Abstract
Expression of the bacterial RecA protein in plants stimulates homologous recombination in tobacco. Here we show that RecA plays a direct role in DNA strand exchange in vivo. The number of sister chromatid exchanges (SCEs) was increased 2.4-fold over wild type in transgenic tobacco plants expressing a nuclear-targeted RecA (nt-RecA) protein and could not be increased further by DNA damage, which caused a doubling of the baseline SCE frequency in wild-type plants. Although gene targeting requires homologous recombination, the number of targeted gene replacements was not increased markedly by the presence of nt-RecA by using Agrobacterium-mediated transformation. However, the number of double-strand breaks that were repaired at both sides by homologous recombination was increased 3.3-fold. Stimulation of SCE and fidelity of double-strand break repair by nt-RecA, but not by gene targeting, suggests that the stimulatory activity of RecA is linked to active DNA synthesis. Therefore, nascent replication-associated single strands may be a prerequisite for RecA action in plant cells.
The RecA protein plays a central role in the homologous recombination pathway of Escherichia coli. RecA alone mediates the search for homology, recognition of sequence similarities, and strand exchange between two DNA molecules. Homologues of this protein have been found in bacteria and archea as well as in lower and higher eukaryotes. The best known RecA homologues are the eukaryotic Rad51 proteins. Biochemical analysis of these proteins demonstrated that they are not only structural but also functional RecA homologues (reviewed in refs. 1–3).
The bacterial protein RecA protein and a derivative carrying a nuclear localization signal (nt-RecA, ref. 4) have been expressed in tobacco plants and shown to stimulate homologous recombination in plants by using intrachromosomal recombination and DNA-damage repair assays (5). Studies in a mammalian system suggest that ectopic expression of the Rad51 protein can have a similar effect (6, 7). A key player in the E. coli recombination pathway is also the Holliday junction resolvase RuvC. Overexpression of this protein also enhances homologous recombination in plants (8). These data suggest that ectopic expression of several components of the homologous recombination pathway can stimulate the homologous recombination reaction.
Another important aspect determining the overall efficiency of recombination is initiation. Induction of site-specific double-strand breaks (DSBs) has been shown to stimulate homologous recombination at the target locus by several orders of magnitude in eukaryotes (for review, see refs. 9, 10). It has been demonstrated that homologous recombination is a significant DSB repair pathway in plants and that both ends of the break are processed independently and in a step-wise fashion (refs. 11–16; for review, see ref. 17). Thus, products with either one (“one-sided”) or two (“two-sided”) homologous junctions can arise during the reaction.
Although several recombination enzymes have been overexpressed and their effect on homologous recombination studied, their effect on foreign DNA integration is still unknown. To analyze whether overexpression of a recombination protein, nt-RecA, can stimulate gene targeting in plants, we performed gene-targeting experiments with two different systems, both relying on Agrobacterium-mediated transformation. Although there was no increase in gene-targeting frequencies in nt-RecA-expressing cells, they showed elevated levels of strand-exchange activity, as indicated by the frequency of sister chromatid exchanges (SCEs), and nt-RecA had a strong influence on the products of DSB repair as well.
Materials and Methods
Artificial Target Loci and Repair Constructs.
The B18/4 target locus was obtained by leaf-disk transformation. SR1 tobacco was transformed with Agrobacterium strain B18 by using standard procedures (18) and selection on methotrexate- (100 μg/liter) containing media. One particular transformant that contained the T-DNA in single copy and that expressed the neomycin phosphotransferase II gene (nptII) in a seed-specific manner was selected and this line designated B18/4. The B18 strain is Agrobacterium GV3101pMP90RK carrying the plasmid pMN018. Plasmid pMN018 was obtained by insertion of an EcoRI–BamHI fragment carrying the high molecular-weight glutenin promoter (HMW) (19) into the corresponding restriction sites of pMN001 (20). The pMN125 repair construct (see Fig. 1) was made in several steps starting from pMN018. These resulted in the replacement of the nptII gene by the D42 deletion mutant (ref. 21; nptII ΔC in Fig. 1) and an insertion of a cauliflower mosaic virus (CaMV) 35S promoter fragment (black arrow in Fig. 1) from pDH51 (22) between the HMW promoter and the D42 nptII gene. Plasmid pMN125 was transformed in Agrobacterium GV3101pMP90RK (18) to yield strain G125.
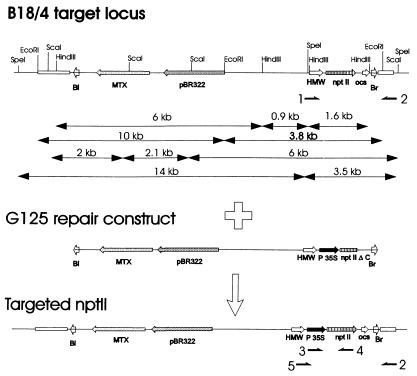
Figure 1.
Gene targeting at the B18/4 locus. Schematic physical maps of the B18/4 target locus, the G125 repair construct, and the recombination product obtained after precise gene replacement are shown. Relevant genes, promoters, and polyadenylation sites are shown as large arrows (MTX, methotrexate resistance gene; pBR322, part of pBR322, including the origin of replication and the Ampicillin resistance gene; ocs, octopine synthase polyadenylation signal; P35S, CaMV 35S promoter; nptII ΔC, carboxyl-terminal deletion mutant of nptII), the T-DNA left (Bl) and right (Br) borders as small arrows, and characterized flanking tobacco DNA sequences as small boxes. The map is not drawn to scale. The size of fragments obtained after restriction of B18/4 target locus DNA with EcoRI, HindIII, ScaI, and SpeI is indicated. The primers used in the PCR analysis of recombination events at the B18/4 locus are shown schematically (1, hmw2404; 2, b18rbin; 3, 5634; 4, 2391; 5, hmw35S).
The I-SceI expression vector pCISceI, the repair construct pRC2, and the target locus pTS in the tobacco line 1–12 were as described (11).
Plant Lines and Transformation Procedures.
To introduce the target loci and nt-RecA gene into the same genetic background, line G63/19 that carries the nt-RecA transgene and a sulfonamide resistance marker (5) was crossed with lines 1–12 and B18/4. To introduce the vector alone, a line carrying a single-copy T-DNA insert of an empty vector carrying a sulfonamide resistance gene, G61/2 (data not shown), was also crossed to line 1–12. Lines uniformly hemizygous for the target loci and nt-RecA transgenes were selected for the gene-targeting experiments. Lines heterozygous for the target locus alone were selected from crosses of G61/2 and line 1–12 and B18/4 and SR1, respectively, by selection for the antibiotic resistance genes carried by the individual lines. The transformation experiments by using the 1–12 target locus were performed by using the seedling transformation method as described (23). The B18/4 lines were transformed by the leaf-disk transformation. Selection was on media containing kanamycin (100 mg/liter) or, for the control transformations in SR1, on methotrexate (100 μg/liter).
DNA Analysis of Transformants.
Isolation of total plant DNA and Southern blotting were as described (24). To analyze recombination events at the 1–12 locus, Southern blots were probed with a HindIII fragment of the plasmid containing parts of the kanamycin gene and a BamHI fragment carrying the hygromycin resistance gene of pTS, which were labeled with α-32P-CTP by using the random primed labeling kit (Megaprime DNA labeling system RPN1607, Amersham). To analyze recombination events of the B18/4 locus, the following probes were used: nptII; a BamHI HindIII fragment from pMN018 carrying the nptII gene, Amp; the plasmid pBR322, Mtx; an Asp-718 fragment from pMN018 carrying the dhfr gene, tobacco DNA flanking the right border; a cloned and sequenced fragment obtained by inverse PCR from B18/4, tobacco DNA flanking the left border; and a BglII EcoRI subfragment containing the flanking genomic DNA of a plasmid obtained by plasmid rescue from B18/4 genomic DNA.
PCR and Sequence Analysis.
Genomic DNA was analyzed via PCR as described (24). For analysis of recombination events of the 1–12 locus, the oligonucleotides pH1 (pCAAGACCGAC CTGTCCGGTG CCCTG) and pR3 (pAGCCGCCGCA TTGCATCAGC C) (see Fig. 2) and in some cases pR1 (pGTGACAACGT CGAGCACAGC TGCG) and pP3 (15) for a second round of amplification were used. The binding sites of the oligonucleotides used for analysis of the B18/4 locus, hmw35S (pCAGTCGGGG ATCAATTCCCA), hmw2404 (pCTCCACAATT TCATCATCACC), b18rb-in (pAGAAGGATCG TTCATGTTGAG), 5634 (pGTGGATTGAT GTGATATCTC C), and 2391(CCGCTCAGAA GAACTCGTCA) are shown in Fig. 1.
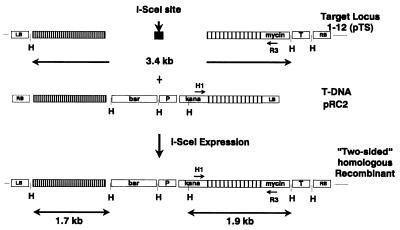
Figure 2.
Gene targeting at the 1–12 locus. Schematic physical maps of the 1–12 target locus (pTS) with a I-SceI-site (black box) and repair construct pRC2 are shown. Homologies with the target locus are indicated. The homologous region within the kanamycin gene (widely shaded) is 1 kb in length, whereas the homology at the other end (narrowly shaded) is 1.3 kb in length. The oligonucleotides H and R3 were used to detect the nptII gene restored by homologous recombination. The nptII gene of the original target locus resides on a 3.4-kb HindIII (H) fragment that is altered to a 1.9-kb fragment by homologous gene replacement. Homologous recombination at both ends of the DSB (“two-sided” recombinants) generates a 1.7-kb fragment carrying the hygromycin resistance gene in addition to the 1.9-kb fragment. The bar gene in pRC2 was used to estimate the absolute numbers of transformants in these experiments (see also Table 2). LB, left border; RB, right border; 35S, CaMV 35S promoter; T, CaMV terminator.
Detection of SCEs.
Root tips of germinating tobacco seeds (lines G61/2 and G63/19) were treated as described for Nicotiana plumbaginifolia (25) and squashes stained by the fluorescent plus Giemsa technique (26). For mutagen induction of SCEs, the root tips were treated for 0.5 h with mitomycin C (5 × 10−6 M) between BrdUrd and dThd incubation. Five root tips were squashed per slide and microscopically inspected.
Results
Systems to Study Gene Targeting.
Two different artificial target loci were used to assess gene targeting in plants, the newly developed B18/4 locus and the 1–12 locus described by Puchta et al. (11). Transformation of leaf disks with Agrobacterium B18 resulted in several transformants; one was selected that contained a T-DNA insert in single copy. This transformant was designated B18/4 (Fig. 1). The 7.9-kb B18 T-DNA insert in this line consists of a methotrexate resistance gene (MtxR) as a selectable marker, a pBR322 replicon, and an nptII (kanamycin resistance) gene under control of the seed-specific HMW promoter (19). The nptII gene is exclusively expressed in seeds, and hence B18/4 plants, leaf disks, and calli are sensitive to kanamycin (data not shown). The B18/4 T-DNA insert was further characterized by sequencing of the tobacco genomic DNA flanking the left and right borders of the T-DNA (data not shown). The G125 repair construct (Fig. 1) is to a large extent identical to the B18/4 target locus T-DNA. However, the former differs from the latter by the presence of a CaMV 35S promoter between the HMW promoter and nptII gene and a carboxyl-terminal deletion in the nptII gene that renders this gene nonfunctional. Targeted replacement results in the insertion of the 35S promoter between the HMW promoter and a functional nptII gene carried by the target locus (Fig. 1) and hence constitutive expression of NPTII. The repair construct provides 6,693 bp of homology from the 35S promoter insertion to the left border and 752 bp of homology from the insertion to the other border of the target locus.
The second system was described by Puchta et al. (11). The tobacco line 1–12 is transgenic for the recombination substrate pTS (Fig. 2). This substrate carries a I-SceI restriction enzyme recognition site between a nonfunctional part of an nptII gene and a hygromycin resistance gene. The repair construct pRC2 is a T-DNA vector containing part of the nptII gene, a glufosinate resistance gene, and a hygromycin resistance gene (Fig. 2). Thus pRC2 is homologous to sequences on both sides of the I-SceI site, and homologous recombination of the repair construct with the target locus restores a functional nptII gene. A DSB can be induced at the target locus by transient expression of the restriction enzyme I-SceI. The binary vector pCISceI (11) contains the I-SceI gene under the control of the CaMV 35S promoter. Coinoculation with two Agrobacterium strains, one carrying pCISceI and the other the repair construct pRC2, could lead to induction of a DSB and to simultaneous homologous recombination with the repair construct.
Gene-Targeting Assays.
For gene-targeting experiments with B18/4, leaf disks derived from the nt-RecA transgenic line G63/19xB18/4 and the non-nt-RecA transgenic line B18/4xSR1 were transformed with Agrobacterium strain G125 harboring the repair construct, and kanamycin-resistant calli were selected. Transformation efficiencies were determined by transformation of SR1 leaf disks with the same Agrobacterium culture under identical conditions and by counting the number of transformants obtained after selection on methotrexate. Kanamycin-resistant calli were analyzed further by PCR and Southern blotting. The results are shown in Table 1. In total, the number of events analyzed equals 6,000 nt-RecA transgenic and 4,000 non-nt-RecA transgenic transformants. These transformants were obtained in three different independent leaf-disk transformation experiments. Among those transformants, four homologous events were found in the nt-RecA transgenic lines, whereas two events were present in the wild-type transformants. Thus, the number of homologous interactions at the target locus was only barely higher in nt-RecA transgenic lines. Although a large number of kanamycin-resistant calli were obtained, only few were caused by homologous interactions at the target locus. Moreover, there was no correlation between the number of kanamycin-resistant calli and the number of gene-targeting events. Because the nptII gene of the B18/4 locus remained unaltered and no intact nptII gene copy was detected in these kanamycin-resistant calli, mechanisms acting in trans are likely to be responsible for the ectopic activation of nptII in these cases. These mechanisms are unclear, but somaclonal variation, eventually leading to transcriptional activation of the HMW promoter, is a possibility.
Table 1.
Gene replacement events at the B18/4 target locus
| Line | Number of transformants | Kanamycin-resistant calli | Precise events (“two-sided”) | Ectopic events (“one-sided”) | Hom. rec. freq. × 10−4 |
|---|---|---|---|---|---|
| B18/4xSR1 | 4,424 | 32 | 2 | 0 | 4.5 |
| B18/4xG63/19 | 6,227 | 129 | 1 | 3 | 6.4 |
Hom. rec. freq., frequency of homologous recombination.
Kanamycin-resistant calli were initially screened by PCR for an intact nptII gene, under 35S promoter control (primers 5634 and 2391; see Fig. 1). Three of the six events found by this screen, all of them found in one particular transformation experiment, were not true gene-targeting events because PCR with primers complementary to the HMW promoter sequences (primer hmw2404) and tobacco genomic sequences flanking the B18/4 T-DNA insert at its 3′end (primer b18rbin) detected a fragment representing the intact target. Therefore, the modified nptII gene had to reside in an additional T-DNA copy in which the incoming T-DNA was converted by the target locus and then integrated elsewhere. This phenomenon was described before for plants (27, 28) and mammals (29) as ectopic targeting. Further PCR analyses showed that two of these modified target locus copies also contained the flanking tobacco DNA sequences of the original B18/4 locus. PCR with the 5′ junction-specific primer (hmw35S), and the 3′ locus-specific primer (B18rbin) yielded a fragment corresponding exactly to the size of the predicted fragment of the modified nptII gene in these two cases. For the true gene-targeting events obtained in the other two transformation experiments, PCR with primers hmw35S and b18rbin also yielded a product that indicated a correctly modified target locus, but no unmodified target locus sequences were detectable with the target locus-specific PCR (primers hmw2404 and b18rbin).
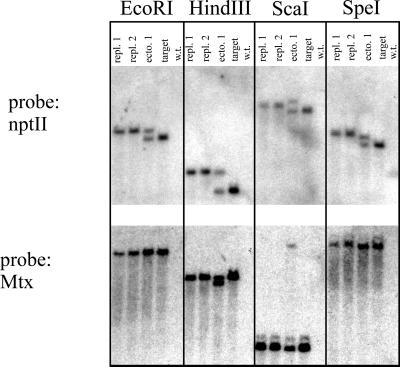
The PCR results were confirmed by Southern blot analysis. Representative results are shown in Fig. 3. Plant DNA was digested with restriction enzymes that release fragments either from within the B18/4 T-DNA insert (HindIII) or from the B18/4 T-DNA insert, including sequenced regions of the flanking tobacco genomic DNA (EcoRI) or from the T-DNA insertion including tobacco genomic sequences from further outside (ScaI and SpeI). This filter was hybridized with an nptII gene-specific probe. This probe detected a fragment representing the unmodified target locus in all digests with ectopic recombination events, and the unmodified target locus was absent in all cases of true gene targeting, as expected from the PCR results. The increase in size of the fragment representing the modified nptII gene was approximately 500 bp, as predicted for the precise insertion of the 35S promoter, with all four restriction digests obtained for true gene-targeting events. However, for ectopic events, the increase in size of the modified nptII gene was 500 bp only for the enzymes cutting in or close to the T-DNA insertion (HindIII and EcoRI), but not for those cutting at a distance (ScaI and SpeI) (Fig. 3). This result confirmed that these modified loci contained B18/4 tobacco flanking DNA but were inserted at a different position in the genome.
Figure 3.
Southern analysis of B18/4 target locus events DNA, digested by the restriction enzymes indicated at the panels, separated by gel electrophoresis, and blotted. The blot was hybridized with the probes indicated (Left). A representative selection of events is shown (repl.1, precise gene replacement event 1; repl. 2, precise gene replacement event 2; ecto1, ectopic recombination event 1; target, B18/4 target locus; wild type, SR1).
This result was confirmed by an analysis of the 5′ends of the target locus. Hybridization of the filter with a methotrexate-resistance gene-specific probe indicated the presence of a second T-DNA copy, which contained the modified gene integrated at a different position in the cases of ectopic recombination. Precise gene replacement was confirmed for the true gene-targeting events (Fig. 3). These results were verified by using additional probes (data not shown).
For analysis of gene targeting and DSB repair by using locus 1–12, 2-wk-old seedlings were coinoculated with a 4:1 mixture of two Agrobacterium strains carrying the plant-specific expression cassette of I-SceI (pCISceI) and the repair construct pRC2 or with the Agrobacterium strain carrying pRC2 alone (15, 23). Transformation efficiencies were estimated by selection on gluphosinate–ammonium (15). Homologous recombination events were selected on kanamycin-containing media. Kanamycin-resistant calli were analyzed by PCR by using primers P2 and R3 to confirm the restoration of the kanamycin resistance gene via homologous recombination (see Fig. 2). The results of the analysis are summarized in Table 2. The frequency of restoration of the kanamycin resistance gene by homologous recombination was about one and a half times higher in the RecA-expressing plant line G63/19x1–12 than in line G61/2x1–12, irrespective of whether a DSB was induced. However, DSB induction itself resulted in an enhancement of the homologous recombination frequency by about one order of magnitude for both lines. To characterize further the nature of the recombination events, a representative number of calli was regenerated and the nature of the homologous interaction analyzed by Southern blotting (see Fig. 2). Probes specific for both ends of the DSB discriminated between correct restoration of both ends of the DSB (two-sided events, appearance of a 1.9-kb kanamycin-specific fragment and a 1.7-kb hygromycin-specific fragment) or only one end (one-sided events, appearance of the 1.9-kb kanamycin-specific fragment but not of the 1.7-kb hygromycin-specific fragment). The results are summarized in Table 3, and representative Southern blots are shown in Fig. 4. Only “one-sided” events were found in the absence of DSB induction, irrespective of the presence of nt-RecA. The original target locus represented by a 3.4-kb kanamycin- and hygromycin-specific band was still present in the recombinants in all three cases obtained in the absence of nt-RecA and in seven of eight cases in its presence (data not shown and Fig. 4, lanes 2–4). Thus, these recombinants very likely represent ectopic targeting events, like those found in the experiments with line B18/4.
Table 2.
Gene replacement events obtained at the 1–12 locus with and without DSB induction by I-SceI expression
| Line | I-SceI | Number of transformants × 103 | PCR + calli | Hom. rec. freq. × 10−4 | Enh. |
|---|---|---|---|---|---|
| G61/2x1–12 | − | 40.3 | 5 | 1.2 | 1.0 |
| G61/2x1–12 | + | 10.6 | 12 | 11.3 | 9.4 |
| G63/19x1–12 | − | 51.3 | 10 | 1.9 | 1.6 |
| G63/19x1–12 | + | 13.6 | 23 | 16.9 | 14.1 |
Hom. rec., homologous recombinants; Enh., enhancement in relation to G61/2 without I-SceI expression.
Table 3.
Compilation of 1–12 recombination events analyzed by Southern blotting
| Line | I-SceI | Analyzed | Homologous recombinants
|
% | |
|---|---|---|---|---|---|
| “One sided” | “Two sided” | ||||
| G61/2x1–12 | − | 3 | 3 | 0 | 0 |
| G61/2x1–12 | + | 7 | 6 | 1 | 14 |
| G63/19x1–12 | − | 8 | 8 | 0 | 0 |
| G63/19x1–12 | + | 13 | 7 | 6 | 46 |
%, percentage of “two-sided” events of all recombinants obtained.
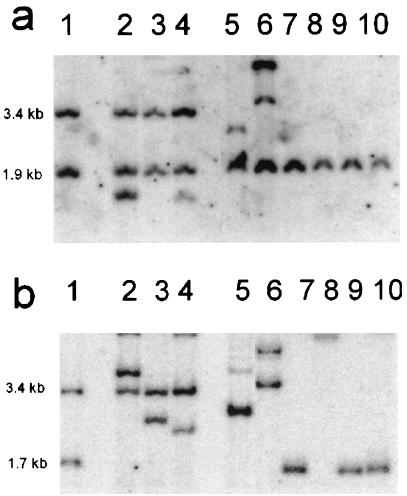
Figure 4.
Southern blot analysis of 1–12 events. A representative selection of events is shown. HindIII-digested plant DNA was probed either with a kanamycin resistance gene-specific (a) or a hygromycin resistance gene-specific (b) probe. Lane 1, recombinant line 1–12c (11) harboring 1.9-kb and 1.7-kb bands indicative of “two-sided” homologous recombination and a 3.4-kb band specific for the target locus; lanes 2–4, recombinant lines G61/2x1–12-1, G63/19x1–12-1, and G63/19x1–12-2 obtained without coexpression of I-SceI. In addition to the 1.9-kb band representing the recombined nptII gene, the 3.4-kb fragment, indicating presence of an unchanged target locus, is still present. lanes 5–10, recombinant lines G61/2x1–12+2, G61/2x1–12+3, G63/19x1–12+1, G63/19x1–12+2, G63/19x1–12+3, and G63/19x1–12+4 obtained with coexpression of I-SceI. The 3.4-kb fragment representing the unmodified target locus is absent in all lines, indicating that homologous recombination occurred at the target locus. The presence of the 1.7-kb hygromycin-specific fragment in lanes 7, 9, and 10 indicates that both ends of the break were repaired by homologous recombination. The extra bands of various sizes present in lanes 2, 4–6 most probably represent further integration events of the repair construct pRC2 elsewhere in the genome.
After DSB induction, 1 of 7 recombinants in the absence and 6 of 13 recombinants in the presence of nt-RecA were “two-sided” (Table 3, Fig. 4, lanes 5–10). In total, there were only four cases (two for each line) in which the 3.4-kb band specific for the target locus was not replaced in the recombination reaction, indicating that in the vast majority of the DSB-induced cases, the target locus was indeed changed via homologous recombination. Several lines (see Fig. 4, lanes 2, 4–6) contain further pRC2-specific fragments that most likely represent extra copies of the repair construct integrated randomly in the genome.
The restored kanamycin resistance gene was stably transferred to the progeny in all lines tested that represented all classes of “one-sided” and “two-sided” recombinants and segregated as a single Mendelian trait (data not shown).
Analysis of SCE.
Seedlings were treated with BrdUrd such as to result in a unifilar substitution of thymidine by this analogue and the number of SCEs evaluated. On average, 0.44 SCEs per chromosome were found in non- nt-RecA transgenic (G61/12) seedlings (Table 4). This number corresponds to 21.2 SCE per cell (tobacco: 2n = 4x = 48). Mitomycin C treatment increased the number of SCEs to 0.74 SCEs per chromosome or 35.3 SCEs per cell. These figures correspond well with the data obtained previously for N. plumbaginifolia under comparable conditions (25). In nt-RecA transgenic seedlings (G63/19), the number of SCEs in the absence of treatment with mitomycin C was already higher than in the non-nt-RecA transgenic line after mutagenic treatment, and this number could not be increased further by treatment with the mutagen (Table 4).
Table 4.
Analysis of SCEs with and without mitomycin C treatment (5 × 10−6 M, 0.5 h)
| Line | MMC | Chromosomes inspected | SCE/ch | SCE/ce | Enh. |
|---|---|---|---|---|---|
| G61/2 | − | 720 | 0.44 | 21.2 | 1.0 |
| G61/2 | + | 480 | 0.74 | 35.3 | 1.7 |
| G63/19 | − | 480 | 1.10 | 51.9 | 2.4 |
| G63/19 | + | 480 | 0.97 | 46.5 | 2.2 |
MMC, mitomycin C; ch, chromosome; ce., cell; Enh., enhancement in relation to G61/2 without MMC induction.
Discussion
Gene targeting was assayed with two different artificial target loci, the B18/4 and the 1–12 loci. Although these loci differ in several features, like their chromosomal location, the general design of the repair constructs, and regions of homology, the overall number of homologous recombination events was in the same order of magnitude (10−3 to 10−4). This number corresponds well to frequencies observed previously (reviewed in refs. 30–33). The number of homologous events observed was barely increased by the presence of nt-RecA with both target loci (B18/4, 1.4; 1–12, 1.6). This picture did not change after induction of a DSB in the 1–12 locus. This suggests that stimulation of homologous recombination by RecA cannot substantially improve gene-targeting frequencies in plants by using Agrobacterium, even if recombination is induced by a DSB.
A detailed analysis of the recombination events revealed that they consisted of both classical gene-targeting events and ectopic recombination events. By using the B18/4 locus, three cases were characterized as classical gene-targeting events, in which both ends of the incoming T-DNA recombined with their homologous counterpart in the plant genome resulting in a “two-sided” homologous recombination event. In the remaining three events, all obtained from one particular experiment in the presence of nt-RecA, only one end of the T-DNA was restored by homologous recombination. In all of these cases, the original target locus remained unaltered, and the integration of the modified repair construct occurred at a different position in the genome. In these cases, genomic sequences residing in the target locus were copied into the repair construct by homologous interactions. This process seemed to be restricted to the target locus itself in one case, whereas in the other two cases also sequences flanking the target locus were included. Similar events have been described before in tobacco (27, 28).
No true gene-targeting event was observed for the target locus 1–12 in the absence of nt-RecA, because the locus remained unchanged in all three cases analyzed. In the presence of nt-RecA, the unchanged hemizygous single-copy target locus is still present in seven of the eight cases analyzed (data not shown). Therefore, ectopic targeting occurred in all 10 events and only 1 event was caused by a one-sided homologous integration of the repair construct at the target locus.
DSB induction at the 1–12 locus enhances homologous recombination about 9-fold, and nt-RecA had no influence on the frequency in this process. However, DSB induction revealed a major effect of nt-RecA on the distribution of recombination product classes. Although the target locus was retained in two of seven recombinants in the absence of nt-RecA, this was the case only for two of thirteen recombinants in the presence of nt-RecA. A complete analysis of the events (Table 3) revealed that the ratio of “two-sided” or true gene-targeting events to ectopic events was shifted from 14% in the absence of nt-RecA to 46% in its presence. Therefore, nt-RecA seems to have an effect on the fidelity of the recombination reaction but not on its initiation in plants.
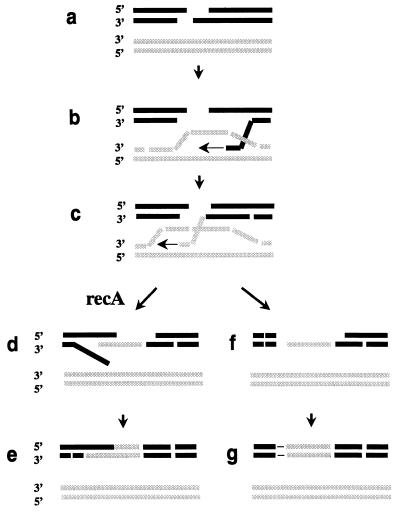
DSB repair by homologous recombination in somatic plant cells mainly proceeds by a one-sided invasion/synthesis-dependent strand-annealing mechanism (14, 16, 17, 33). According to our model (Fig. 5), acceptor DNA derived from the DSB invades as a single strand into the homologous donor sequence and initiates DNA synthesis. This process appears to be frequently aborted in somatic plant cells leading to “one-sided” events, or ectopic recombination, depending on the mode of resolution. Recombination at the other end of the break seems to be independent. Single-stranded DNA is an obligatory substrate for RecA-mediated strand-exchange reactions in vitro (for reviews, see refs. 1–3). Therefore, RecA protein is completely inactive in the absence of single-stranded DNA. It is conceivable that free single-stranded DNA is mostly not available during the invasion process in Agrobacterium transformation. The single-stranded T-DNA is likely to be masked by the single-stranded DNA-binding protein virE from Agrobacterium tumefaciens (for review, see ref. 34), and the ends of the invading DNA may be blocked by the copying process on the donor sequences. Presumably RecA is inactive in this situation because it cannot find a substrate to act on. However, free single-stranded DNA may be generated during repair and replication processes, thus generating the substrate for RecA. In yeast, DSB repair involves replication-like intermediates (35). RecA then could find homologies between a single-stranded loop of the newly synthesized DNA, the ends of which are blocked and the acceptor molecule, which is not accessible to RecA because it is double stranded. This would explain why RecA preferentially acts on a later step and leads to improvement of the fidelity of the recombination reaction. In accordance with this finding, DNA synthesis is not a prerequisite for initial annealing in gene targeting (reviewed in ref. 9). Although the proposed model easily explains why nt-recA exclusively affects the second step of DSB repair, there are alternative explanations. nt-RecA could play a more passive role and simply could prevent factors involved in illegitimate recombination from binding and resolving break intermediates. In addition, it cannot be excluded that the presence of nt-RecA favors the invasion of both ends of the break into the donor sequence, resulting in a different independent repair mechanism according to the DSB repair model of recombination (36).
Figure 5.
Model of the involvement of nt-RecA in DSB repair in somatic plant cells. Exonucleases process the DSB in the acceptor molecule to produce 3′ single-stranded overhangs (a). A free 3′-end invades the double-stranded donor forming a D-loop, and repair synthesis commences (b). The invading end of the single strand is blocked by replication (c). The intermediate is processed further, and the break is sealed either by homologous (“two-sided,” d and e) or by illegitimate recombination (“one-sided,” f and g). Free single-stranded DNA is generated by replacement of one of the strands of the acceptor molecule during DNA synthesis that can act as a substrate for nt-RecA (d). nt-RecA promotes strand exchange with the homologous strand, and the break is sealed via homologous recombination at both ends (e). In contrast, in the absence of nt-RecA, the enzymatic machinery promoting illegitimate recombination present in somatic plant cells dominates, single strands are not paired or abandoned (f), and the break is sealed via illegitimate recombination (g).
Whatever the precise mechanism might be, the results presented in this study suggest that stimulation of homologous recombination by RecA is limited to processes that transiently produce nascent single strands, as during DNA synthesis. SCE represents a homologous recombination reaction linked to DNA replication. The frequency of SCE can be increased by mitomycin C treatment. In nt-RecA transgenic seedlings, the frequency of SCE was 2.4 times higher than in wild-type seedlings. Mitomycin C treatment under conditions that led to an approximate doubling of the SCE frequency in wild-type tobacco and in several other plant species (25, 37, 38) did not cause a further increase of SCE frequency in these plants. SCE are interpreted as the microscopically detectable fraction of recombinative bypass repair of replication-blocking DNA lesions during DNA replication (39). Therefore, a base-line recombination between sister chromatids in the nt-RecA transgenic line is already higher than after mitomycin C treatment of wild-type seedlings and might be sufficient to cope with mitomycin C-induced DNA damage. The nt-RecA-mediated increase of homologous recombination between sister chromatids during S-phase agrees with the observation that eukaryotic homologues of RecA are involved in SCE formation in vertebrate cells (40). The necessity for single-stranded DNA or DNA synthesis in RecA-mediated processes also explains why intrachromosomal recombination and resistance to mitomycin C was increased by nt-RecA (5), because both processes seem to be at least partially linked to DNA replication.
Acknowledgments
We thank Charles White for discussions on mechanisms and Christa Fricke and Barbara Hildebrandt for excellent technical assistance. This study was partially funded by grant BIO4 CT97 2028 of the European Union Biotechnology Program to B.R. and H.P.
Abbreviations
- nt-RecA
nuclear-targeted RecA
- DSB
double-strand break
- SCEs
sister chromatid exchanges
- HMW
high molecular-weight glutenin promoter
- CaMV
cauliflower mosaic virus
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050582797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050582797
References
- 1.Cox M M. Prog Nucleic Acid Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 2.Camerini-Otero R D, Hsieh P. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- 3.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss B, Kosak H, Klemm M, Schell J. Mol Gen Genet. 1997;253:695–702. doi: 10.1007/s004380050373. [DOI] [PubMed] [Google Scholar]
- 5.Reiss B, Klemm M, Kosak H, Schell J. Proc Natl Acad Sci USA. 1996;93:3094–3098. doi: 10.1073/pnas.93.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vispe S, Cazaux C, Lesca C, Defais M. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnaudeau C, Helleday T, Jenssen D. J Mol Biol. 1999;289:1231–1238. doi: 10.1006/jmbi.1999.2856. [DOI] [PubMed] [Google Scholar]
- 8.Shalev G, Sitrit Y, Avivi-Ragolski N, Lichtenstein C, Levy A A. Proc Natl Acad Sci USA. 1999;96:7398–7402. doi: 10.1073/pnas.96.13.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasin M. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 11.Puchta H, Dujon B, Hohn B. Proc Natl Acad Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiurazzi M, Ray A, Viret J-F, Perera R, Wang X-H, Lloyd A M, Singer E R. Plant Cell. 1996;8:2057–2066. doi: 10.1105/tpc.8.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shalev G, Levy A A. Genetics. 1997;146:1143–1151. doi: 10.1093/genetics/146.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin E, Levy A A. Mol Cell Biol. 1997;17:6294–6302. doi: 10.1128/mcb.17.11.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puchta H. Plant J. 1998;13:331–339. [Google Scholar]
- 16.Puchta H. Genetics. 1999;152:1173–1181. doi: 10.1093/genetics/152.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbunova V, Levy A A. Trends Plant Sci. 1999;4:263–269. doi: 10.1016/s1360-1385(99)01430-2. [DOI] [PubMed] [Google Scholar]
- 18.Koncz C, Schell J. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 19.Colot V, Robert L S, Kavanagh T A, Bevan M W, Thompson R D. EMBO J. 1987;6:3559–3564. doi: 10.1002/j.1460-2075.1987.tb02685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiss B, Koncz C, Moore I, Schell J. Plant Physiol (Life Sci Adv) 1994;13:143–149. [Google Scholar]
- 21.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 22.Pietrzak M, Shilito R D, Hohn T, Potrykus I. Nucleic Acids Res. 1986;14:5857–5869. doi: 10.1093/nar/14.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puchta H. Methods Mol Biol. 1999;113:447–451. doi: 10.1385/1-59259-675-4:447. [DOI] [PubMed] [Google Scholar]
- 24.Salomon S, Puchta H. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert I. Biol Zent Bl. 1994;113:487–491. [Google Scholar]
- 26.Schubert I, Sturelid S, Döbel P, Rieger R. Mutat Res. 1979;59:27–38. [Google Scholar]
- 27.Offringa R, Franke-van Dijk M E, de Groot M J A, van den Elzen P J M, Hooykaas P J J. Proc Natl Acad Sci USA. 1993;90:7346–7350. doi: 10.1073/pnas.90.15.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risseeuw E, Offringa R, Franke-van Dijk M E, Hooykaas P J. Plant J. 1995;7:109–119. doi: 10.1046/j.1365-313x.1995.07010109.x. [DOI] [PubMed] [Google Scholar]
- 29.Dellaire G, Lemieux N, Belmaaza A, Chartrand P. Mol Cell Biol. 1997;17:5571–5580. doi: 10.1128/mcb.17.9.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vergunst A C, Hooykaas P J. Crit Rev Plant Sci. 1999;18:1–31. [Google Scholar]
- 31.Mengiste T, Paszkowski J. Biol Chem. 1999;380:749–758. doi: 10.1515/BC.1999.095. [DOI] [PubMed] [Google Scholar]
- 32.Hohn B, Puchta H. Proc Natl Acad Sci USA. 1999;96:8321–8323. doi: 10.1073/pnas.96.15.8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puchta H. Trends Plant Sci. 1998a;3:77–78. [Google Scholar]
- 34.Rossi L, Tinland B, Hohn B. In: The Rhizobiaceae. Spaink H, Hooykaas P, Kondorosi A, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 303–320. [Google Scholar]
- 35.Holmes A M, Haber J E. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 36.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 37.Schubert I, Heindorff K. Mutat Res. 1989;211:301–306. doi: 10.1016/0027-5107(89)90013-4. [DOI] [PubMed] [Google Scholar]
- 38.Schubert I, Rieger R. Mutat Res. 1994;323:137–142. doi: 10.1016/0165-7992(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 39.Lindenhahn M, Schubert I. Mutat Res. 1983;108:301–316. doi: 10.1016/0027-5107(83)90128-8. [DOI] [PubMed] [Google Scholar]
- 40.Sonoda E, Sasaki M S, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]