Abstract
Although the concept of domain merging and shuffling as a major force in protein evolution is well established, it has been difficult to demonstrate how domains coadapt. Here we show evidence of coevolution of the Sinorhizobium meliloti NifA (SmNifA) domains. We found that, because of the lack of a conserved glycine in its DNA-binding domain, this transactivator protein interacts weakly with the enhancers. This defect, however, was compensated by evolving a highly efficient activation domain that, contrasting to Bradyrhizobium japonicum NifA (BjNifA), can activate in trans. To explore paths that lead to this enhanced activity, we mutagenized BjNifA. After three cycles of mutagenesis and selection, a highly active derivative was obtained. Strikingly, all mutations changed to amino acids already present in SmNifA. Our artificial process thus recreated the natural evolution followed by this protein and suggests that NifA is trapped in a restricted sequence space with very limited solutions for higher activity by point mutation.
Contemporary proteins are often assemblages of functionally and evolutionarily independent domains (1, 2). This modular architecture has conferred great flexibility for new specificities, altered recognition properties, and modified functions to flourish with a strikingly limited set of structurally different domains (3–6). But how the domains coadapt to achieve an optimal fitness is poorly understood. It is anticipated that appropriate domain interfaces and balanced activities need to be attained by a complex combinatorial optimization process (7) that is constantly in operation but limited by the evolutionary constrains inherent to the protein folds. Moreover, the optimal fitness needs to be dynamically maintained by compensatory mutations in a changing environment.
Earlier work showed that the enhancer-binding proteins (EBP), as the majority of transactivator proteins, are modular regulators with evolutionarily distinct DNA-binding, transcriptional activation, and regulatory domains (8–10). The DNA-binding and positive control functions of NifA, a member of the EBP family that controls nitrogen fixation gene expression in eubacteria, have been separated (11). The DNA-binding function resides at the C-terminal domain, whereas the activation domain is located at the center of the protein.
The EBP bind to remote DNA sites, functionally similar to the eukaryotic enhancers, and activate transcription by contacting the σ54 form of the RNA polymerase, bound at the promoter, in a process that requires nucleoside triphosphate hydrolysis (12–15). The binding of the EBP at the enhancers may help to increase the local concentration of the activator in the vicinity of the promoter and to direct the central domain to interact in the correct orientation with Eσ54. Thus, the level of expression of a given promoter results from both the DNA-binding affinity and the intrinsic activation activity of the EBP.
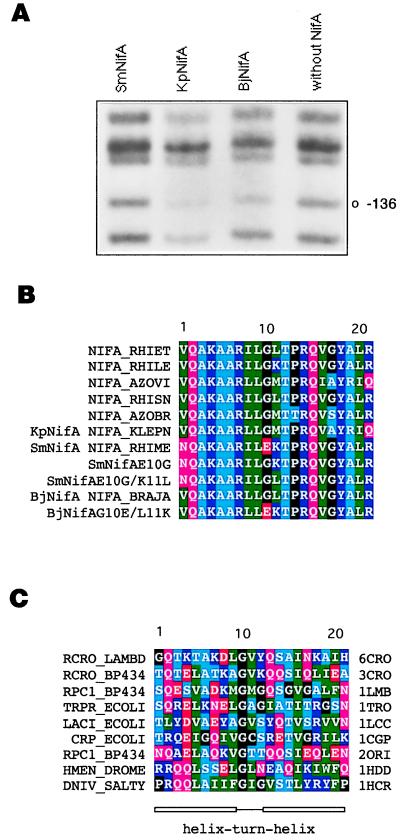
The C-terminal region of several EBP is predicted to form a helix–turn–helix (hth) supersecondary structure (8). Mutagenesis (16), spectroscopic (17), and NMR (P. Ray, K. J. Smith, R. A. Dixon, and E. I. Hyde, personal communication) studies of NifA support this interpretation. This motif is present in a wide range of site-specific DNA-binding proteins (18). When we compared the hth of several different NifA proteins we observed that the protein of Sinorhizobium meliloti NifA (SmNifA) has a glutamic acid (Fig. 1B) in a position where glycine (G10) is highly conserved (18) (Fig. 1C). In this study, we report the detrimental effect of the lack of the glycine on DNA binding, which, in SmNifA, was compensated by developing a highly efficient activation domain, in a process we named reciprocal domain evolution.
Figure 1.
DNA-binding properties of different NifA proteins and amino acid sequence of various hth motifs. (A) In vivo DMS footprinting of the K. pneumoniae nifH enhancer with different NifA proteins, as indicated. Protection from methylation of guanine-136 by KpNifA and BjNifA is indicated. This residue is part of the TGT-N10-ACA nifH enhancer. (B) Sequence alignment of the hth motif of several NifA and the SmNifA and BjNifA mutant proteins constructed in this work. The SwissProt name for each protein is indicated. (C) Alignment of the hth motifs of proteins whose structures have been solved as cocrystals with their DNA-binding sites. SwissProt (first column) and Protein Data Bank (last column) names are specified. 6CRO, lambda cro; 3CRO, 434 cro; 1LMB, lambda repressor; 1TRO, trp repressor; 1LCC, lac repressor; 1CGP, catabolite activator protein (cap); 2ORI, 434 repressor; 1HDD, Drosophila homeodomain protein; 1HCR, Hin recombinase. A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Materials and Methods
Bacterial Strains and Plasmids.
Escherichia coli JM101 strain was used for all experiments except for activation in trans, where ET8894ΔglnAntrBC was used to avoid cross-activation of the nifH promoter by NtrC, as described (19). Plasmid pRJ7511 (20) carries the Bradyrhizobium japonicum nifA gene or its derivatives. Plasmid pACYCNifA carries the B. japonicum nifA gene constitutively expressed from the cat promoter. This plasmid was constructed by inserting a BamHI–PstI fragment from pRJ7511 into pACYC177. Plasmids pKKnifH and pVB007 are derivatives of pKK232–8 (21) and carry the cat gene under the control of the Klebsiella pneumoniae and S. meliloti (deleted of the enhancer) nifH promoters, respectively. Plasmids pRT22 (22) and pMB210 (23) carry the K. pneumoniae and S. meliloti nifH promoter regions, respectively, fused to lacZ gene. Plasmid pUCNifA was constructed by subcloning the entire S. meliloti nifA gene, or its mutant derivatives, into pUC19. Plasmid pCU101(19) carries the S. meliloti nifH promoter fused to the lacZ gene, and pSU003 is a derivative from pCU101 without the enhancer.
β-Galactosidase Assays.
Strains carrying the different plasmids were grown in modified NFDM medium as described (20), at 30°C in aerobic or microaerobic conditions until they reached an optical density of 0.4–0.6 at 600 nm, as described (19).
In Vivo Dimethyl Sulfate (DMS) Footprinting.
The accessibility of K. pneumoniae nifH promoter DNA to DMS was performed in vivo as described (19). A 32P-5′-labeled synthetic oligonucleotide priming upstream of the nifH promoter was extended with 0.5 unit of the Klenow fragment of DNA polymerase for 10 min at 50°C, and the products were analyzed on sequencing gels.
Immunoblotting Techniques.
E. coli cells, expressing the nifA mutant derivatives, were cultured as described for β-galactosidase. Cells were pelleted, suspended in SDS sample buffer, and incubated for 10 min at 90°C. Detection was carried out as described previously (19). Two peptides, corresponding to the sequences of the hth of BjNifA (QAKAARLLGLTPRQVGY) and SmNifA (QAKAARILEKTPRQVGY), were synthesized and conjugated to BSA. New Zealand rabbits were immunized with these products to produce the specific polyclonal antibodies.
Site-Directed and PCR Mutagenesis.
When required, single mutant oligonucleotides were synthesized to generate the specific mutants by PCR site-directed mutagenesis or by replacing the appropriated restriction fragment, as described (19). PCR mutagenesis was performed essentially as described (24). The PCR products were ligated into pACYCNifA, pRJ7511, or pUCNifA plasmids, which were restricted with the appropriate enzymes to replace the wild type with mutant fragment.
Selection and Screening Systems.
E. coli JM101 cells harboring pKKnifH plasmid were electrotransformed with pools of mutant nifA genes in pACYCNifA. Expression of the cat gene in the former plasmid is activated by the BjNifA derivatives coded in the latter plasmid. The minimal inhibitory concentration (MIC) of chloramphenicol (Cm) for cells that carried the parental gene was determined, and a higher concentration was used in plates to select for mutants with enhanced transcriptional activity, as indicated below.
Sensitivity of NifA Mutant Proteins to Oxygen.
E. coli cultures with the appropriate plasmids were grown in NFDM under microaerobic conditions. When cultures reached an OD of 0.4–0.6 at 600 nm, they were shifted to aerobic conditions, as described (20). Every 20 min a 2-ml aliquot was taken to measure β-galactosidase activity and the amount of the NifA derivatives by immunoblotting.
Results and Discussion
G10 of the hth Is Critical for DNA Binding.
We compared the DNA-binding properties of several NifA proteins by in vivo footprinting. We observed that the protein of S. meliloti interacts very weakly with the enhancer of the nifH gene, compared with B. japonicum NifA (BjNifA) and K. pneumoniae NifA (KpNifA), such that it protected very weakly guanine-136 of the enhancer from methylation by DMS (Fig. 1A). To assess whether the lack of the conserved glycine G10 in the hth causes the weak binding, we replaced the glutamic acid with glycine by site-directed mutagenesis, resulting in SmNifAE10G (Fig. 1B). Additionally, a second derivative with a lysine-to-leucine substitution at position 11 (SmNifAE10G/K11L) was constructed to make the hth motif more similar to that of the other NifA proteins (Fig. 1B). In vivo footprinting analysis showed that both mutant proteins protected the nifH enhancer from methylation, indicating that they now bind strongly to the DNA (Fig. 2A) and that, indeed, the lack of G10 impairs this function. In the cocrystal structure of several hth proteins with their DNA-binding sites, G10, the first amino acid of the turn, is facing opposite to the DNA (25) (Fig. 1C). Thus, it is unlikely that residue 10 of the hth motif of NifA could make a direct contact with the DNA; the weak binding of SmNifA perhaps is due to an indirect effect, such as a more rigid or improperly oriented motif.
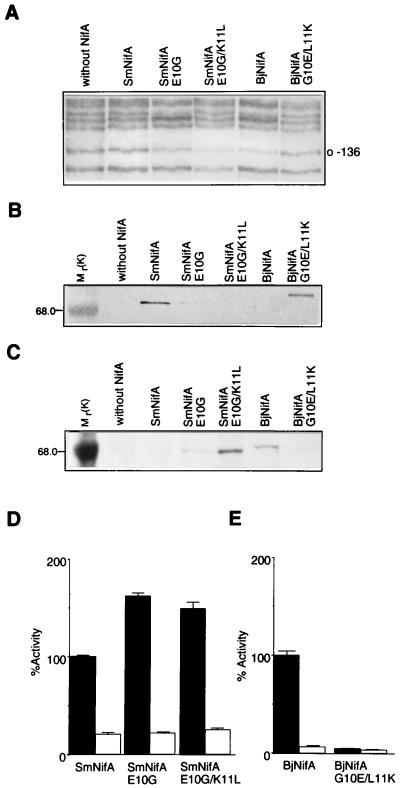
Figure 2.
Analysis of DNA binding, protein stability, and transcriptional activation of SmNifA, BjNifA, and their mutant derivatives. (A) Footprinting in vivo of the K. pneumoniae nifH enhancer with different NifA proteins and mutant derivatives, as indicated. Guanine-136 of the nifH enhancer is marked. (B and C) Immunodetection of the SmNifA and BjNifA (see Materials and Methods) proteins and their mutant derivatives. Antibodies raised against synthetic peptides corresponding to the hth of SmNifA or BjNifA were used for the immunodetection shown in B and C, respectively. Mr values of control proteins are denoted. Note that the anti-SmNifA hth peptide antibodies did not react against either BjNifA or SmNifAE10G/K11L and only weakly against SmNifAE10G, but efficiently recognized BjNifAG10E/L11K. Conversely, antibodies raised against BjNifA hth peptide did not recognize either SmNifA or BjNifAG10E/L11K and reacted weakly against SmNifAE10G, but efficiently recognized SmNifAE10G/K11L. (D and E) Transcriptional activation of a nifH-lacZ fusion carrying (solid bars) or deleted of (open bars) the enhancer by different NifA proteins and their mutant derivatives, as indicated. Because SmNifA and BjNifA are coded in different vectors and expressed from different promoters, it is not feasible to correlate actual β-galactosidase activities. Values represent the mean ± SD of three independent experiments and are expressed as percentage of β-galactosidase activity in Miller units.
Transcriptional Activation Activity of BjNifA and SmNifA.
Notwithstanding the weak DNA binding, SmNifA activates efficiently nif gene expression (23) (it is a natural protein after all), and the SmNifAE10G and SmNifAE10G/K11L mutants showed a very clear but moderate increase in transcriptional activation when strongly bound at the enhancer (Fig. 2D). These results suggest that SmNifA has a highly efficient transcriptional activation function that allows nif gene expression even when weakly bound to the enhancers. To test this hypothesis we carried out two different experiments.
In the first experiment we altered the hth motif of BjNifA to make it as that of SmNifA by constructing a G10-to-E and L11-to-K (BjNifAG10E/L11K) derivative (Fig. 1B). We rationalized that if BjNifA has a similar transcriptional activation activity as SmNifA, impairing its DNA-binding properties would result only in a moderate reduction of nifH expression, whereas if it is less active, then it should be more dependent on a strong interaction with the enhancers. As anticipated, the mutations strongly impaired the binding to the nifH enhancer (Fig. 2A) and, in contrast to SmNifA, the mutant protein failed to activate nifH gene expression (Fig. 2E). Immunodetection of the NifA derivatives showed that the mutations in the hth motif did not significantly affect the stability of any of the proteins (Fig. 2 B and C). Interestingly, the mutations selectively inverted the immunoreactivity of the SmNifA and BjNifA derivatives to antibodies raised against synthetic peptides comprising the hth of SmNifA (Fig. 2B) and BjNifA (Fig. 2C).
In the second experiment we directly compared the activation function of each protein by assaying their ability to activate transcription from solution, that is, to activate promoters devoid of the enhancer. Promoters that form strong, closed complexes with the RNA polymerase σ54, such as the S. meliloti nifH (SmnifH), can be activated partially by NifA when deleted from the enhancers (26). If SmNifA is less dependent on the binding to the DNA, we rationalized that it would be able to activate at higher levels than BjNifA, a SmnifH promoter lacking the enhancer. Fig. 2D shows that SmNifA significantly activates this promoter (at about 25% of the intact promoter), in sharp contrast to BjNifA, which was almost totally dependent on the enhancer (Fig. 2E). This result indicates that, although SmNifA is still dependent on the binding to the enhancers for full activation [required for fidelity of activation (27)], indeed it has a higher transcriptional activation activity than BjNifA.
Directed Evolution of the Central Domain of BjNifA.
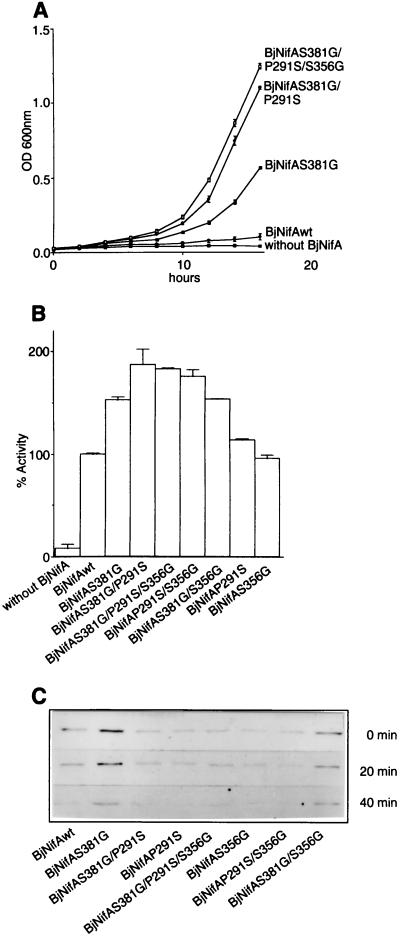
How did SmNifA achieve its higher transcriptional activity? Perhaps this was selected to compensate for the weak DNA binding caused by the G10-to-E mutation, although it is also possible that a lower DNA-binding activity was selected in response to an enhanced activation activity detrimental to the cell. Reconstructing the sequence of ancestral proteins by using sequence information from closely related extant proteins has helped to identify residues responsible for enhanced activity (28). Alternatively, directed evolution (29) can provide information about relevant positions critical for enhanced activity. To explore possible paths that lead SmNifA to its higher transcriptional activity, we subjected BjNifA to a directed evolution process. We developed a positive selection scheme for NifA mutants displaying enhanced ability to activate the expression of a Cm resistance gene under the control of a KpnifH promoter. The activation domain of NifA was mutagenized by random, error-prone PCR, and the derivatives conferring increasing resistance to Cm were selected. About 1 × 105 clones were analyzed in each cycle. The majority of the mutations impaired the function, as observed for other proteins (30), about 30% retained some activity (as assayed for their ability to activate a nifH-lacZ fusion), and only very few grew on Cm. In the first cycle three clones were selected on 10-μg/ml Cm (the MIC of the BjNifA wild type is 8 μg/ml), and the DNA fragment corresponding to their activation domains sequenced. Remarkably, all had an amino acid replacement in common, S381 to G. Two of them had other synonymous substitutions (BjNifAS381G/A377A/L342L). Because the activity of the mutant derivatives was very similar, the clone with the single mutation (BjNifAS381G) was chosen and subjected to a second cycle of mutagenesis and selection on 15-μg/ml Cm plates. Only one colony grew faster than the parental strain. This clone, BjNifAS381G/P291S, which had an additional P291-to-S substitution, was mutagenized further, and only a single clone of the third generation (BjNifAS381G/P291S/S356G) grew on plates containing 20 μg/ml Cm. Sequencing of their entire activation domain showed that it had S356 to G as the only additional substitution. Fig. 3A shows growth curves of the strains with the wild type and each of the mutant BjNifA proteins, which successively attained a faster growth in aerobic liquid cultures containing Cm, as higher expression of a nifH-lacZ fusion (Fig. 3B).
Figure 3.
Transcriptional activity, stability, and sensitivity to oxygen of the BjNifA derivatives. (A) Growth curves of strains carrying the wild type or the directly evolved BjNifA derivatives, as indicated. Cells were grown on NFDM medium with Cm (40 μg/ml) under aerobic conditions. The level of expression of the cat gene in pVB007, under the S. meliloti nifH promoter (deleted of the enhancer), and, therefore, the resistance to Cm is dependent on the activity of each BjNifA derivative. Values represent the mean ± SD of three independent experiments. (B) Transcriptional activation of a nifH-lacZ fusion by the BjNifA mutant derivatives. (C) Stability and sensitivity of BjNifA mutant proteins to oxygen. To analyze whether the mutations affected the stability of the protein or any intrinsic function leading to a higher transcriptional activation, we grew strains carrying each of the mutant BjNifA proteins in microaerobic cultures, and the amount of the protein was detected after being shifted to heavily aerated flasks. Samples were taken at time 0 min, 20 min, and 40 min, and the BjNifA derivatives were immunodetected in soluble cell extracts with an antibody raised against a polypeptide comprising part of the central domain. All mutant proteins had similar stability and oxygen sensitivity because they decreased at the same rate as the wild type after being shifted to air.
To assess the contribution of each mutation to the enhanced activity we segregated them and constructed the combination of all double mutants by different molecular genetic tech- niques. Their ability to activate the expression of a nifH pro- moter fused to lacZ was determined as β-galactosidase activity in microaerobic cultures in NFDM. In this assay the second- and the third-generation mutants (BjNifAS381G/P291S and BjNifAS381G/P291S/S356G) showed similar activities (about twice the wild type), whereas the mutant of the first generation (BjNifAS381G) was more active than any of the other two single mutants (BjNifAP291S and BjNifAS356G), which themselves had a similar activity as the wild type. The double mutant, BjNifAS381G/P291S, was slightly more active than BjNifAP291S/S356G and BjNifAS381G/S356G. These results are consistent with the additive effect of the mutations (Fig. 3A) and with the fact that the S381-to-G mutation was the only one selected in the first generation appearing in three independent clones.
Oxygen Stability of the Mutant BjNifA Derivatives.
To test whether the mutations affected the stability or a catalytic property of the mutant proteins, the relative concentration of the NifA derivatives was determined by immunoblotting. As shown in Fig. 3C, the mutant proteins were present at levels similar to the wild-type NifA. Our previous studies on the regulation of NifA activity showed that BjNifA is oxygen-labile (20). To determine the oxygen sensitivity of the mutant proteins, cultures harboring each of the mutant BjNifA derivatives were grown overnight microaerobically and shifted to heavily aerated conditions. Samples were taken at 20-min intervals and assayed for the fraction of protein remaining. Fig. 3C shows that the mutant BjNifA derivatives decreased at about the same rate as the wild type. Thus, this experiment indicates that the mutations did not significantly affect the oxygen stability of the mutant BjNifA derivatives.
NifA Is Trapped in a Restricted Sequence Space.
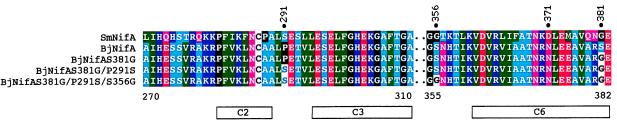
Despite the inexorable limitation of any attempt to explore the sequence space of a protein, the mutagenesis strategy and the number of independent BjNifA derivatives analyzed here allowed us to screen a great number of mutants, including all possible single and a considerable proportion of double mutants in the central domain. Therefore, we were surprised to find that, from the many possible substitutions, all those with higher activity changed to amino acids that are already present in the highly active SmNifA (Fig. 4) and normally are the most frequent amino acids at these positions in other EBP (9, 10). Thus, our directed evolution process led to the same solutions for enhanced activity as SmNifA.
Figure 4.
Multiple sequence alignment of SmNifA, BjNifA, and its mutant derivatives. Only regions near the mutated residues on BjNifA are shown. Dots indicate discontinuity of the alignment. Numbers above the sequences indicate amino acid positions mutated in BjNifA. Numbers below the alignment show amino acid positions of BjNifA. Boxes indicate the highly conserved regions in the EBP proteins (9, 10). The central domains of BjNifA and SmNifA are 68% identical, whereas the degree of identity of the whole family is around 40%.
Our attempts to further increase the activity of the central domain by the same mutagenic procedure were unsuccessful. Segregation by in vitro recombination of all individual mutations [gene shuffling (31)], with further mutagenesis, resulted only in the regeneration of the triple-mutant clone (with one additional mutation that did not affect the activity) after selecting on 25-μg/ml Cm plates. Thus, it is likely that with these three mutations the protein attained a local optimum in a sequence space accessible by point mutation [one-mutant distance neighborhood (32)].
That we found the same solutions for higher activity as natural proteins suggests that NifA is trapped in a very narrow activity “peak” on its fitness landscape. Is it possible to move NifA to a different activity “peak”? It has been proposed that multiple mutations can make a protein “jump beyond” to a new activity “peak” in its fitness landscape (32). We currently are searching for suppressor derivatives of stable mutants of BjNifA with residual activity in an attempt to explore new solutions on its sequence space or, following the metaphor of the fitness landscape (32), to move BjNifA to different activity “peaks.”
Conclusions
The results presented here show that SmNifA has found a novel optimum fitness, perhaps by increasing its activation function in response to a mutation that impaired DNA binding. Alternatively, a weak DNA binding could have been selected in response to an unfavorable high transcriptional activation function. Whatever the order of the events was, it is clear that neither this protein nor BjNifA is at its maximum ability to activate gene expression. Taken together, our results show that a modular protein can stay around its optimal fitness by dynamically balancing the activities of its different domains, a process we called reciprocal domain evolution. We propose that continuous compensatory mutations allow the exploration of the sequence space, which occasionally might result in novel activities. Thus, it is likely that reciprocal domain evolution has been a strong force in the sharpening of the contemporary proteins.
Acknowledgments
We are grateful to M. Olvera, R. Hernández, and S. Caro for technical assistance, P. Gaytan and E. López for oligonucleotides synthesis, T. Olamendi for polypeptides synthesis, E. Mata and G. Cabeza for assistance in rabbit immunizations, B. Valderrama for pVB007 plasmid, and X. Soberón, A. Alagón, A. Garciarrubio, M. Huynen, and P. Bork for fruitful discussions. E.M. thanks the Consejo Nacional de Ciencia y Tecnología and Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México, for support for a sabbatical leave. K.J. was a recipient of a Consejo Nacional de Ciencia y Tecnología scholarship. E.M. was an Alexander von Humboldt fellow. This research was supported in part by grants from Consejo Nacional de Ciencia y Tecnología (G0030-N9607) and The European Commission (CI1*-CT94–0060).
Abbreviations
- EBP
enhancer-binding proteins
- SmNifA and BjNifA
NifA from Sinorhizobium meliloti and Bradyrhizobium japonicum, respectively
- MIC
minimal inhibitory concentration
- DMS
dimethyl sulfate
- hth
helix–turn–helix
- Cm
chloramphenicol
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060444897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060444897
References
- 1.Henikoff E S, Greene A, Pietrokovski S, Bork P, Attwood T K, Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle R F. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle R F, Bork P. Sci Am. 1993;269:50–56. doi: 10.1038/scientificamerican1093-50. [DOI] [PubMed] [Google Scholar]
- 4.Netzer W J, Hart F U. Nature (London) 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 5.Chothia C. Nature (London) 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 6.Orengo C A, Jones D T, Thornton J M. Nature (London) 1994;372:631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 7.Kauffman S A, Weinberger E D J. Theor Biol. 1989;141:211–245. doi: 10.1016/s0022-5193(89)80019-0. [DOI] [PubMed] [Google Scholar]
- 8.Drummond M, Whitty P, Wootton J. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osuna J, Soberón X, Morett E. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H S, Berger D K, Kustu S. Proc Natl Acad Sci USA. 1993;90:2266–2270. doi: 10.1073/pnas.90.6.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kustu S, Santero E, Keener J, Popham D, Weiss D. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merrick M J. Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 14.Rippe K, von Hippel P H, Langowski J. Trends Biochem Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 15.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 16.Morett E, Buck M. Proc Natl Acad Sci USA. 1988;85:9401–9405. doi: 10.1073/pnas.85.24.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Missaillidis S, Jaseja M, Ray P, Chittock R, Wharton C W, Drake A F, Buck M, Hyde E I. Arch Biochem Biophys. 1999;361:173–182. doi: 10.1006/abbi.1998.0980. [DOI] [PubMed] [Google Scholar]
- 18.Wintjens R, Rooman M. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- 19.González V, Olvera L, Soberón X, Morett E. Mol Microbiol. 1998;28:55–67. doi: 10.1046/j.1365-2958.1998.00772.x. [DOI] [PubMed] [Google Scholar]
- 20.Morett E, Fischer H M, Hennecke H. J Bacteriol. 1991;173:3478–3487. doi: 10.1128/jb.173.11.3478-3487.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosius J. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 22.Tuli R, Merrick M J. J Gen Microbiol. 1988;134:425–432. doi: 10.1099/00221287-134-2-425. [DOI] [PubMed] [Google Scholar]
- 23.Better M, Ditta G, Helinski D. EMBO J. 1985;4:2419–2424. doi: 10.1002/j.1460-2075.1985.tb03950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tintut Y, Gralla J D. J Bacteriol. 1995;177:5818–5825. doi: 10.1128/jb.177.20.5818-5825.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Susuki M, Brenner S, Gerstein M, Yagi N. Protein Eng. 1995;8:319–328. doi: 10.1093/protein/8.4.319. [DOI] [PubMed] [Google Scholar]
- 26.Buck M, Cannon W. Mol Microbiol. 1992;6:1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- 27.Morett E, Buck M. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 28.Opitz J G, Ciglic M I, Haugg M, Trautwein-Fritz K, Raillard S A, Jermann T M, Benner S A. Biochemistry. 1998;37:4023–4033. doi: 10.1021/bi9722047. [DOI] [PubMed] [Google Scholar]
- 29.Kushner O, Arnold F H. Trends Biotechnol. 1997;12:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 30.Suckow J, Markiewicz P, Kleina L G, Miller J, Kisters-Woike B, Muller-Hill B. J Mol Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 31.Stemmer W P. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 32.Kauffman S, Levin S. J Theor Biol. 1987;128:11–45. doi: 10.1016/s0022-5193(87)80029-2. [DOI] [PubMed] [Google Scholar]