Abstract
Dozens of PCR-based methods are available for chromosome walking from a known sequence to an unknown region. These methods are of three types: inverse PCR, ligation-mediated PCR and randomly primed PCR. However, none of them has been generally applied for this purpose, because they are either difficult or inefficient. Here we describe a simple and efficient PCR strategy—T-linker-specific ligation PCR (T-linker PCR) for gene or chromosome walking. The strategy amplifies the template molecules in three steps. First, genomic DNA is digested with 3′ overhang enzymes. Secondly, primed by a specific primer, a strand of the target molecule is replicated by Taq DNA polymerase and a single A tail is generated on the 3′ unknown end of the target molecule, and then a 3′ overhang-T linker (named T-linker) is specifically ligated onto the target. Thirdly, the target is amplified by two rounds of nested PCR with specific primers and T-linker primers. T-linker PCR significantly improves the existing PCR methods for walking because it uses specific T/A ligation instead of arbitrary ligation or random annealing. To show the feasibility and efficiency of T-linker PCR, we have exploited this method to identify vector DNA or T-DNA insertions in transgenic plants.
INTRODUCTION
PCR isolation of the DNA fragments adjacent to known sequences is a favorite strategy for chromosome walking. Several modified PCR methods are available for this purpose: (i) inverse PCR (1–7); (ii) ligation-mediated PCR (LM-PCR), including biotin-capture PCR (8–13) and adaptor/cassette/linker ligation PCR (11,12,14–20); and (iii) randomly primed PCR (RP-PCR), including hemispecific or one-sided PCR (21–25) and TAIL-PCR (26–28). Although all the mentioned PCR methods, including their improved versions such as vectorette PCR, claim to be efficient, their inherent limitations limit their usefulness to a subset of unknown sequences. Inverse PCR is primarily limited by uneven restriction site distribution, and non-specific amplification is the main problem of LM-PCR, predominantly due to ligating an adaptor or cassette to the genomic DNA fragments. The non-specific annealing of arbitrary primer in RP-PCR often results in excessive accumulation of non-specific molecules. Up to now, none of the existing methods can perform a successive chromosome walk. We report a new universal method, i.e. T-linker-specific ligation PCR (T-linker PCR), for successive chromosome or gene walking.
Principle of T-linker PCR
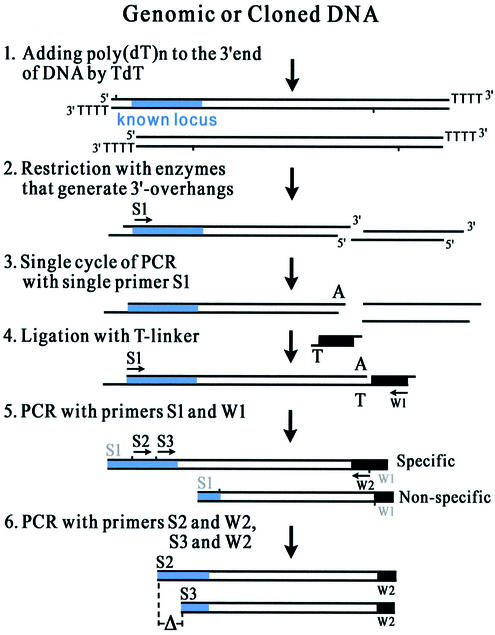
The basic principle of T-linker PCR is outlined in Figure 1. It consists of the following six steps: (i) Add poly(dT)n to the 3′ ends of genomic DNA molecules by terminal deoxynucleotidyl transferase (TdT) to prevent the genomic DNA ends from obtaining a single A in the reaction of step (iii); (ii) digest the poly(dT)n-tailed DNA with restriction enzymes, which generate 3′ overhangs but not a single A; (iii) use the specific S1 primer to replicate a strand of the target molecule and generate a single A tail on the 3′ end of the molecule by Taq DNA polymerase; (iv) ligate a T-linker specifically onto the 3′ end of the target molecule using T4 DNA ligase; (v) perform a first round of nested PCR to amplify the target molecule, primed by the outer pair of primers (S1 and W1); and (vi) perform a second round of the nested PCR to amplify the product of the first round, primed by the second pair of primers (S2 and W2). Meanwhile, primers S3 and W2 are used to amplify the product of the first round for identification of the specifically amplified molecules by the length difference (ΔS2–S3) of S2–W2 and S3–W2 products.
Figure 1.
The scheme of T-linker PCR for genome walking. S1, S2 and S3: the specific primers binding to the known sequence (the blue area within the double line) of the target molecule. W1 and W2: the walking primers binding to the T-linker sequence (the black area within the double line). A or T: the A tail of the target molecule or the T nucleotide of the T-linker. Δ: the anticipated difference in amplified products with the specific primers S2 and S3 in the separated second round reactions.
MATERIALS AND METHODS
Template DNA
Genomic DNA was extracted from rice mutant S8-1, from the variety ‘Nante’ and from Arabidopsis mutants as PCR templates for T-linker PCR amplifications.
Genomic DNA of the rice mutant S8-1, the semi-dwarfism plant generated by biolistic transformation for resistance to rice dwarf virus (RDV), was used to identify the insertions of the integrated S8 genes (29).
The gibberellin 20-oxidase gene (GA5) in genomic DNA of the rice variety ‘Nante’ was employed to obtain the gene’s promotor sequence.
Genomic DNA of Arabidopsis mutants, with the plants tagged by dipping-flower transformation with activation-tagging vector pSki015 (30), was used to identify their T-DNA insertions.
Restriction enzymes and genomic DNA digestion
Restriction enzymes that generate 3′ overhangs, such as HhaI (GCG↓C), Hsp92II (CATG↓), BseNI (ACTGGN↓), BseGI (GGATGNN↓), PvuI (CGAT↓CG), PstI (CTGCA↓G), KpnI (GGTAC↓C), etc., were chosen to cut genomic DNA. The DNA was either completely digested with 6 bp cutters to obtain large fragments flanking the known sequence, or incompletely digested with 4 or 5 bp cutters. Complete digestion was achieved using 10–20 U of restriction enzymes to cut 1 µg of genomic DNA in 50 µl for 12–16 h, and incomplete digestion was done (20–50% restriction sites to be cleaved) with 1–2 U of 4 or 5 bp cutters for 30–45 min.
Oligonucleotide primers
Three specific primers for each walking step were designed based on the known sequence. The specific primers, T-linker and two T-linker primers sequences are as follows. Primers for S8-1: dw1 (S1) 5′-CTATACATTGAACTCGATAACTC AGAC-3′; dw2 (S2) 5′-CATCGATTGATGATTTCGATGT GTCTGA-3′; dw3 (S3) 5′-GTGGAGGAAATGCCCGGT CCTGA-3′. Primers for the GA5 gene: UP479 (S1) 5′-CTA CGAGCTTCCGCACCAGGT-3′; UP415 (S2) 5′-CACGA CAGCGTCTCCTTCCAC-3′; UP237 (S3) 5′-GTGTAACC ACCAGGAAGAAGCCGT-3′. Primers for Arabidopsis-integrated T-DNA: DL1 (S1) 5′-GACAACATGTCGAGGCTC AGCAGG-3′; DL2 (S2) 5′-TGGACGTGAATGTAGAC ACGTCGA-3′; DL3 (S3) 5′-GCTTTCGCCTATAAATA CGACGG-3′. T-linker: 5′-GTAGGTGTGTGGAGGAT GCTGAGCGAGGTA-3′, 3′-TCATCCACACACCTCCTAC GACTCGCTCCA-5′. T-linker primer (W1): 5′-ACCTCGC TCAGCATCCTCCAC-3′ and T-linker primer (W2): 5′-TC AGCATCCTCCACACACCTACT-3′.
PCR procedure
Step A. Add poly(dT)n to template DNA by TdT. (i) The reaction: 1–5 µg of genomic DNA, 10–50 U of TdT, 5 µl of 10× TdT buffer, 5 µl of 10 mM dTTP, 10 µl of 0.1% bovine serum albumin. Bring the total volume to 50 µl with deionized water. Incubate the reaction at 37°C for 30 min. (ii) Add ethanol mix (100 µl of ethanol + 5 µl of 3 M sodium acetate, pH 5.2) to precipitate the DNA, then re-dissolve in sterile water (20 µl/µg DNA).
Step B. Incompletely digest the poly(dT)n-tailed DNA with 3′ overhang restriction enzymes. (i) Digestion reaction: 1 µg of poly(dT)n DNA, 1.5 U of restriction enzyme, 5 µl of 10× buffer. Bring the total volume to 50 µl with deionized water. Incubate the reaction at 37°C for 20–30 min. (ii) Precipitate the digested DNA as described in step A.ii and re-dissolve in 11 µl of sterile water.
Step C. Add a single A to the 3′ end of the target molecule specifically by LA Taq polymerase. (i) A-tailing reaction: 8 µl of 2.5 mM dNTPs (the mixture of dATP, dTTP, dGTP, dCTP, each 2.5 mM), 2 U of LA Taq polymerase, 25 µl of 2× GC buffer I, 20 pmol of primer S1, 10 µl of digested DNA from step B.ii. Bring the total volume to 50 µl with deionized water. Then run single cycle PCR: denaturing at 94°C for 3 min, annealing at 55–60°C for 2 min with extension at 72°C for 6–8 min. Immediately place on ice. The LA Taq polymerase and the 2× GC buffer I are parts of a kit from TaKaRa Biotechnology Co. Ltd. (ii) Precipitate and re-dissolve the template DNA in 8 µl of sterile water.
Step D. Specific ligation between target molecule and T-linker. Ligation reaction: 7.5 µl of A-tailed DNA, 2 pmol of T-linker, 3 U of T4 DNA ligase (Weiss), 1 µl of 10× ligation buffer. Bring the total volume to 10 µl with deionized water. Ligate at 16°C for 12–16 h.
Step E. First round of nested PCR. (i) Reaction I: 4 µl of 2.5 mM dNTPs, 1.25 U of LA Taq, 12.5 µl of 2× GC buffer I, 10 pmol of primer S1, 10 pmol of primer W1, 1.5 µl of template DNA (from step D). Bring the total volume to 25 µl with deionized water. (ii) Thermal cycling condition: (a) Pre-denaturing at 94°C for 3 min; (b) denaturing at 94°C for 1 min; (c) annealing at 55–60°C for 1 min; (d) extension at 72°C for 3 min; (e) repeat for 34 cycles; (f) final extension at 72°C for 10 min. (iii) Dilute 1 µl of PCR product of the first round in 20–50 µl of sterile water.
Step F. Second round of nested PCR. (i) Reaction II: 4 µl of 2.5 mM dNTPs, 1.25 U of LA Taq, 12.5 µl of 2× GC buffer I, 10 pmol of primer S2, 10 pmol of primer W2, 1 µl of the diluted PCR product from the first round (step E.iii), sterile water up to 25 µl. (ii) Reaction III: 4 µl of 2.5 mM dNTPs, 1.25 U of LA Taq, 12.5 µl of 2× GC buffer I, 10 pmol of primer S3, 10 pmol of primer W2, 1 µl of the diluted PCR product from the first round (step E.iii), sterile water up to 25 µl. (iii) Thermal cycling condition for reactions II and III: the same as the first round (step E.ii).
Cloning and sequencing of PCR products
The PCR products of reactions II and III were separated by electrophoresis (5 V/cm) on an agarose gel to discern the specific bands. These bands, which show an anticipated difference between II and III, are supposed to be the specific molecules. The molecules were then cloned and sequenced with an ABI PRISM™ 377XL DNA Sequencer from TaKaRa Biotechnology (Dalian) Co. Ltd.
RESULTS
Amplifying genomic sequences flanking multi S8 genes in the S8-1 rice mutant by T-linker PCR
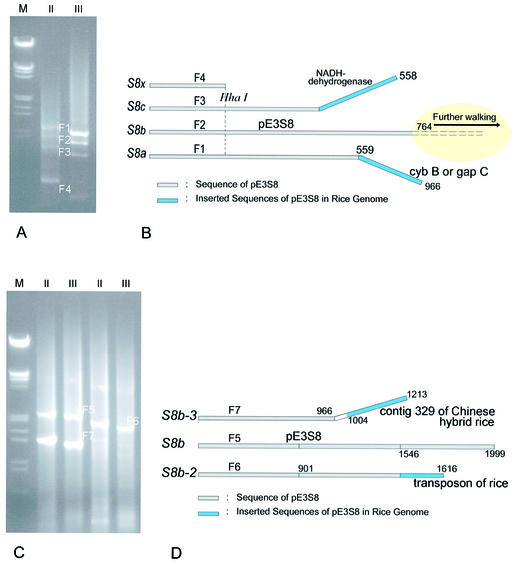
The RDV S8 genes were transformed into rice by biolistic bombardment. Biolistic transformation will usually allow several S8 gene copies to integrate into different genomic sites with different vector borders. We amplified sequences flanking the S8 transgenes by T-linker PCR and obtained four specific fragments (Fig. 2A). Sequencing showed that the 3′ ends of fragments F1 and F3 are inserted sequences of pE3S8 in the rice genome (Fig. 2B), and fragments F2 and F4 are parts of pE3S8. Based on the 3′ ends of F2, we conducted a further walking step and obtained three specific fragments (Fig. 2C). Sequencing results showed that F5 belongs to pE3S8, whereas the 3′ ends of F6 and F7 are beyond pE3S8 (Fig. 2D). In conclusion, the 3′ ends of fragments F1, F3, F6 and F7 are inserted sequences of the RDV S8 transgene in the mutant S8-1, The 3′ end of F6 encodes putative ATPase of rice (3709.t00007|OJ1118_G04) while the end of F7 encodes a hypothetical protein of rice (3293.t00016|OSJNBa0061G20).
Figure 2.
Chromosome walking downs the RDV S8 gene from rice mutant S8-1 by T-linker PCR. (A) Four fragments (F1, F2, F3 and F4) are obtained from the rice mutant S8-1 by T-linker PCR with the restriction enzyme HhaI (GCG↓C) and two specific primers [dw2 (for reaction II) and dw3 (for reaction III)]. Δdw2–dw3 is 136 bp. M: λ DNA EcoRI–HindIII markers. (B) Sequencing of the fragments F1, F2, F3 and F4, and alignments with vector pE3S8. F1, F2 and F3 were obtained from the S8 copies S8a, S8b and S8c, respectively, whereas F4 was from any of them (S8x). (C) At the end of the F2 fragment, we obtained a further three fragments (F5, F6 and F7) in the second walking step by T-linker PCR with the enzymes SphI (GCAT↓GC) and BseNI (ACTGGN↓), and specific primers dw9 (for reaction II, 5′-TACCTTTCCAGAGTCCTTGTCAGATTC-3′) and dw10 (for reaction III, 5′-GGCTGCCATTTTTGGGGTGAGG-3′), with Δdw9–dw10 of 154 bp. M: λ DNA EcoRI–HindIII markers. (D) Sequencing of the fragments F5, F6 and F7, and alignments with vector pE3S8. We deduced that F5, F6 and F7 were obtained from the S8 copies S8b, S8b-2 and S8b-3, respectively.
Walking single copy sequence by T-linker PCR
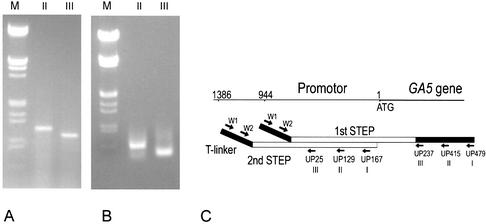
The GA5 gene, a single copy sequence of the rice genome, was used as T-linker PCR template to obtain the gene’s promotor sequence by successive walking. In the first step, we acquired a 1.1 kb DNA fragment with the specific primer UP237 (III), and a larger one was obtained with the primer UP415 (Fig. 3A). Sequencing confirmed that the fragment contained a GA5 944 bp promotor region (Fig. 3C). In the next round of T-linker PCR walking, we obtained a 0.5 kb fragment with primer UP129 and a smaller one with primer UP25 (Fig. 3B), and sequencing showed that the promotor region had been extended by 442 bp (Fig. 3C). The walking results prove the feasibility of T-linker PCR walking for single copy sequence of the genome.
Figure 3.
Chromosome walking by T-linker PCR for the GA5 gene promoter. (A) T-linker PCR amplification produced a specific band of 1.1 kb with primer UP237 (for reaction III) and of 1.3 kb with primer UP415 (for reaction II) in the first walking step. ΔUP415–UP237 is 178 bp. M: λ DNA EcoRI–HindIII markers. (B) The second step produced a band of ∼0.5 kb with primer UP25 (for reaction III, 5′-CTGTGCCACCACCAGATTGATGAC-3′) and a band of ∼0.56 kb with primer UP129 (for reaction II, 5′-GACGCTATGACGAGATTTAGACAACCA-3′). ΔUP129–UP25 is 104 bp. M: λ DNA EcoRI–HindIII markers. (C) Cloning and sequencing results of the fragments obtained in the first and second walking steps. ATG: the translation start code of the GA5 gene.
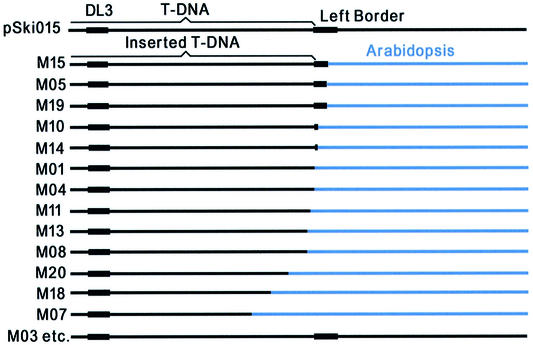
Identification of T-DNA insertions in Arabidopsis
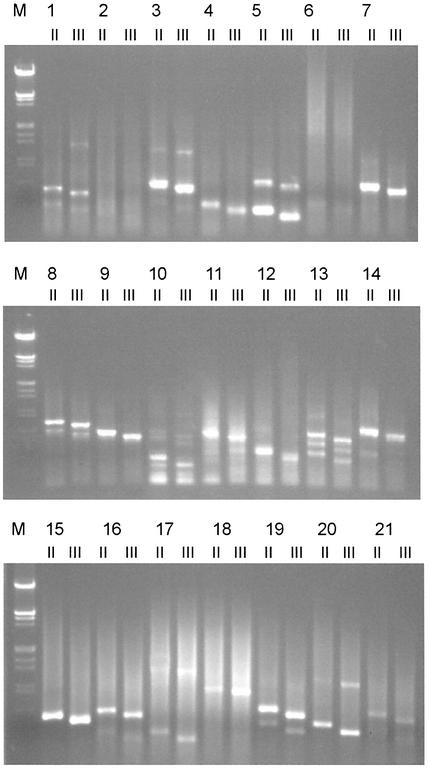
Genomic DNA of Arabidopsis mutant 1–mutant 21 was used as template to identify the T-DNA left borders inserted into Arabidopsis plants. We identified specific bands from all mutants except numbers 2 and 6 (Fig. 4). Subsequent sequencing of the specific bands showed that the inserted T-DNA left ends in 13 mutants (mutants 1, 4, 5, 7, 8, 10, 11, 13, 14, 15, 18, 19 and 20) that are adjacent to Arabidopsis genomic DNA (Fig. 5), whereas sequences of other mutants (mutants 3, 12, 16, 17 and 21) remain intact near their left border. These results showed the complexity of T-DNA insertions in plants and supported De Buck’s hypothetic mode of T-DNA insertion: in most cases (>60%), the T-DNA integrations are guided by the vector borders, but in some cases (∼30%), called read-through (31), they did not obey the borders’ guide.
Figure 4.
Identification of T-DNA-inserted sequences of Arabidopsis mutants 1–21 by T-linker PCR with the restriction enzyme HhaI (GCG↓C) and two specific primers: DL2 (for reaction II) and DL3 (for reaction III)]. ΔII–III is 56 bp (Fig. 1).
Figure 5.
Sequence comparison between T-DNA of vector pSki015 and the inserted T-DNA in the Arabidopsis mutants.
Statistics of the 3′ overhang restriction sites in rice and Arabidopsis genomes
Employing rice and Arabidopsis genomic sequences (>3 Mb), we counted the distribution of restriction sites of six 3′ overhang-generating restriction enzymes (Table 1). The percentages of fragments shorter than 1.5 and 3 kb represent the probabilities of obtaining the specific fragments of <1.5 and 3.0 kb, respectively, by the PCR method. In the complete digestion conditions, the T-linker PCR could amplify the fragment between the specific primer site and the nearest restriction site, with the mean size of ‘the mean length’ in Table 1. In the incomplete digestion conditions (20–50% of restriction sites were cleaved), the T-linker PCR could produce several fragments between the specific primer site and the nearest first, second, third, etc., restriction sites.
Table 1. The mean size and size distribution of genomic DNA fragments digested by 3′ overhang-generating restriction enzymes.
| Restriction enzymes and recognition sites | Genome | Mean length (kb) | Percentage of fragments <1.5 kb | Percentage of fragments <3 kb |
|---|---|---|---|---|
| Hsp92II CATG↓ | Rice | 0.1889 | 99.77 | 99.99 |
| Arabidopsis | 0.2370 | 99.59 | 99.99 | |
| HhaI GCG↓C | Rice | 0.3362 | 88.46 | 93.57 |
| Arabidopsis | 1.4236 | 70.39 | 84.45 | |
| BseNI ACTGGN↓ | Rice | 1.4709 | 63.92 | 85.11 |
| Arabidopsis | 1.8629 | 56.00 | 79.66 | |
| BsGI GGATGN↓ | Rice | 0.9046 | 79.42 | 94.58 |
| Arabidopsis | 1.2885 | 66.80 | 88.54 | |
| NsiI ATGCA↓T | Rice | 1.7320 | 58.58 | 79.40 |
| Arabidopsis | 2.4772 | 50.46 | 71.28 | |
| PstI CTGCA↓G | Rice | 3.2440 | 41.37 | 60.42 |
| Arabidopsis | 4.7246 | 36.52 | 50.15 |
DISCUSSION
PCR strategies for chromosome walking: the technical development from inverse PCR to T-linker PCR
Scientists have tried three kinds of PCR strategies for chromosome walking: inverse PCR (1–7), LM-PCR (8,10,12,14,16–20) and RP-PCR (22–24,26–28). Inverse PCR was the first generation PCR method for chromosome walking. Now it is used less for chromosome walking because of no available restriction sites in the unknown/known region or poor circularization of the template molecule. LM-PCR and RP-PCR are more popular for chromosome walking. The latter is the simplest and most popular method for identification of T-DNA or transposon insertions (26–28), but its amplified products are generally small (<1 kb). To obtain larger fragments (>1 kb) or to walk step by step, we usually resort to LM-PCR. However, LM-PCR is an inefficient strategy because in most cases non-specific amplification accounts for the major proportion of the final PCR products. To resolve this problem, scientists improved the method time after time with different adoptors/casssettes, and several improved LM-PCRs have been reported (13,18,32). However, none of the improved methods could eliminate this drawback completely. T-linker PCR resolves the problem thoroughly by ‘specific ligation’ between the specific target molecule and the T-linker (Fig. 1). Therefore, T-linker PCR gives higher amplification efficiency for chromosome walking than the other existing methods.
The results reported here have experimentally illustrated the high efficiency and feasibility of T-linker PCR.
The restriction enzymes employed by T-linker PCR
As T-linker PCR is based upon LM-PCR, the restriction enzyme site is an important factor to consider. To cut genomic DNA, we can employ 6, 5 and 4 bp cutters which generate 3′ overhangs. When the 4 bp cutters are used, we digest the genomic DNA partially (20–50% restriction sites to be cleaved) to obtain fragments larger than ‘the mean length’ in Table 1. The incomplete digestion worked very well in our experiments (see Results). For example, we amplified specific bands from 90% of the total transgenic Arabidopsis plants and at least 50% of them have two bands. The statistics of the restriction sites in rice and Arabidopsis also showed a high probability to amplify fragments (<1.5 kb or <3.0 kb) with the 4 and 5 bp cutters (Table 1). For example, employing Hsp92II (CATG↓), T-linker PCR could produce specific bands (<3.0 kb) with a probability of 99.9% in rice and Arabidopsis. These results illustrated that T-linker PCR should not be significantly limited by restriction site availability. Therefore, it is a reliable and sensitive PCR strategy for chromosome walking, whereas there is no report of incomplete digestion in the existing LM-PCR thus far.
T-linker PCR advantages in comparison with existing PCRs
Compared with the other existing PCR methods for walking, T-linker PCR has many advantages: (i) high specificity; (ii) a high positive rate of a ‘specific band’; (iii) long length of a walking step; and (iv) a high success rate to walk down in one direction. The comparison of T-linker PCR and other PCRs is outlined in Table 2.
Table 2. The comparison between T-linker PCR and other existing PCRs.
| T-linker PCR | Inverse PCR (10) | Ligation-mediated PCR (8,10) | Biotin-capture PCR (13) | One-sided PCR (24,25) | TAIL-PCR (27) | |
|---|---|---|---|---|---|---|
| Adaptor ligation or primer annealing | Specific ligation | Inverse ligation | Non-specific ligation | Non-specific ligation or low-temperature annealing | Low-temperature annealing | Low-temperature annealing |
| Number of adaptor/casset. required | 1 (T-linker) | 0 | Dozens | Dozens | Several arbitrary primers | Arbitrary degenerated primers |
| Restriction enzymes required | 3–6 (3′-overhang enzymes) | Dozens | Dozens | Dozens | – | – |
| PCR specificity | High | High | Low | Low | Low | Medium |
| Specific positive rate | 50–99% | Low | Low | Low | Low | 70–80% |
| Length of a walking step | 0–3 kb | 0–3 kb | 0–3 kb | 0–3 kb | Small | Small |
| Cost | Low | Low | High | High | Low | Low |
The potential applications of T-linker PCR in genome-related research
The T-linker PCR strategy is an optimal method for amplification of DNA segments adjacent to known sequences. We believe that it is promising in genome-related research for the following experiments: (i) consecutively walking along chromosome to identify important genes that control the traits of human, animals and plants using a molecular marker or known gene (sequence) as a starting pad; (ii) to identify T-DNA or transposon insertion sites in known or unknown genomes for genome or gene function assay. T-linker-PCR is especially effective in identifying multi-copy insertions; (iii) to obtain non-conservative parts of genes in new species according to the conservative sequence of reported genes; (iv) to fill in gaps or unknown chromosome regions in genome sequencing; and (v) to isolate the ends of phage P1, yeast and bacterial artificial chromosomes for construction of the physical map of the genome.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Qin Gen-Ji and Kang Ding-Ming for providing Arabidopsis materials, Dr Li Yi for providing transgenic rice, and Dr Daniel J. Kliebenstein (Department of Vegetable Crops, University of California, Davis) for critical review of the manuscript. This work was supported by the National Special Project of Trasgenic Plants (Grant No. J00-A-005), the National Key Basic Science ‘973’ Program (Grant No.G2000016204), the State ‘863’ High Technology R&D Program (Grant No.2001AA214191) and the National Natural Science Foundation Program (Grant No.39980003) of the Chinese Government.
REFERENCES
- 1.Triglia T., Peterson,M.G. and Kemp,D.J. (1988) A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res., 16, 8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ochman H., Gerber,A.S. and Hartl,D.L. (1988) Genetic applications of an inverse polymerase chain reaction. Genetics, 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman G.A., Ye,R.D., Pollock,K.M., Sadler,J.E. and Korsmeyer,S.J. (1989) Use of yeast artificial chromosome clones for mapping and walking within human chromosome segment 18q21.3. Proc. Natl Acad. Sci. USA, 86, 7485–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huckaby C.S., Kouri,R.E., Lane,M.J., Peshick,S.M., Carroll,W.T., Henderson,S.M., Faldasz,B.D., Waterbury,P.G. and Vournakis,J.N. (1991) An efficient technique for obtaining sequences flanking inserted retroviruses. GATA, 8, 151–158. [DOI] [PubMed] [Google Scholar]
- 5.Arveiler B. and Porteous,D.J. (1991) Amplification of end fragments of YAC recombinants by inverse polymerase chain reaction. Technique, 3, 24–28. [Google Scholar]
- 6.Huang S.H. (1994) Inverse polymerase chain reaction: an efficient approach to cloning cDNA ends. Mol. Biotechnol., 2, 15–22. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama K., Watanabe,H., Tsukada,S. and Sasai,H. (2000) A novel method for constructing gene-targeting vectors. Nucleic Acids Res., 28, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenthal A. and Jones,D.S.C. (1990) Genomic walking and sequencing by oligo-cassette mediated polymerase chain reaction. Nucleic Acids Res., 18, 3095–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenthal A., MacKinnon,R.N. and Jones,D.S.C. (1991) PCR walking from microdissection clone M54 identifies three exons from the human gene for the neural cell adhesion molecule L1 (CAM-L1). Nucleic Acids Res., 19, 5395–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenthal A. (1992) PCR amplification techniques for chromosome walking. Trends Biotechnol., 10, 44–48. [DOI] [PubMed] [Google Scholar]
- 11.Espelund M. and Jakobsen,K.S. (1992) Cloning and direct sequencing of plant promotors using primer adapter mediated PCR on DNA coupled to a magnetic solid phase. Biotechniques, 13, 74–81. [PubMed] [Google Scholar]
- 12.Warshawsky D. and Miller,L. (1994) A rapid genomic walking technique based on ligation-mediated PCR and magnetic separation technology. Biotechniques, 16, 792–798. [PubMed] [Google Scholar]
- 13.Sterky F., Holmberg,A., Alexandersson,G., Lundeberg,J. and Uhlen,M. (1998) Direct sequencing of bacterial artificial chromosomes (BACs) and prokaryotic genomes by biotin-capture PCR. J. Biotechnol., 60, 119–129. [DOI] [PubMed] [Google Scholar]
- 14.Mueller P.R. and Wold,B. (1989) In vivo footprinting of a muscle specific enhancer by ligation-mediated PCR. Science, 246, 780–786. [DOI] [PubMed] [Google Scholar]
- 15.Pfeifer G.P., Steigerwald,S.D., Mueller,P.R., Wold,B. and Riggs,A.D. (1989) Genomic sequencing and methylation analysis by ligation mediated PCR. Science, 246, 810–813. [DOI] [PubMed] [Google Scholar]
- 16.Riley J., Butler,R., Ogilvie,D., Finniear,R., Jenner,D., Powell,S., Anand,R., Smith,J.C. and Markham,A.F. (1990) A novel, rapid method for the isolation of terminal sequence from yeast artificial chromosome (YAC) clones. Nucleic Acids Res., 19, 5395–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity P.A. and Wold,B.J. (1992) Effects of different DNA polymerase in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl Acad. Sci. USA, 89, 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones D.H. and Winistorfer,S.C. (1992) Sequence specific generation of a DNA panhandle permits PCR amplification of unknown flanking DNA. Nucleic Acids Res., 20, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer G.P. and Riggs,A.D. (1996) Genomic sequencing by ligation-mediated PCR. Mol. Biotechnol., 5, 281–288. [DOI] [PubMed] [Google Scholar]
- 20.Dai S.-M., Chen,H.-H., Chang,C., Riggs,A.D. and Flanagan,S.D. (2000) Ligation-mediated PCR for quantitative in vivo footprinting. Nat. Biotechnol., 18, 1108–1111. [DOI] [PubMed] [Google Scholar]
- 21.Frohman M.A., Dush,M.K. and Martin,G.R. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA, 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh E.Y., Elliot,J.F., Cwirla,S., Lanier,L. and Davis,M.M. (1989) Polymerase chain reaction with single-sided specificity: analysis of T cell receptor δ chain. Science, 243, 217–220. [DOI] [PubMed] [Google Scholar]
- 23.Ohara O., Dorit,R.E. and Gilbert,W. (1989) One-sided polymerase chain reaction: the amplification of cDNA. Proc. Natl Acad. Sci. USA, 86, 5673–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shyamala V. and Ames,G.F. (1989) Genome walking by single-specific-primer polymerase chain reaction: SSP-PCR. Gene, 84, 1–8. [DOI] [PubMed] [Google Scholar]
- 25.Parker J.D., Rabinovitch,P.S. and Burmer,G.C. (1991) Targeted gene walking polymerase chain reaction. Nucleic Acids Res., 19, 3056–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y.-G. and Whittier,R.F. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics, 25, 674–681. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y.-G., Mitsukawa,N., Oosumi,T. and Whittier,R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J., 8, 457–463. [DOI] [PubMed] [Google Scholar]
- 28.Terauchi R. and Kahl,G. (2000) Rapid isolation of promoter sequences by TAIL-PCR: the 5-flanking regions of Pal and Pgi genes from yams (Dioscorea). Mol. Gen. Genet., 263, 554–560. [DOI] [PubMed] [Google Scholar]
- 29.Zheng H.H., Li,Y., Yu,Z.H., Li,W., Chen,M.Y., Ming,X.T., Casper,R. and Chen,Z.L. (1997) Recovery of transgenic rice plants expressing the rice dwarf virus outer coat protein gene (S8). Theor. Appl. Genet., 94, 522–527. [Google Scholar]
- 30.Weigel D., Ahn,J.H., Blazquez,M.A., Borevitz,J.O., Christensen,S.K., Fankhauser,C., Ferrandiz,C., Kardailsky,I., Malancharuvil,E.J., Neff,M.M. et al. (2000) Activation tagging in Arabidopsis. Plant Physiol., 122, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Buck S., De Wilde,C., Van Montagu,M. and Depicker,A. (2000) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol. Breed., 6, 459–468. [Google Scholar]
- 32.Arnold C. and Hodgson,I.J. (1991) Vectorette PCR: a novel approach to genomic walking. PCR Methods Appl., 1, 39–42. [DOI] [PubMed] [Google Scholar]