Abstract
Although APC mutations occur at a high frequency in colorectal cancers, few studies have performed a comprehensive analysis by screening the whole gene for mutations and assessing allelic loss. APC seems to act as a tumor-suppressor gene in a “nonclassical” fashion: data from familial adenomatous polyposis (FAP) show that the site of the germ-line mutation determines the type of “second hit” in FAP tumors, and simple protein inactivation is selected weakly, if at all. In this study, we screened the entire coding region of APC for mutations and assessed allelic loss in a set of 41 colorectal cancer cell lines. Of 41 cancers, 32 (83%) showed evidence of APC mutation and/or allelic loss. We identified several APC mutations and found a “hotspot” for somatic mutation in sporadic colorectal tumors at codon 1,554. Our results suggest that APC mutations occur in the great majority of colorectal cancers, the exceptions almost all being RER+ tumors, which may substitute for altered APC function by mutations in β-catenin and/or at other loci. When combined with previously published data, our results show that there is interdependence of the “two hits” at APC in sporadic colorectal tumors as well as in FAP. APC mutations in the “mutation cluster region,” especially those close to codon 1,300, are associated with allelic loss, whereas tumors with mutations outside this region tend to harbor truncating mutations. The causes of this phenomenon are probably selection for retained N-terminal and lost C-terminal APC functions, effects on β-catenin levels, and APC protein stability.
Mutations in the APC gene can be detected in many colorectal cancers, and these mutations are likely to be the initiating events in tumorigenesis (1). Although most colorectal tumors are thought to follow a genetic pathway involving APC, the proportion of tumors harboring APC mutations is unclear and varies among studies. The reasons for this inconsistency are 4-fold. First, there are difficulties inherent in screening any gene for mutations in tumors, especially early lesions. Second, the APC gene is unusually large, making comprehensive screening time consuming and difficult, especially when studying archival material. Third, it has been reported that most somatic APC mutations occur within a small part of the gene—the so-called “mutation cluster region” (ref. 2; MCR; codons 1,286–1,513)—and studies have understandably tended to concentrate on this region to the exclusion of other parts of the APC gene. Fourth, allelic loss (loss of heterozygosity or LOH) at APC has been assessed relatively infrequently in parallel with mutation screening. Estimates of the frequency of allelic loss in colorectal carcinomas vary widely, but many studies report 30–40% LOH.
APC is frequently cited as a paradigm of how mutations at tumor suppressor loci cause cancer. Two protein-inactivating mutations at APC often occur in sporadic tumors of the colorectum (3, 4). In common with other tumor suppressors, one APC mutation usually leads to a truncated protein, and the other is either a similar mutation or a change resulting in allelic loss. In the Mendelian disorder familial adenomatous polyposis (FAP), characterized by multiple colorectal tumors, a truncating APC mutation is inherited (5–7), and in adenomas, the other allele either harbors a similar mutation or, more rarely, is lost (8). Homozygous deletions of APC seem to be very rare or absent.
We have previously shown that, contrary to classical tumor suppressor theory, the type of “second hit” at APC in FAP patients' tumors depends on the site of the germ-line mutation (9). Patients with germ-line mutations around codon 1,300 tend to acquire their second hit by allelic loss and to suffer more severe disease. Other FAP patients tend to acquire their second hit by a truncating mutation in the MCR. These results suggest that APC mutations are not selected simply for loss of function but that the balance between lost C-terminal functions and retained N-terminal functions is important. Some mutant APC genotypes, moreover, may provide cells with a greater selective advantage than others, as further evidenced by the finding of a third hit—loss of the germ-line mutant—in some FAP tumors, which do not show allelic loss as a second hit (4, 9, 10).
Selection of advantageous APC mutants in the FAP colon is likely to be more stringent than in the normal colon. In FAP, multiple tumors are initiated and grow relatively early in life. Thus, the tumors sampled have grown rapidly and have outgrown their peers. Tumors with the fittest genotypes will be larger and hence more likely to be sampled. In sporadic cancer, disease occurs later in life, and lesions are usually solitary. Thus, competition is less severe, and a wider range of APC genotypes, with widely varying selective advantages, will be seen in sampled tumors. This factor aside, the selective parameters in the normal colon may otherwise be very similar to those in the FAP colon. Thus, it is reasonable to test the hypothesis that the “two hits” at APC are associated with one another in sporadic colorectal tumors as well as in FAP. We have screened the entire coding region of APC for mutations and also assessed allelic loss in a set of 40 colorectal cancer cell lines. These samples were chosen, primarily because they avoid the problem of contaminating normal tissue. Cell lines also provide a ready source of high-quality DNA and, in many of the lines we studied, mRNA and protein. The data resulting from this study (i) provide an accurate estimate of the proportion of colorectal cancers that harbor APC mutations; (ii) identify the positions of those mutations and the frequency of allelic loss; (iii) test, in sporadic colorectal cancers, whether mutations near codon 1,300 are associated with allelic loss and whether mutations elsewhere in the gene are associated with truncating mutations; and (iv) determine the frequency of colorectal cancers that harbor neither APC mutations nor β-catenin mutations and may therefore progress along a distinct genetic pathway.
Methods
The colorectal cancer cell lines were derived from public sources, from kind gifts, and from tumors cultured locally. DNA was prepared by using standard methods. mRNA was purified from the cell lines by using the Quick prep mRNA purification kit (Amersham Pharmacia). cDNA for the protein truncation test (PTT) was then made by using the First Strand cDNA synthesis Kit (Amersham Pharmacia). APC exon 15 was screened for mutations by using the protein truncation test (PTT) on genomic DNA as described (11). Exons 1–14 and exon 15 regions G–I (6) were screened by single-strand conformation polymorphism (SSCP) analysis by using published oligonucleotides and PCR conditions (6); from cell lines with mRNA available, the PTT was also used to screen exons 1–14 as described (11). For any samples with bandshifts in the PTT or SSCP analyses, the appropriate region or regions of the gene were reamplified in the PCR and sequenced in forward and reverse orientations by using the ABI377 semiautomated sequencer (Applied Biosystems). Certain samples were cloned into the pGEM TEasy vector system (Promega), if necessary, to define the mutation.
Allelic loss at APC could not be assessed directly in the cell lines, owing to the absence of constitutional DNA. We therefore initially genotyped each cell line at five highly polymorphic microsatellite loci (D5S2055, D5S2001, D5S659, D5S656, and D5S489) mapping close to APC (http://www.cedar.genetics.soton.ac.uk) by using standard protocols, dye-labeled oligonucleotides, and the ABI377 sequencer. Cell lines apparently homozygous at four or five of the microsatellite loci were considered to show allelic loss, because the probability of four of five or five of five loci being homozygous in the germ line is less than about 0.001, as determined by using Centre d'Étude du Polymorphisme Humain (Paris)/Genethon allele frequencies (http://www.genethon.fr). In all cases, allelic loss was confirmed wherever possible by (i) inspection of sequencing results (for absence of wild-type nucleotides in regions of truncating mutation) and (ii) the absence of normal protein on PTT gels that showed the truncated allele. Although both these phenomena can arise by mechanisms other than allelic loss, they are unlikely to produce false-negative indications of loss when studying cell line samples. In fact, every case assessed as having allelic loss by the microsatellites was consistent with the sequencing and/or PTT findings.
For mutation screening of β-catenin, exon 3 of each cancer cell line was amplified with the following oligonucleotides: 5′-ATTTGATGGAGTTGGACATGGC-3′ and 5′-CCAGCTACTTGTTCTTGAGTGAAGG-3′. PCR was performed by using standard conditions and 50 mM KCl/10 mM Tris⋅HCl, pH 9.0/0.1% Triton X-100/1.5 mM MgCl2. A denaturing cycle of 94°C for 4 min was followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. SSCP analysis was then used to screen for mutations by using precast 20% acrylamide gels on the Amersham Pharmacia Phast system. After electrophoresis, the gels were stained with silver nitrate. Samples that showed altered mobility patterns were sequenced in forward and reverse orientations by using newly generated PCR products. Microsatellite instability in the cell lines was determined as described (12).
Data on APC mutations were also obtained from the APC Mutation Database, version 5.0 (http://perso.curie.fr/Thierry.Soussi/APC.html).
Results
Of the 40 colorectal cancer cell lines (Table 1 and Fig. 1), 32 (83%) of them showed evidence of an APC mutation. In 29 of 33 (88%) of these cancers, two hits were found. Of these 29 cancers, 17 (59%) showed allelic loss plus truncating mutation; 10 (34%) harbored two truncating mutations; 1 had a truncating mutation plus putative splice variant; and 1 had a truncating mutation plus missense change (E1317Q). In the remaining four cases, only one truncating APC mutation or allelic loss was found in each cancer. The overall frequency of allelic loss at APC was 48% (19 of 40 cancers). In the seven cell lines without any evidence of APC changes, four (10% of total) had β-catenin mutations in exon 3. In the remaining three cell lines, we found neither APC nor β-catenin mutations.
Table 1.
Summary of results from colorectal cancer cell lines studied
| Cell line | Age | Sex | Site | Dukes' stage | Differentiation | LOH | APC mutation 1 | Nucleotide change | APC mutation 2 | Nucleotide change | β-Catenin mutation | RER status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C32 | 66 | F | Colon | C | Well | L | 776 | TATAGACAAT to TATAAT | − | |||
| C106 | 78 | F | Rectum | A | Moderate | NL | 1,238 | ACAGAGTAG to ACAGTAG | 1,490 | TACATTTTGC to TACATTTTTGC | − | |
| C125 | L | |||||||||||
| C10 | 71 | M | Descending colon | B | Moderate | L | − | |||||
| LS180 | 58 | F | Colon | B | Well | NL | Codon 45 | + | ||||
| COLO678 | L | 1,554 | GAAAAAACT to GAAAAAAACT | |||||||||
| GP2D | F | Recurrence of colon cancer | B | Poor | L | 1,444 | CTCAAACAGC to CTCAGC | + | ||||
| C99 | 69 | M | Rectum | C | Moderate | L | 1,367 | CAG to TAG | − | |||
| C70 | 69 | F | Sigmoid colon | B | Moderate | L | 1,309 | TAAAAGGAAAGA to TAAAATAAAAG | − | |||
| COLO741 | 69 | F | Colon, pelvic wall metastasis | D | NL | (+) | ||||||
| GP5D | F | Recurrence of colon cancer | B | Poor | L | 1,444 | CTCAAACAGCT to CTCAGCT | + | ||||
| VACO5 | 78 | F | Caecum | C | Poor | NL | 1,554 | GAAAAAACT to GAAAAAAACT | 1,499 | GGA to TGA | + | |
| HT55 | NL | 1,131 | CAA to TAA | 1,303 | CAA to TAA | − | ||||||
| C75 | 56 | M | Sigmoid colon | D | Moderate | NL | 811 | TCA to TGA | 1,486 | GATACTTTATTA to GATA | − | |
| C84 | 67 | M | Caecum | C | Poor | NL | 1,451 | − | ||||
| CX-1 | NL | 853 | GAG to TAG | 1,554 | GAAAAAACT to GAAAAAAACT | ? | ||||||
| C80 | 69 | M | Rectum | D | NL | − | ||||||
| COLO205*† | 70 | M | Caecum, ascitic fluid | D | Poor | NL | 1,554 | GAAAAAACT to GAAAAAAACT | − | |||
| CACO2† | 72 | M | Colon | Well | L | 1,367 | CAG to TAG | − | ||||
| SW480 | 50 | M | Colon | B | Moderate | L | 1,338 | CAG to TAG | − | |||
| SW403 | 51 | F | Colon | B | NL | 1,197 | CATTTTCATTCTCAAA to CATTTTCATTTTAAA | 1,278 | TCA to TAA | − | ||
| SW948 | 81 | F | Colon | C | NL | 1,114 | CGA to TGA | 1,429 | CAA to TAA | − | ||
| LOVO | 56 | M | Colon, distant lymph node | D | NL | 1,114 | CGA to TGA | 1,430 | AAACCAT to AAACAT | + | ||
| SW837 | 53 | M | Rectum | C | Poor | L | 1,450 | CGA to TGA | (+) | |||
| SW1222 | Colon | C | Moderate | L | 1,306 | GAA to TAA | (+) | |||||
| SW48 | 83 | F | Colon | C | Poor | NL | Codon 33 | + | ||||
| COLO320 | 55 | F | Sigmoid colon | C | Moderate | L | 811 | TCA to TGA | − | |||
| LS411 | 32 | M | Caecum | B | Poor | NL | 789 | CAG to TAG | 1,556 | GAGGCAGAAA to GAGGCAGGAAA | + | |
| SKCO1‡ | 65 | M | Colon | NL | 1,443 | CCACCTCC to CCACTCC | 1,317 | GAA to CAA (E to Q missense) | − | |||
| HCT15 | M | Colon | C | L | 1,417 | GTGGCATT to GTGGATT | + | |||||
| VACO4A | 59 | M | Rectum | D | Moderate | L | 1,354 | GAATTTTCTTC to GAATTC | − | |||
| LS174T | 58 | F | Colon | B | Well | NL | Codon 45 | + | ||||
| T84 | 72 | M | Colon, lung metastasis | L | 1,488 | ACTTTATTA to ACTTTTTA | − | |||||
| HCA7 | 58 | F | Hepatic flexure | B | Moderate | NL | + | |||||
| HRA19 | 66 | M | Rectum | B | Well | L | 1,450 | CGA to TGA | − | |||
| LS1034 | 54 | M | Caecum | C | Moderate/poor | L | 1,309 | GAAAAGATT to GATT | − | |||
| HCT116 | M | Colon | NL | Codon 45 | + | |||||||
| HCA46†§ | 53 | F | Sigmoid colon | C | Poor | NL | 213 | CGA to TGA | −8 intron 7 | TTAATTTTT to TTAGTTTTT | − | |
| HT29 | 44 | F | Colon | Moderately well | NL | 853 | GAG to TAG | 1,555 | GAAAAAACT to GAAAAAAACT | − | ||
| SW1417 | 53 | F | Colon | C | L | 1,450 | CGA to TGA | − |
The Table shows clinicopathological data (where known), allelic loss at APC (L, loss; NL, no loss), codon at which truncating (and missense) APC mutations occur, sequence change of each mutation, presence of β-catenin mutations in exon 3, and RER status [+, high level; (+), low level; −, no ].
The 1,554 frameshift mutation in COLO205 seems to be homozygous/hemizygous, and no normal product is seen with the PTT in the region of the mutation. Microsatellites suggest, however, that this tumor does not show allelic loss. It is possible that another type of mutation, such as a small deletion, is the other hit.
† Exons 5 and 6 of β-catenin were not screened systematically for mutations in this study but have been reported in COLO205, CACO2, and HCA46 (22).
‡ SKCO1 harbors a E1317Q missense change, hypothesized by White et al. (23) and Frayling and coworkers (9) to act as a colorectal tumor predisposition allele when present in the germ line. E1317Q was found as somatic mutation in one colorectal tumor by White et al. (23). We are unable to determine whether this change in SKCO1 is germ-line or somatic, but the absence of allelic loss or other detectable APC mutation in this line suggests that E1317Q may be pathogenic.
§ Unique among colorectal cancer cell lines, HCA46 has no APC protein as determined by Western blotting. The intron 7/exon 8 845–848 A → G splice acceptor mutation is predicted to lead to abnormal mRNA splicing by comparison with a germ-line change found by Pedemonte et al. (24) close by in intron 7 (845–817 A → G) in an almost identical repeated sequence. The mutation of Pedemonte et al. led to the identification of an aberrant APC mRNA. We found no evidence of alternatively spliced mRNA by using reverse transcription–PCR in HCA46, suggesting mRNA instability but leaving open the possibility that the mutation is nonpathogenic. HCA46 was shown by Ilyas et al. (22) to have a β-catenin exon 5 (codon 183) mutation.
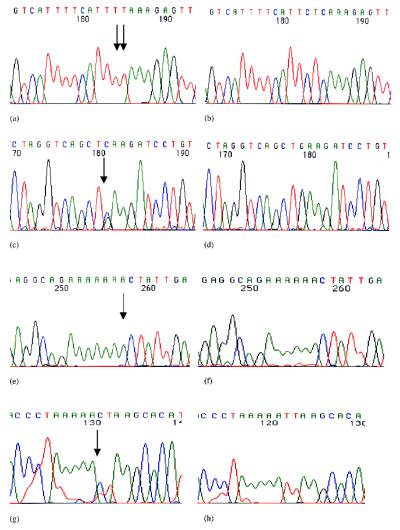
Figure 1.
Examples of mutations in the APC gene. (a) TTCTC to TTTT change in the colorectal cell line SW403 at codon 1,197 (cloned forward sequence). (b) Normal APC sequence from the same region. (c) E1317Q (GAA to CAA) change in the colorectal cell line SKCO1 (forward sequence). (d) Normal APC sequence from the same region. (e) GAA ins1a in the colorectal cell line COLO678 at codon 1,554 (forward sequence). (f) Normal APC sequence from the same region. (g) The TTAA to TTAG sequence change in intron 7 of HCA46 (reverse sequence). (h) Normal APC sequence from the same region.
The genetic changes found at APC comprised several known and some previously unidentified mutations. All mutations occurred 5′ to the SAMP repeats (13). We found a “hotspot” for somatic mutation in colorectal tumors at codon 1,554 (Fig. 1), 3′ to the MCR. The mutation at codon 1,554 was the most common non-LOH change detected, comprising 6 of 41 (15%) of all truncating mutations. Of these 41 truncating mutations, 23 (56%) occurred in the MCR, in agreement with previous studies (2). In those cancers with only one identified hit at APC, we presume that most did harbor a second hit but that this hit either was missed by the methods used or was in a noncoding region; possibilities include truncating mutations not detected by SSCP analysis, mutations in introns or gene control regions, whole exon deletion, or missense mutations outside exon 15 regions G–I. Transcriptional silencing of one or both APC alleles is also possible (14). With the possible exception of line HCA7 (Table 1), our preliminary analyses have shown little evidence of APC promoter methylation that is consistent with a second hit (data not shown). No cell line showed evidence of homozygous deletion of APC.
Of the seven tumors with no detected APC mutations or allelic loss, all bar one were RER+, including four tumors with β-catenin mutations (Table 1). It is not known whether the RER+, APC-wild-type lines originated from hereditary nonpolyposis colon cancer (HNPCC) patients or from sporadic tumors, although these cases had unremarkable ages of presentation and location in the colon, and it is therefore most unlikely that all were from HNPCC families (Table 1). The difference between APC mutation frequencies in RER+ and RER− cancers was significant (P < 0.005, Fisher's exact test). In the RER+ cell lines with one or more mutations at APC, there was no trend away from allelic loss as a second hit (P = 1.00, Fisher's exact test) or toward frameshift rather than nonsense truncating mutations (P > 0.5, Fisher's exact test), as some studies have reported (15). Our data are therefore consistent with the notion that APC mutations occur before mismatch repair gene changes in most cases of sporadic colorectal tumorigenesis (16).
In FAP adenomas, the likelihood that a tumor shows allelic loss depends strongly on the site of the germ-line mutation (9). In sporadic colorectal tumors, it is not possible to determine which hit was first and which was second. Nevertheless, our data suggest that the first hit has determined the second hit in these tumors as well. On the assumption that none of the colorectal cancer cell lines is derived from an FAP patient, we have combined our data with those from previous studies that have screened sporadic colorectal adenomas and carcinomas both for truncating mutations and for allelic loss; these data were derived from the APC Mutation Database. Our studies of FAP had shown that allelic loss was associated with germ-line mutation between codons 1,194 and 1,392 (at most). The combined data from the sporadic colorectal cancers show that tumors that harbor a truncating mutation before codon 1,194 have a significantly lower frequency of allelic loss (χ22 = 24.12; P < 0.001) than tumors with a truncating mutation after codon 1,194 (Table 2). (This analysis relies on the assumption that homozygous deletions are very rare or absent at the APC locus, which has been suggested by other workers and which is supported by our failure to detect any such deletions in our set of cell lines.) Sporadic colorectal cancers also resemble FAP tumors with truncating mutations between codons 1,194 and 1,392 in that they have a significantly higher frequency of allelic loss (Table 3) than tumors with mutations 3′ to this region (χ21 = 4.10; P < 0.05).
Table 2.
Association between the sites of the two APC mutations in sporadic colorectal tumors: Is allelic loss associated with truncating mutations after codon 1,192?
| Sites and types of mutation
|
Total | ||||||
|---|---|---|---|---|---|---|---|
| LOH | <1,194 | >1,194 | <1,194 | <1,194 | >1,194 | ||
| LOH | LOH | LOH | <1,194 | >1,194 | >1,194 | ||
| Observed | 0 | 5 | 36 | 3 | 16 | 8 | 68 |
| Expected | 6.3 | 8.1 | 20.7 | 2.6 | 13.3 | 17.0 | 68 |
Data are derived from the cell lines analyzed in this study and sporadic colorectal tumors in the APC Mutation Database. Only tumors with two hits identified are included. A caveat must be added; namely, the analysis makes the necessary assumption that all of the data are unbiased by incomplete screening of tumors for truncating mutations and/or LOH. Numbers of tumors are shown, categorized according to their mutations, which are classified as three types: (i) allelic loss (LOH), which is assumed to be equivalent to homozygous deletion if present on both alleles; (ii) truncating mutations before codon 1,194; and (iii) truncating mutations after codon 1,194. Expected numbers of tumors are calculated on a multiplicative basis, assuming independence of the two hits. In addition to the expected deficiency of homozygous deletions, the excess of LOH is apparent in tumors with truncating mutations after codon 1,194. χ2 = 24.12; df = 2; P < 0.001.
Table 3.
Association between the sites of the two APC mutations in sporadic colorectal tumors: By comparison with FAP (9), is allelic loss more common in sporadic tumors with a truncating mutation between codons 1,194 and 1,392 than in tumors with truncating mutations after codon 1,392?
| Sites and types
of mutation
|
Total | ||
|---|---|---|---|
| 1,194–1,392 | >1,392 | ||
| LOH | 18 | 16 | 34 |
| No LOH | 7 | 19 | 26 |
| Total | 25 | 35 | 60 |
As for Table 2, data are derived from the cell lines analyzed in this study and sporadic colorectal tumors in the APC Mutation Database, and only tumors with two hits identified are included. Numbers of tumors are shown, categorized according to their mutations, which are classified as (i) allelic loss (LOH) versus no loss (that is, an identified truncating mutation) and (ii) truncating mutations between codons 1,194 and 1,392 versus truncating mutations after codon 1,192. A problem in assessing these data is how to categorize tumors with one mutation between 1,194 and 1,392 and the other mutation after codon 1,392. We have taken an essentially conservative approach, which is to count these tumors twice in the table. Expected numbers of tumors (not shown) are then calculated on a multiplicative basis, assuming independence of the two hits. LOH is more strongly associated with tumors with truncating mutations between codons 1,194 and 1,392 (χ2 = 4.10; df = 1; P < 0.05). If tumors with one mutation between 1,194 and 1,392 and the other mutation after codon 1,392 are counted once (that is, half in each category), the association is stronger (χ2 = 4.53).
No associations were found (details not shown) between any of the available clinicopathological data (Table 1) and any of the molecular data (presence, site, and type of APC mutation, β-catenin mutation, and RER status). Clinicopathological details were, however, lacking for some of the cancers, and this lack undoubtedly reduced the power of the study to detect clinicopathological–molecular associations.
Discussion
We have shown that the great majority of colorectal cancers harbor APC mutations and that a thorough screen of all coding regions of the gene and analysis of allelic loss can detect two hits at APC in over 80% of colorectal cancers under favorable conditions, such as the analysis of tumor cell lines. These conclusions rely on the set of colorectal cell lines that we have studied being broadly representative of sporadic colorectal carcinomas; the case for this assumption, although plausible as regards APC mutations at least, is unproven. The much higher detection rate of APC mutations in this study than in some others—including some of our own work—probably originates from three problems inherent in the other studies: (i) failure to screen the whole of the gene; (ii) contaminating normal tissue or the presence of multiple clones, particularly in early adenomas in FAP; and (iii) the unproven hypothesis that APC mutations are selected less often or at a later stage in some colorectal cancers, such as “flat” lesions. Another possibility is that some colorectal adenomas grow without APC mutations (17) but that these lesions regress and do not become cancerous.
We have found frequent frameshift mutations at codon 1,554 of APC in colorectal tumors; 6 of 41 (15%) of all non-LOH mutations were at this site. This region comprises an (A)6 tract that is presumably prone to replication mispairing and slippage. Mutations at this site occurred in both RER+ and RER− tumors and were found by using independent PCR products for the PTT and sequencing, strongly suggesting that the tumors did not result from a PCR artifact. Mutations at codons 1,554 have been reported to occur frequently in upper gastrointestinal cancers (18) and in occasional colorectal tumors in FAP (10). It is possible that reliance on the MCR in colorectal tumors has created a bias in mutation screening at APC and consequently has underestimated the frequency of codon 1,554 mutations. Perhaps the MCR should be extended 3′ to include this codon and indeed all codons 5′ to the first SAMP repeat.
The colorectal cancer cell lines without any evidence of APC mutation were predominantly RER+ tumors; the clinicopathological features of these tumors suggest that most were of sporadic origin (see Table 1). The accumulated data suggest that a minority of RER+ cancers—whether from HNPCC families or of sporadic origin—do initiate tumorigenesis by mutations at a locus other than APC. The mismatch repair genes (in HNPCC) and β-catenin (in sporadic cancers) are good candidates, but other possibilities include axin/conductin (13) and TCF4 (19). It remains puzzling as to why more tumors do not have β-catenin mutations when only one hit is required, compared with two needed at APC. Even if it is assumed that there are 4,500 susceptible and selected nucleotides within APC and only 3 susceptible and selected nucleotides within β-catenin, the probability of two mutations at APC is much lower than one mutation at β-catenin. It is possibly the case that APC mutations have important functions other than their effects on β-catenin levels and that additional mutations are necessary for β-catenin mutations to have a tumorigenic effect.
Our analysis of data from this study and from the APC Mutation Database shows that, as in FAP, the two APC mutations in sporadic colorectal tumors are not independent of each other and are not selected for simple loss of protein function. Allelic loss is observed much more often in sporadic tumors with truncating APC mutations 3′ to codon 1,194 than in tumors with truncating mutations before this codon. Truncating mutations between codons 1,194 and 1,392 are strongly associated with allelic loss; mutations after codon 1,392 are less strongly associated with loss. The data from sporadic tumors generally agree with those from FAP, although allelic loss was not found in FAP tumors from patients with germ-line APC mutations after codon 1,392 (9). The difference between the observations in FAP and in sporadic cancers probably results either from some allelic loss being undetected in the FAP tumors—as a result, for example, of polyclonal tumor origin (20)—or from less stringent selection for APC mutations in sporadic tumors, as theory predicts (see above). The overall frequency of APC allelic loss is higher in the colorectal cancer cell lines (60% in lines with two hits identified at APC) than in FAP adenomas (about 25% in unselected cases) (10). This difference primarily reflects the higher frequency of allelic loss in sporadic cancers with mutations 3′ to codon 1,392. There is also a deficiency of APC mutations before codon 750 in the sporadic cancers relative to the FAP germ line, although the reasons for this deficiency are unclear.
Taking into account the data from FAP and sporadic cancers (3, 4, 9, 10), we can hypothesize the following as general rules for two hits at APC. First, there is no absolute requirement for particular mutant APC genotypes, and a variety of genotypes can be selected in tumors. Some mutations are, however, more advantageous than others, with a mutation near codon 1,300 plus allelic loss probably being most strongly selected. Second, third hits at APC—such as loss of the germ-line mutant—can be found if the resulting genotype is more strongly selected. Third, certain typical APC genotypes in colorectal tumors seem to occur. One of these comprises a truncating mutation in the MCR plus allelic loss; another is a mutation 5′ to codon 1,190 and a mutation late in or 3′ to the MCR; and another, in sporadic tumors, is a mutation late in the MCR and allelic loss. Fourth, if the first hit, whether germ-line or somatic, occurs close to codon 1,300, then allelic loss usually follows, because it occurs at a relatively high spontaneous frequency and it is strongly selected. If the first hit is 5′ to about codon 1,190, then allelic loss rarely occurs, and the second hit tends to occur late in the MCR. If the first hit is late in the MCR, then allelic loss or another truncating mutation may occur. Two hits in the MCR are rarely seen, probably because allelic loss is a more common spontaneous event; two hits early in the APC gene are rarely seen, because they are selected only weakly.
The functional sequelae of the genetic changes in APC remain to be determined. A wealth of data shows that almost all colorectal tumors with APC mutations lose the SAMP (connexin/actin/β-catenin binding) repeats and all but one or two of the seven β-catenin binding/degradation sites. The reason for the retention of one or more of the β-catenin repeats is not clear; perhaps β-catenin levels can be superoptimal for colorectal tumorigenesis. Selection of lost C-terminal and retained N-terminal APC functions is complicated by the issue of stability of truncated APC proteins. It has been reported that the APC protein truncated at codon 1,309 is very stable relative to N- and C-terminal mutants (21). Perhaps a contributing factor to the fact that allelic loss is strongly associated with such mutants is that 1,309-mutant homodimers are very stable and can therefore exert their effects (through loss of C-terminal functions) very effectively. Mutants away from the region around codon 1,300 may be further from optimal stability as homodimers. Truncating second hits in particular regions of the gene may provide greater stability to the compound mutant APC dimer. It is even possible that, although effects of APC mutations on β-catenin protein remain important, selection of APC mutations is primarily for loss of the SAMP repeats combined with stability of the mutant protein. Finally, it is clear that the effects of APC mutations—and possibly those of other tumor suppressor genes—should be considered at the level of the genotype, rather than that of the individual mutant allele.
Acknowledgments
We are grateful to the Equipment Park (Imperial Cancer Research Fund) for important help with genotyping and sequencing. The APC Mutation Database (http://perso.curie.fr/Thierry.Soussi/APC.html) was invaluable for part of this study.
Abbreviations
- FAP
familial adenomatous polyposis
- MCR
mutation cluster region
- LOH
loss of heterozygosity
- PTT
protein truncation test
- SSCP
single-strand conformation polymorphism
- HNPCC
hereditary nonpolyposis colon cancer
References
- 1.Kinzler K, Vogelstein B. Cell. 1997;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 3.Miyaki M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Konishi M. Crit Rev Oncol Hematol. 1995;19:1–31. doi: 10.1016/1040-8428(94)00129-h. [DOI] [PubMed] [Google Scholar]
- 4.Miyaki M, Konishsi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, Muraoka M, Takahashi H, Amada Y, Fukayama M, et al. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 5.Bodmer W F, Bailey C J, Bodmer J, Bussey H J, Ellis A, Gorman P, Lucibello F C, Murday V A, Rider S H, Scambler P, et al. Nature (London) 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 6.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler K W, Nilbert M C, Vogelstein B, Bryan T M, Levy D B, Smith K J, Preisinger A C, Hamilton S R, Hedge P, Markham A, et al. Science. 1991;251:1366–1370. doi: 10.1126/science.1848370. [DOI] [PubMed] [Google Scholar]
- 8.Ichii S, Takeda S, Horii A, Nakatsuru S, Miyoshi Y, Emi M, Fujiwara Y, Koyama K, Furuyama J, Utsunomiya J, et al. Oncogene. 1993;8:2399–2405. [PubMed] [Google Scholar]
- 9.Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, Frayling I, Efstathiou J, Pack K, Payne S, et al. Nat Med. 1999;5:1071–1075. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 10.Spirio L, Samowitz W, Robertson J, Robertson M, Burt R, Leppert M, White R. Nat Genet. 1998;20:385–388. doi: 10.1038/3865. [DOI] [PubMed] [Google Scholar]
- 11.van der Luijt R, Khan P M, Vasen H, van Leeuwen C, Tops C, Roest P, den Dunnen J, Fodde R. Genomics. 1994;20:1–4. doi: 10.1006/geno.1994.1119. [DOI] [PubMed] [Google Scholar]
- 12.Wheeler J, Beck N, Kim H, Tomlinson I, Mortensen N, Bodmer W. Proc Natl Acad Sci USA. 1999;96:10296–10301. doi: 10.1073/pnas.96.18.10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behrens J, Jerchow B-A, Würtele M, Grimm J, Asbrand C, Wirtz R, Kühl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 14.Hiltunen M, Alhonen L, Koistinaho J, Myohanen S, Paakkonen M, Marin S, Kosma V, Janne J. Int J Cancer. 1997;70:644–648. doi: 10.1002/(sici)1097-0215(19970317)70:6<644::aid-ijc3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 15.Olschwang S, Hamelin R, Laurent-Puig P, Thuille B, De Rycke Y, Li Y, Muzeau F, Girodet J, Salmon R, Thomas G. Proc Natl Acad Sci USA. 1997;94:12122–12127. doi: 10.1073/pnas.94.22.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlinson I, Novelli M, Bodmer W. Proc Natl Acad Sci USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samowitz W, Powers M, Spirio L, Nollet F, van Roy F, Slattery M. Cancer Res. 1999;59:1442–1444. [PubMed] [Google Scholar]
- 18.Toyooka M, Konishi M, Kikuchi-Yanoshita R, Iwama T, Miyaki M. Cancer Res. 1995;55:3165–3170. [PubMed] [Google Scholar]
- 19.Duval A, Gayet J, Zhou X, Iacopetta B, Thomas G, Hamelin R. Cancer Res. 1999;59:4213–4215. [PubMed] [Google Scholar]
- 20.Novelli M R, Williamson J A, Tomlinson I P M, Elia G, Hodgson S V, Talbot I C, Bodmer W F, Wright N A. Science. 1996;272:1187–1190. doi: 10.1126/science.272.5265.1187. [DOI] [PubMed] [Google Scholar]
- 21.Dihlmann S, Gebert J, Siermann A, Herfarth C, von Knebel Doeberitz M. Cancer Res. 1999;59:1857–1860. [PubMed] [Google Scholar]
- 22.Ilyas M, Tomlinson I P M, Rowan A, Pignatelli M, Bodmer W F. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White S, Bubb V J, Wyllie A H. Genes Chromosomes Cancer. 1996;15:122–128. doi: 10.1002/(SICI)1098-2264(199602)15:2<122::AID-GCC7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 24.Pedemonte S, Sciallero S, Gismondi V, Stagnaro P, Biticchi R, Haeouaine A, Bonelli L, Nicolo G, Groden J, Bruzzi P, et al. Genes Chromosomes Cancer. 1998;22:257–267. doi: 10.1002/(sici)1098-2264(199808)22:4<257::aid-gcc1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]