35% for level 1, respectively. The response surface design suggested 16 different reaction mixtures [central composite design, according to the statistical software JMP (SAS, Heidelberg, Germany)].
Microarray hybridization
Purified, dye-labeled and pooled cDNA was mixed with 120 µl of UltraHyb hybridization buffer (Ambion, Huntingdon, UK), preheated to 50°C, and applied to pre-heated (50°C) microarrays mounted in a GeneTAC Hybridization Station (Genomic Solutions, Ann Arbor, USA). Hybridizations were performed for 16 h at 50°C with gentle agitation. Thereafter, the slides were automatically washed at room temperature with: (i) 0.5× SSC, 0.1% (w/v) SDS for 5 min; (ii) 0.05× SSC, 0.1% (w/v) SDS for 3 min; (iii) 0.05× SSC for 2 min; (iv) 70% (v/v) ethanol for 30 s; (v) 100% ethanol for another 30 s, and finally, air dried for 5 min.
Hybridizations
Twenty micrograms of total RNA from HL60 cells was labeled and hybridized against RNA from TPA induced HL60 cells on the array type which was produced for the initial screening of spotting buffer components. The result of this experiment was validated by hybridizing 10 or 50 µg of total RNA from 3T3 cells against 10 or 50 µg of total RNA from F9 cells, respectively. Each amount of RNA was hybridized six times on each slide type (i.e. different surface chemistries).
Ten micrograms of total RNA from HL60 cells was hybridized against the same amount of RNA from TPA differentiated HL60 cells for the initial comparison of reverse transcriptases. Each labeling variant was tested with three slides. The same type of RNA was hybridized for the following response surface design and the corresponding validation experiment. The 16 different reaction mixtures were characterized using 2-fold replication, with the only difference of inverted dye labeling for the repeated experiment. Thus, a total of 32 experiments were conducted. To validate the predictions of the resulting model, each labeling condition (Omniscript, Fluoroscript and Fluoroscript modified according to the model prediction) was tested with six slides.
Data acquisition, processing and analysis
Hybridized microarrays were scanned and analysed using a GenePix 4000B microarray scanner (Axon Instruments, Union City, USA). All relevant data were measured for each slide using the software GenePix Pro 3.0 (Axon).
Concerning the optimization of the spotting buffer, the background corrected intensity was selected as the performance measure. The hybridization results were divided into different result vectors, each vector containing the corrected intensities of all spots of one gene in the Cy3- or Cy5-specific channel. Separation of genes and channels was necessary since some of the genes were differentially expressed. With each gene being dissolved in 64 different buffers and spotted in triplicate, the process resulted in a matrix of 192 × 8. As a second measure of interest, spot size was processed in the same way.
A similar performance measure was calculated for the optimization of RNA labeling. The median signal/background ratio for all spots of an array was determined for each channel and sorted into result vectors, according to the type of fluorescent marker of the HL60 control cDNA. Thus, having two separate color channels and 16 experiments, replicated once with reverse labeling, the final result matrix was 16 × 4.
To obtain a mathematical model of the system, the data resulting from the experimental series were combined with the initial experimental design and the assumptions about the underlying relationship between influencing factors and result. The analysis yields the strength of influence for each factor and for the interactions between factors on each measured result variable as well as a P-value for these influences. A similar P-value can be calculated for the whole resulting model. Often, the significance of this model can be improved by removing interactions between factors that have an insignificant P-value and sometimes even by excluding complete factors in case they have no determinable effect on the system. The model can be used to identify factors with strong influence and to make predictions about the response of the system.
To analyse the resulting matrices in combination with the corresponding experimental design, multiple analysis of variance (MANOVA) analysis was performed using functions provided by the R programming language (http://www.r-project.org) (21) or JMP (SAS).
In the validation experiments the data of different arrays were rendered comparable by automatically filtering all data sets from individual hybridizations as specified below. All data sets for spots not recognized by the GenePix analysis software were excluded from further consideration in all experiments. The remaining data sets were ranked according to spot homogeneity (as assayed by the ratio of median and mean fluorescence intensities), ratio of spot to local background intensity and the variance of the logarithmic ratios of replicates. Data points ranking among the lower 20% were removed before further analyses were conducted.
The signal/background ratio for either the median of a group of spots (spotting buffer validation) or one chip (labeling optimization) was chosen as a qualified performance measure. Here, data were analyzed using Excel and Access (Microsoft).
RESULTS AND DISCUSSION
Design of a spotting buffer
The screening design of eight buffer components was arranged in a way that would allow the determination of primary effects as well as all first-order interactions between factors. Even though several of the first-order interactions were indicated as being significant, it became apparent that including them would make the model far too complex for a screening of components. Thus, interaction effects were removed from the model and the analysis was focused on the primary effects. The results are presented in Table 1. The arraying technology allowed the use of many replicate data points, thus enabling the development of a statistically highly significant model. The simplified model still has a significance of P < 1E–16.
DMSO, as well as betaine, have a reliable effect on the resulting intensity (P < 1E–10). In both cases, the summed estimate shows the positive effect on signal intensity, with betaine having a two times stronger effect. Addition of formamide also increases spot intensities and the effect is still detectable with high reliability (P < 1E–4). Comparable in effect are nitrocellulose and Tris as spotting buffer components (P < 0.01). Both have a positive influence on signal intensity. SSC and SDS probably have an effect as well, but the P-values are clearly larger and less significant (P < 0.05). Whereas SSC leads to increased intensities, SDS seems to have a strong deteriorating effect on signal strength. KCl showed no detectable influence.
Analysis of the spot size revealed that DMSO has the most reliable effect on this value (P < 1E–16). It increases the spot size to a significantly higher degree than all other buffer components (summed estimate of 164 compared to values below 40 for all other components).
In conclusion, the three components with the highest P-value (excluding DMSO) were chosen as spotting buffer components at the concentrations employed in the screening design. The selection was limited to three to avoid making the composition too complex, assuming that a higher number would allow for more interactions between the different components and possibly lead to a less predictable system.
The results concerning spot size made it necessary to exclude DMSO from the spotting buffer. Whereas it may have a positive effect on the intensity, this is outweighed by the negative (enlarging) influence on spot size. Concerning low density arrays, a buffer consisting of DMSO, formamide and betaine could be a good spotting solution, but it is inappropriate for higher density arrays.
The final buffer composition was chosen to be 0.5 M betaine, 25% (v/v) formamide and 0.5 µg/µl nitrocellulose. This buffer has been named FBNC buffer.
DNA constitution on a glass surface
Since cDNA probes are usually spotted onto the glass surface as double-stranded molecules, they need to be denatured on the slide later. This is usually achieved by treatment with NaOH (1), contact with boiling water (16) or rehydration and snap drying (15). Here, the screening design for component selection provided further insight into the hybridization conditions on a glass surface. The three components (DMSO, betaine, formamide) with the highest significance (P < 0.001) all have a denaturing effect on DNA. This suggests that it is advantageous to spot DNA in its single-stranded state on the substrate. It can be assumed that methods to denature DNA on a glass surface are unable to access or act on the entire DNA of a spot once it has been delivered and attached to the substrate as a double strand.
It is another common feature of the three mentioned components that they all prevent or at least decelerate spot dehydration. As a side effect this improves spot homogeneity. Any fast drying buffer has the tendency to create spots with ring structures (‘donuts’) having increased intensities towards their borders (22). The FBNC buffer prevents this condition and leads to very smooth and circular spots. At the same time, one can speculate that DNA, if dried in its double-stranded state, will only be partially converted back to its single-stranded form by the denaturing methods given above. In contrast to that, DNA that is kept in a fully denatured state during the dehydration process will necessarily keep that state even if fully dried. Thus, the amount of DNA that is actually accessible during hybridization is largely increased, as indicated by our results concerning the FBNC buffer.
Influence of spotting buffer and substrate surface on the average signal intensity
The different substrates tested in the verification experiment can be considered as a representative snapshot of the products available today, comprising many common surface chemistries (aminosilane, epoxysilane, aldehyde, poly-l-lysine; Table 2). The spotting buffers cover both a variety of commercial products, two of the most commonly used non-commercial mixtures and FBNC (Table 3).
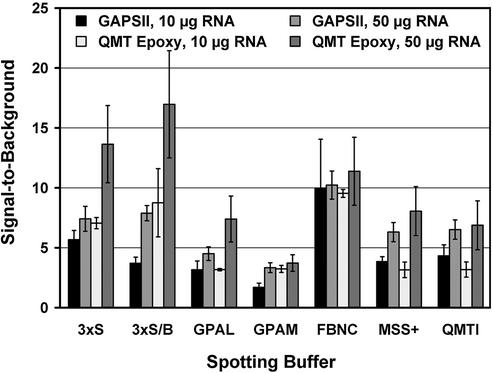
The chosen combination of spotting buffer and substrate has a potent impact on the achievable average signal intensity. Of all tested combinations, GAPSII and QMT Epoxy slides clearly showed the best performance, but with a strong influence of the respective spotting buffer (Fig. 1). 3× SSC, 3× SSC + betaine and FBNC generally yield stronger signals compared to the other buffers, starting both at 10 and 50 µg of total RNA, with FBNC being the best choice for low quantities of total RNA. In contrast, SSC-based buffers performed slightly better on epoxysilane substrates in combination with higher quantities of RNA.
Figure 1.
Signal-to-background versus spotting buffer versus amount of RNA. The signal-to-background ratio was calculated as the average over all spots printed with a certain combination of substrate and spotting buffer. This value is defined here as the ratio of foreground to background signal, adding up Cy3 and Cy5 signals, respectively. Hybridizations using 10 and 50 µg of total RNA per sample were analysed separately to estimate the effect of a limited amount of target. The given standard deviation of the mean represents the chip to chip variation. 3xS, 3× SSC; 3xS/B, 3× SSC + 1.5 M betaine; GPAL, GSSA Aldehyde Spotting Solution; GPAM, GSS Amine Spotting Solution; FBNC, formamide, betaine, nitrocellulose buffer; MSS+, Micro Spotting Solution Plus; QMTI, QMT Spotting Solution I.
Of the combinations surveyed here, combining QMT Epoxy substrates and FBNC as the spotting buffer seems to be the best compromise in terms of spot diameter, signal intensity and sensitivity. This configuration allowed a density of up to 70 000 spots per slide.
Optimization of RNA labeling
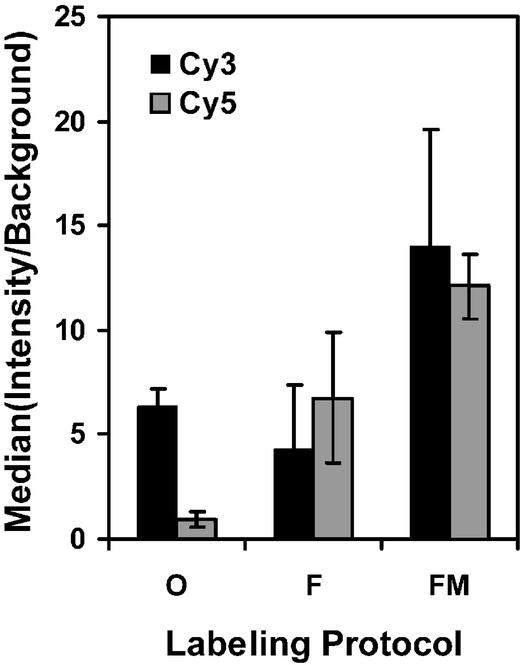
Since the choice of the reverse transcriptase was considered to be critical, a simple comparison between several enzymes was conducted, and the selected system was subsequently characterized with a response surface design. The Superscript enzyme performed slightly worse than the Omniscript enzyme while the Superscript II enzyme had the same efficiency (data not shown). The comparison suggested the use of Fluoroscript since this enzyme incorporates Cy5-dUTP approximately five times better than Omniscript (see validation, Fig. 2).
Figure 2.
Comparison of three alternative labeling protocols. The median of intensity/background ratio in one channel (Cy3 or Cy5) over all spots of an array was calculated for each experiment. The figure shows the mean values, averaged over several experiments of the same type (O, Omniscript, n = 3; F, Fluoroscript, n = 6; FM, modified Fluoroscript protocol, n = 6).
The complete model with all interactions showed no significance and so the model was further reduced to the four most significant effects (RT, dNTP, RT*dNTP, dNTP*dNTP; *, denotes an interaction between two factors and dNTP*dNTP denotes the quadratic dNTP effect). The P-value for this model was determined to be 0.026. The major factor found with this approach was a negative quadratic effect of the dNTP concentration (summed estimate of –1.47 and a P-value of 0.0024). The effect of enzyme concentration, dNTP concentration as well as the effect of their interaction are positive (RT, 0.09; dNTP, 0.53; RT*dNTP, 0.55). Effects of cyanine-labeled dUTP were completely removed from the model since they had no detectable effect in the range surveyed.
The negative quadratic effect for dNTP concentration was not unexpected since a concentration that is too small will result in a lower amount of cDNA being produced, whereas a high concentration will reduce the amount of fluorescently marked dUTP incorporated into the cDNA. Thus, the model only suggests a slight increase in concentration. It is surprising to see that the amount of Cy dye has no detectable effect in the concentration range we examined. This would even allow to decrease the use of Cy dye by at least 30%, an option that we are now examining in more detail.
The given model was used to predict optimal settings for the concentration of Fluoroscript and dNTPs. This prediction suggested to increase the enzyme concentration by 50%. For cost reasons this increase was limited to 20% (0.9 U/µl), also considering the rather small primary effect. The amount of dNTPs was increased by 30% (1.3/20 of the stock solution) as suggested by the prediction.
The comparison of Omniscript, Fluoroscript and modified (according to the values given above) Fluoroscript labeling shows a 5-fold increase of signal intensity for the Cy5-dUTP labeling with the standard Fluoroscript protocol, as compared to the same reaction carried out with Omniscript (Fig. 2). The optimized Fluoroscript protocol shows a 3-fold increase in intensities for Cy3 labeling and a 10-fold increase for Cy5 labeling as compared to the Omniscript protocol.
CONCLUSIONS
We have successfully applied DOE to microarray protocol optimization. It reduces the number of experiments and still provides a large amount of information. In combination with standard approaches it provides an excellent strategy to find good and stable experimental conditions. The results suggest a more widespread use of such methods concerning complex protocols used in biological sciences. Based on the DOE approach, a new spotting buffer has been developed and the spotting conditions have been optimized for high density arrays.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Daniela Bodemer for experimental support. This study was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF; FKZ 01 KW 9937) as well as the Nationales Genomforschungsnetzwerk (NGFN, 01 GR 0101).
REFERENCES
- 1.Duggan D.J., Bittner,M., Chen,Y., Meltzer,P. and Trent,J.M. (1999) Expression profiling using cDNA microarrays. Nature Genet., 21, 10–14. [DOI] [PubMed] [Google Scholar]
- 2.Hegde P., Qi,R., Abernathy,K., Gay,C., Dharap,S., Gaspard,R., Hughes,J.E., Snesrud,E., Lee,N. and Quackenbush,J. (2000) A concise guide to cDNA microarray analysis. Biotechniques, 29, 548–550, 552,–554, 556. [DOI] [PubMed] [Google Scholar]
- 3.Cheung V.G., Morley,M., Aguilar,F., Massimi,A., Kucherlapati,R. and Childs,G. (1999) Making and reading microarrays. Nature Genet., 21, 15–19. [DOI] [PubMed] [Google Scholar]
- 4.Naef F., Lim,D.A., Patil,N. and Magnasco,M. (2002) DNA hybridization to mismatched templates: a chip study. Phys. Rev. E Stat. Nonlin. Soft Matter Phys., 65, 040902. [DOI] [PubMed] [Google Scholar]
- 5.Peterson A.W., Heaton,R.J. and Georgiadis,R.M. (2001) The effect of surface probe density on DNA hybridization. Nucleic Acids Res., 29, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilipauskas D.R. (1999) Can the time from synthesis design to validated chemistry be shortened? Med. Res. Rev., 19, 463–474. [DOI] [PubMed] [Google Scholar]
- 7.Cochran W.G. and Cox,G.M. (1992) Experimental Designs, 2nd Edn. John Wiley and Sons, New York, NY.
- 8.Wildsmith S.E., Archer,G.E., Winkley,A.J., Lane,P.W. and Bugelski,P.J. (2001) Maximization of signal derived from cDNA microarrays. Biotechniques, 30, 202–206, 208. [DOI] [PubMed] [Google Scholar]
- 9.Armstrong R.A., Eperjesi,F. and Gilmartin,B. (2002) The application of analysis of variance (ANOVA) to different experimental designs in optometry. Ophthalmic Physiol. Opt., 22, 248–256. [DOI] [PubMed] [Google Scholar]
- 10.Festing M.F. (1994) Reduction of animal use: experimental design and quality of experiments. Lab. Anim., 28, 212–221. [DOI] [PubMed] [Google Scholar]
- 11.Sniderman P.M. and Grob,D.B. (1996) Innovations in experimental design in attitude surveys. Annu. Rev. Sociol., 22, 377–399. [Google Scholar]
- 12.Kerr M.K., Martin,M. and Churchill,G.A. (2000) Analysis of variance for gene expression microarray data. J. Comput. Biol., 7, 819–837. [DOI] [PubMed] [Google Scholar]
- 13.Dobbin K. and Simon,R. (2002) Comparison of microarray designs for class comparison and class discovery. Bioinformatics, 18, 1438–1445. [DOI] [PubMed] [Google Scholar]
- 14.Fritz B., Schubert,F., Wrobel,G., Schwaenen,C., Wessendorf,S., Nessling,M., Korz,C., Rieker,R.J., Montgomery,K., Kucherlapati,R. et al. (2002) Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res., 62, 2993–2998. [PubMed] [Google Scholar]
- 15.Schena M., Shalon,D., Heller,R., Chai,A., Brown,P.O. and Davis,R.W. (1996) Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 17.Collins S.J. (1987) The HL-60 promyelocytic leukemia cell line: proliferation, differentiation and cellular oncogene expression. Blood, 70, 1233–1244. [PubMed] [Google Scholar]
- 18.Todaro G.J. and Green,H. (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol., 17, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berstine E.G., Hooper,M.L., Grandchamp,S. and Ephrussi,B. (1973) Alkaline phosphatase activity in mouse teratoma. Proc. Natl Acad. Sci. USA, 70, 3899–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porfiri E., Hoffbrand,A.V. and Wickremasinghe,R.G. (1989) Induction of macrophage-like differentiation of HL60 human myeloid leukemia cells by phorbol myristate acetate triggers an early decline in inositol lipid breakdown. Exp. Hematol., 17, 344–350. [PubMed] [Google Scholar]
- 21.Ihaka R. and Gentleman,R. (1996) R: A language for data analysis and graphics. J. Comp. Graph. Stat., 5, 299–314. [Google Scholar]
- 22.Diehl F., Grahlmann,S., Beier,M. and Hoheisel,J.D. (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res., 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 24.Pinkel D., Segraves,R., Sudar,D., Clark,S., Poole,I., Kowbel,D., Collins,C., Kuo,W.L., Chen,C., Zhai,Y. et al. (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet., 20, 207–211. [DOI] [PubMed] [Google Scholar]