Abstract
The helical junction region of a −1 frameshift stimulating hairpin-type mRNA pseudoknot from sugarcane yellow leaf virus (ScYLV) is characterized by a novel C27·(G7–C14) loop 2–stem 1 minor groove base triple, which is stacked on a C8+·(G12–C28) loop 1–stem 2 major groove base triple. Substitution of C27 with adenosine reduces frameshifting efficiency to a level just twofold above the slip-site alone. Here, we show that the global structure of the C27A ScYLV RNA is nearly indistinguishable from the wild-type counterpart, despite the fact that the helical junction region is altered and incorporates the anticipated isostructural A27·(G7–C14) minor groove base triple. This interaction mediates a 2.3-Å displacement of C8+ driven by an A27 N6–C8+ O2 hydrogen bond as part of an A(n−1)·C+·G-Cn base quadruple. The helical junction regions of the C27A ScYLV and the beet western yellows virus (BWYV) pseudoknots are essentially superimposable, the latter of which contains an analogous A25·(G7–C14) minor groove base triple. These results reveal that the global ground-state structure is not strongly correlated with frameshift stimulation and point to a reduced thermodynamic stability and/or enhanced kinetic lability that derives from an altered helical junction architecture in the C27A ScYLV RNA as a significant determinant for setting frameshifting efficiencies in plant luteoviral mRNA pseudoknots.
Keywords: pseudoknot, frameshifting, RNA structure, base triple, imino proton exchange
INTRODUCTION
Our knowledge of the many complex functions that RNA plays in biology has increased tremendously over the last decade. In particular, the complex process of protein synthesis can now be considered at atomic resolution due to the many high-resolution crystal and cryoelectron microscopic structures of the ribosome in various functional states coupled with detailed kinetics and biochemical experiments (Wilson et al. 2002; Wilson and Nierhaus 2003). For example, selection of cognate tRNAs is now known to be achieved by intricate A-minor hydrogen-bonding interactions between the ribosome and the codon/anticodon base pairs and the tRNA itself plays a direct role in recognition by controlling the on and off rates at the A (aminoacyl) site of the ribosome (Cochella and Green 2005; Ogle and Ramakrishnan 2005). Translocation of the mRNA is accomplished by a ratcheting motion of the ribosome, with EF-G binding and GTP hydrolysis providing the force necessary to move the message one codon at a time (Frank and Agrawal 2000). Despite recent progress, many mechanistic issues relative to normal mRNA decoding as well as myriad recoding events (Namy et al. 2004), e.g., translational readthrough (Orlova et al. 2003), frameshifting (Giedroc et al. 2000; Harger et al. 2002), and incorporation of nonstandard amino acids (Krzycki 2005; Sauerwald et al. 2005; Theobald-Dietrich et al. 2005), remain poorly understood at the molecular level.
Of interest to this study is the translational recoding process of −1 programmed ribosomal frameshifting (−1 PRF) (Giedroc et al. 2000; Baranov et al. 2002). −1 PRF is the process whereby the reading frame is changed from the reference or “0” frame to the “−1” frame. This process requires three elements to be efficient: a heptanucleotide slip site (XXXYYYZ), a 6–8 nucleotide (nt) linker, and an RNA structural element, which in most, but not all cases (Gaudin et al. 2005; Staple and Butcher 2005) is a hairpin (H)-type RNA pseudoknot (ten Dam et al. 1990; Giedroc et al. 2000; Baranov et al. 2002). In several cases, the absence or destabilization of a stable RNA pseudoknot has been shown to eliminate efficient frameshift stimulation (Chen et al. 1995; Kim et al. 1999, 2000; Cornish et al. 2005). This has led to the development of the torsional restraint model (Plant and Dinman 2005). This model hypothesizes that efficient frameshifting is induced because the formation of the pseudoknot-forming stem 2 (S2) prevents the unfettered unwinding of the hairpin forming stem 1 (S1). If the RNA pseudoknot is positioned at the mRNA entry tunnel of the ribosome (Giedroc et al. 2000; Plant et al. 2003; Pallan et al. 2005), the pseudoknot would then function as a roadblock to mRNA movement during the elongation cycle, which could be released by either −1 frameshifting or unfolding of the pseudoknot before frameshifting. Indeed, recent cryoelectron microscopic images of a mammalian ribosome-mRNA pseudoknot complex thought to be an intermediate in −1 frameshifting reveal that the mRNA pseudoknot physically blocks the mRNA entrance channel; this leads to a “springlike” deformation of the P-site-bound transfer RNA, which in competition with the translocase activity of elongation factor 2 (eEF2; EF-G in the bacterial system), breaks the P-site tRNA–mRNA interaction (Namy et al. 2006). Subsequent relaxation of the complex allows repairing of the P-site tRNA in the new −1 frame. These structural studies therefore support a mechanical model for pseudoknot-stimulated ribosomal frameshifting during translocation (Namy et al. 2006).
Another model of ribosomal frameshifting suggests that one or more of the entry tunnel proteins of the ribosome, corresponding to bacterial proteins S3 (rpS3), S4 (rpS9), and S5 (rpS2), possess RNA helicase or unwinding activity (Yusupova et al. 2001; Takyar et al. 2005). Pseudoknots could represent poor substrates for the ribosomal helicase, which would induce ribosomal pausing, and in some cases, −1 frameshifting.
Although mechanical models provide a plausible explanation as to why frameshifting efficiencies are higher for a pseudoknots versus an RNA hairpin, they do not fully explain how structurally similar pseudoknots produce vastly different levels of frameshifting. Several groups have shown that mutations introduced at the helical junction region of a wide variety of H-type pseudoknots often have a large effect on frameshift stimulation both in vivo and in vitro, particularly in those pseudoknot systems with relatively short helical stems (Chen et al. 1995; Kim et al. 1999, 2000; Liphardt et al. 1999; Nixon et al. 2002b; Cornish et al. 2005). In some of these cases, companion thermodynamic studies have revealed small diminutions in global stability as measured by thermal unfolding (Theimer and Giedroc 1999, 2000; Nixon and Giedroc 2000; Nixon et al. 2002a; Cornish et al. 2005), and in one case, apparently large changes in global structure, of functionally compromised pseudoknots (Chen et al. 1995, 1996). Unfortunately, the structures of mutant pseudoknots with low functional activities have not been determined to sufficiently high resolution to convincingly determine the extent to which “ground-state” structure influences frameshift stimulation.
Here we present the solution structure of a sugarcane yellow leaf virus (ScYLV) RNA containing a single nucleotide substitution, C27A, that was previously shown to severely compromise frameshift stimulation (Cornish et al. 2005). These structural findings reveal that, despite readily detected changes in the hydrogen bonding interactions at the junction of the two helical stems, the global solution structures of the wild-type and C27A ScYLV RNAs are essentially identical at this level of resolution. Solvent exchange rates (at 10°C) of select base-paired imino and amino protons are, however, slightly elevated in the C27A versus wild-type RNA pseudoknots, but only for those exchangeable protons closest to the site of the substitution. These findings mirror previous thermodynamic studies of hydrogen bond coupling interactions across the helical junction region (Cornish and Giedroc 2006) and suggest that relatively small, localized perturbations in pseudoknot stability and/or kinetic lability may have a large influence on frameshift stimulation, at least for plant luteoviral mRNA pseudoknots. As we discuss, these findings argue in favor of a mechanical model for pseudoknot-stimulated ribosomal frameshifting (Namy et al. 2006), rather than a purely structural model.
RESULTS
Structure determination of the C27A ScYLV RNA
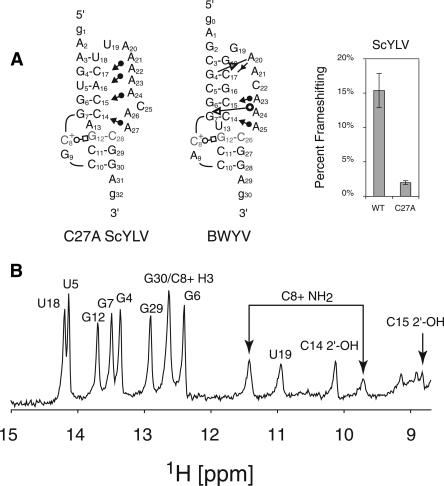
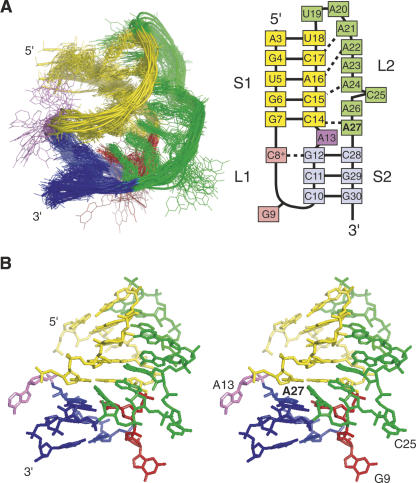
The sequence used for NMR studies here (Fig. 1A) is identical to the sequence used in the previous study for the wild-type RNA except for the C27A substitution (Cornish et al. 2005). Figure 1B shows the downfield region of a jump–return echo 1D NMR spectrum. Similar to previously published luteoviral pseudoknots, the C27A ScYLV RNA has eight Watson–Crick base pairs and a trans Watson–Crick/Hoogsteen base pair indicated by downfield-shifted amino protons and imino proton of C8+. Also observed in this spectrum are the 2′-OH protons of both C14 and C15, which are now known to be diagnostic for the formation of minor groove base triple interactions (Nixon et al. 2002a, b; Giedroc et al. 2003; Cornish et al. 2005, 2006).
FIGURE 1.
(A) The C27A ScYLV RNA sequence used for NMR investigation compared to a 2D representation of the BWYV RNA. The symbols are representative of tertiary interactions as described (Leontis et al. 2002). Of interest in this study are the cis Watson–Crick/sugar-edge tertiary interaction (filled circle and arrow) and a trans Watson–Crick/Hoogsteen tertiary base pair (open circle and square). Also displayed are the relative frameshifting efficiencies for the wild-type and C27A ScYLV RNA pseudoknots as previously determined (Cornish et al. 2005) (B) Jump–return echo 1D NMR spectra (10°C) of the C27A ScYLV mRNA pseudoknot.
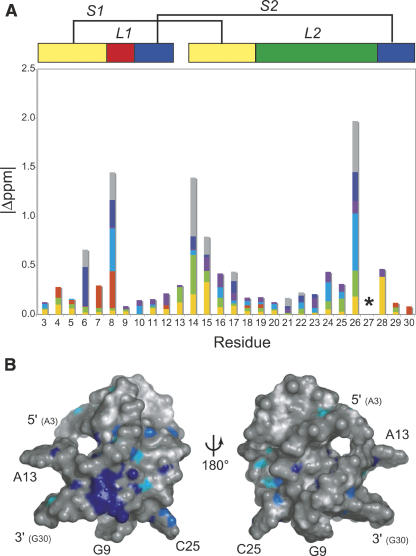
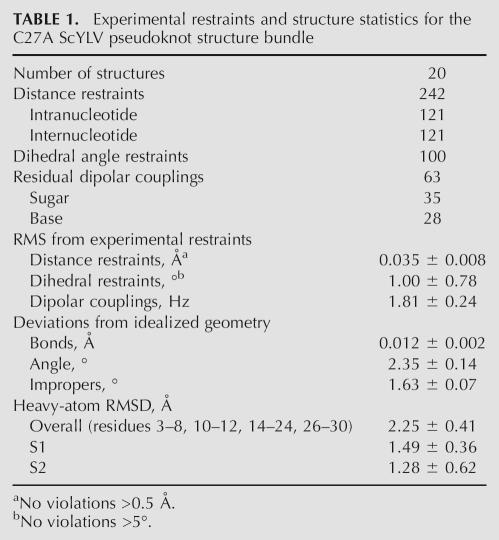
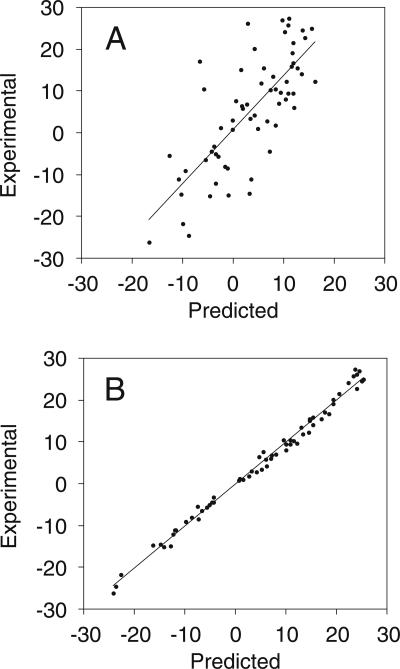
Inspection of the 1H chemical shift perturbation map (Δ ppm = |δWT − δC27A|) (Fig. 2A) superimposed on the structure of wild-type ScYLV RNA (Fig. 2B) suggests that structural differences do not propagate far from the helical junction, i.e., the immediate microenvironment of the C27A substitution. This fact greatly facilitated resonance assignments of the C27A ScYLV RNA at natural isotopic abundance. The solution structure was solved starting from random coordinates subjected to simulated annealing using 242 NOE distance restraints, followed by a one-step rdc-based refinement method using 63 1DCH residual dipolar coupling restraints measured at natural isotopic abundance (see Materials and Methods). The structure statistics are compiled in Table 1. Figure 3A,B shows plots of the predicted versus experimental residual dipolar coupling restraints (RDCs) for the NOE bundle. While poor (r = 0.643 ± 0.062; Fig. 3A), the correlation improves dramatically with a one-step refinement against the experimental RDCs (r = 0.998 ± 0.001; Fig. 3B). The heavy-atom RMSD of the resulting C27A ScYLV RNA structure bundle, excluding unpaired nucleotides is 2.25 ± 0.41 Å (Table 1). Structures of the C27A RNA were also calculated using the wild-type ScYLV RNA bundle (Cornish et al. 2005) as the starting coordinates and are characterized by a heavy-atom RMSD of 2.02 ± 0.48 Å. The refined average structures of the C27A RNA calculated from each method are statistically indistinguishable from one another (data not shown).
FIGURE 2.
Proton chemical shift differences (Δ ppm) between wild-type and C27A ScYLV RNAs. (A) A graphical representation of the absolute value of proton chemical shift differences (Δ ppm) grouped by proton type (H1′, yellow; H2′, green; H1/H3, red; H2/H5, blue; H6/H8, purple; H21/H41/H61, dark blue; and H22/H42/H62, gray) versus residue number. A schematic of the pseudoknot is shown above the plot color coded as in the other figures where yellow, blue, red, and green rectangles represent S1, S2, L1, L2 nucleotides; A13 is between 3′ S2 and 5′ S1 segments, respectively. (*) Residue position of C27/A27 in the wild-type/C27A RNAs. Not shown are |Δppm| values for the 2′ OH protons of C14 (1.6 ppm) and C15 (0.2 ppm). (B) Surface representation of the wild-type ScYLV pseudoknot (Cornish et al. 2005). The left view has the same orientation as Figure 4A and the right view represents a 180° rotation about an arbitrary vertical axis. C27 and all protons with a chemical shift change ≥0.15 ppm are colored in dark blue, 0.1–0.15 ppm in light blue, 0.05–0.1 in cyan, and <0.05 ppm in white. Gray represents all atoms for which there are no data.
TABLE 1.
Experimental restraints and structure statistics for the C27A ScYLV pseudoknot structure bundle
FIGURE 3.
Graphical depictions of experimental versus predicted residual dipolar couplings. Predicted values were calculated using the program DC (Delaglio et al. 1995) from the C27A ScYLV NOE bundle (A) and RDC refined structures (B).
C27A ScYLV RNA structure
Figure 4A (left panel) shows a global overlay of the 20 lowest energy structures while Figure 4B shows a stereoview of the average minimized structure. This RNA clearly adopts a fold that is similar to the wild-type RNA with G9 (red), A13 (violet), and C25 (green) extruded into solvent and is composed of intricate L2–S1 hydrogen bonding interactions that position nearly all of loop L2 (green) in the minor groove of the upper stem S1 (shaded yellow). As found previously for the wild-type ScYLV RNA, A21, A22, and A24 form cis Watson–Crick/sugar edge interactions with C17, A16, and C15, respectively (Cornish et al. 2006). As anticipated (Cornish et al. 2005), A27 forms a cis Watson–Crick/sugar edge base pair with C14 where the N1 accepts a hydrogen bond from the 2′-OH of C14 (δ ≈ 10.2 ppm), a chemical shift similar to that of the PEMV-1 (Nixon et al. 2002a,b) and beet western yellows virus (BWYV) (Cornish et al. 2005) RNAs. For the PEMV-1 RNA, scalar coupling across this hydrogen bond was detected (Giedroc et al. 2003); through-bond coupling was also observed for the analogous C27 NH-C14 2′OH interaction in the wild-type ScYLV RNA (Cornish et al. 2006). In fact, the orientation of A27 in the C27A ScYLV RNA is fully consistent with the formation of an A(n−1)·C+·G-Cn base quadruple motif (unconstrained A27 N6 to C8+ O2 distance of ≈3.2 Å) previously observed in the BWYV RNA (see Fig. 5A,C; Su et al. 1999; Egli et al. 2002).
FIGURE 4.
The solution structure of the C27A ScYLV pseudoknot. (A, left panel) Overlay of the 20 lowest energy structures generated from random coordinates compared to a secondary structure schematic (right panel). S1 is in yellow, S2 is in blue, L1 is in red, L2 is in green, and the extrahelical nucleotide A13 is in violet. (B) Stereo representation of the average solution structure of the C27A ScYLV RNA calculated from the lowest energy bundle, with the same coloration as in panel A.
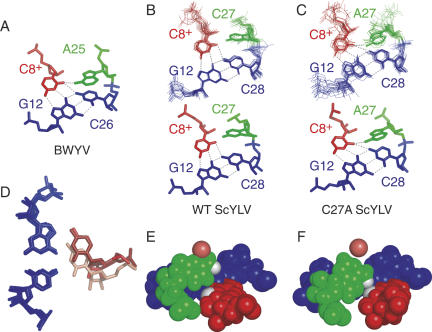
FIGURE 5.
Structural representation of the base quadruple interaction at the helical junction for (A) BWYV, (B) wild-type ScYLV, and (C) C27A ScYLV RNA pseudoknots. The lowest energy structure bundle superposition (n = 20; 0.99 ± 0.17 Å RMSD for residues 7, 8, 12, 14, 27, and 28 in wild-type RNA; 1.27 ± 0.40 Å RMSD for same residues in C27A RNA) (top) and resulting average minimized structures (bottom) are shown for both (B) wild-type and (C) C27A RNAs. The G12–C28 (C26 in BWYV) base pair is in blue, with C8+ forming a trans Watson–Crick/Hoogsteen base pair colored red. The 3′ terminal residue in L2 is in green; A25 in BWYV, C27 in wild-type ScYLV, and A27 in C27A ScYLV. (D) Superposition of the G12–C28 (G12–C26 in the BWYV RNA) base pair (blue) of the BWYV, wild-type ScYLV, and C27A ScYLV structures. C8+ is shaded pink (wild-type ScYLV), light red (C27A ScYLV), and red (BWYV). Space-filling representations of the (E) wild-type and (F) C27A ScYLV structures. The G12–C28 base pair is in blue with C8+ in red. C27 of wild-type ScYLV and A27 of C27A ScYLV RNAs are shaded green. C8+ O2 and C27 H41 atoms of wild-type ScYLV RNA and the A27 H61 atom of the C27A ScYLV are shaded white in both panels; the O4′ atom of C15 is also shown (shaded light red).
Comparison of the structures of the wild-type ScYLV, C27A ScYLV, and BWYV RNAs
C8+ in the wild-type ScYLV RNA is horizontally displaced by ∼2.3 Å relative to the accepting G12–C28 and G12–C26 base pairs in C27A ScYLV and BWYV RNAs, respectively (Fig. 5A–C). This is more clearly observed when the G12–C28 base pairs of the wild-type and C27A ScYLV RNAs and the G12–C26 base pairs of the BWYV pseudoknot are superimposed (Fig. 5D). A space-filling model of the wild-type and C27A ScYLV RNA pseudoknots (Fig. 5E,F) show that an adenosine at the terminal position of L2 (A27) sterically blocks the horizontal displacement of C8+ in the C27A ScYLV RNA in order to maintain an A27 N6 to C8+ O2 hydrogen bond as part of a BWYV-like A(n−1)·C+·G-Cn base quadruple motif (Fig. 5F). In the wild-type ScYLV RNA, the smaller C27 base allows C8+ to come closer and essentially slide under C27 (Fig. 5E); as a result, the analogous C(n−1)·C+·G-Cn base quadruple is not formed (C27 N4–C8+ O2 distance of 5.1 Å). However, a putative C27 N4-H4•••C15 O4′ hydrogen bond is present in all 20 of the lowest energy structures of the wild-type RNA (Fig. 5E). This interaction may help drive this small reorganization of the junction nucleotides of the wild-type ScYLV RNA relative to the C27A mutant. Exocyclic adenosine N6-O4′ hydrogen bonding interactions are found in the small ribosomal subunit (Wimberly et al. 2000) and in an RNA quadruplex (Liu et al. 2002), with at least one other documented cytidine N4-O4′ interaction found on the edge of the large ribosomal subunit (Ban et al. 2000).
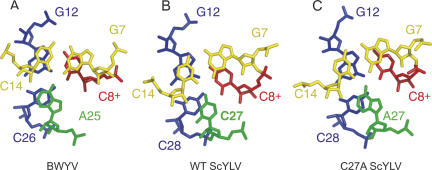
Figure 6A–C displays the two stacked triple base pairs that make up the helical junction regions for the BWYV, wild-type ScYLV, and C27A ScYLV RNAs, respectively. The vertical rotation or helical twist between successive Watson–Crick base pairs at the helical junction is defined by the relative orientations of the closing S1 base pair shown in yellow (G7–C14) relative to the upper base pair of S2 shown in blue (G12–C26 or G12–C28). For canonical A-form RNA, this rotation angle is 32.7°. Here, the two ScYLV RNAs are identical to one another and are strongly overrotated by 79° ± 2.6° and 79° ± 5.4° for the wild-type and ScYLV structure bundle (for ≈102° total helical twist), respectively, with the BWYV RNA characterized by somewhat less overrotation at 48° (≈80° total) (Su et al. 1999).
FIGURE 6.
Helical junction regions viewed down the helix axis of the (A) BWYV, (B) wild-type ScYLV, and (C) C27A ScYLV RNAs. The coloration is the same as in Figure 5, with the G7–C14 base pair in yellow, the G12–C28 (G12–C26 in BWYV) base pair in blue, C8+ in red, and A25, C27, and A27 of BWYV, wild-type ScYLV, and C27A ScYLV, respectively, shaded green.
Figure 7A–C displays stick-and-ribbon representations of the BWYV, wild-type ScYLV, and C27A ScYLV RNAs, respectively, which enables ready comparisons of the global structures of each RNA. As can be seen, L1 crosses the major groove of S2, and L2 crosses the minor groove side of S1, forming a nearly continuous triple-stranded structure. Also, all three RNAs have an extrahelical nucleotide in L1 (G9 in the wild-type and C27A ScYLV RNAs and A9 in BWYV) and an extrahelical nucleotide between S1 and S2 (A13 in wild-type and C27A ScYLV RNAs and U13 in BWYV); the latter allows for efficient stacking of the two helical stems and significant overrotation of the helical junctions.
FIGURE 7.
Stick-and-ribbon representations of the (A) BWYV, wild-type (B) ScYLV, and (C) C27A ScYLV structures. S1 is in yellow, S2 is in blue, L1 is in red, L2 is shaded green, and the extrahelical nucleotide A13 is colored violet. These figures were made using nuccyl 1.5.2 and the program PyMOL (DeLano 2002).
Quantitative measurement of the interhelical bend angle for these RNAs is complicated by the internal bend of each stem toward the major groove, previously shown to be 13° and 7.5° in S1 and S2 for the BWYV pseudoknot, respectively (Su et al. 1999). This helical distortion allows efficient crossing of the lower stem S2 by a single nucleotide L1, as well as for significant penetration of L2 into the minor groove of S1. In any case, the estimated helical bend angle for the BWYV pseudoknot is ≈25° (Su et al. 1999), and is measurably larger for the wild-type and C27A ScYLV RNAs, at 50° ± 14°, and 46° ± 16°, respectively. This difference in bend angle between BWYV and ScYLV pseudoknots may be due to the more extensive base stacking of L2 nucleotides in the S1 minor groove in both ScYLV RNAs, which is further enhanced by one additional L2 nucleotide in the L2 stack relative to the BWYV pseudoknot. The parallel increases in helical overrotation, horizontal displacement (Cornish et al. 2005), and interhelical bend angle noted for the ScYLV RNAs relative to the BWYV pseudoknots are fully consistent with expectations from simple modeling experiments (Michiels et al. 2001). The main point, however, is that the global structures of all three pseudoknots are quite similar.
Comparison of the imino/amino proton solvent exchange rates of wild-type versus C27A ScYLV RNAs
In an effort to detect differences in the dynamics of the wild-type and C27A ScYLV pseudoknots, imino proton solvent exchange rates (k ex) for Watson–Crick and Hoogsteen base-paired regions were determined under standard solution conditions (10°C, 5 mM Mg2+, 0.1 M KCl at pH 6.0) with a simple jump–return magnetization transfer experiment (Fig. 8; Dhavan et al. 1999). Since the imino protons exchange only slowly from the closed base pair, these conditions are likely reporting on intrinsic exchange catalysis by the nitrogen of the accepting base pair, in a process involving one or several water molecules (Gueron et al. 1987; Snoussi and Leroy 2001; Varnai et al. 2004). In order to obtain insight into C27A RNA-specific perturbations of solvent exchange rates that might be linked to diminished functional activity of the C27A RNA, we measured imino and amino proton solvent exchange rates for wild-type, C27A, and ΔC25 ScYLV RNAs, the latter of which stimulates high levels of ribosomal frameshifting (Fig. 8B; Cornish et al. 2005). C25 is flipped out of the triple helical stack and makes very few contacts with the rest of the RNA (see Fig. 4).
FIGURE 8.
Imino/amino proton solvent exchange rates (k ex) for the wild-type, C27A and ΔC25 ScYLV RNA pseudoknots. (A) Schematic representation of the ScYLV pseudoknot highlighting the positions of Hoogsteen (C8+) and Watson–Crick base-paired imino protons in stem S1 (yellow) and stem S2 (purple). (B) Graphical representation of k ex for the wild-type (dark gray bars) versus C27A (light gray bars) RNAs (upper panel), and wild-type (dark gray bars) versus ΔC25 (light gray bars) RNAs (lower panel). The upfield C8+ amino proton (C8+ H41) is represented by the left pairs of bars, with data for the downfield proton (C8+ H42) on the far right. (*) k ex could not be unambiguously measured for the C8+/G30 (C27A RNA) and U18/U5 protons (ΔC25 RNA) imino protons due to resonance overlap. (C) Structure of the C27A ScYLV RNA highlighting the positions of the G7 H1, G12 H1, and C8+ H41 protons relative to A27 (in stick). Conditions: 10 mM potassium phosphate, 100 mM KCl, and 5 mM MgCl2 (pH 6.0), 10°C.
Although the differences are small, k ex is specifically larger for G7 (by 14%), G12 (17%), and the upfield C8+ amino proton (H41; 25%) in the C27A RNA relative to the wild-type RNA; the exchange rates of these three residues are completely unaffected by the ΔC25 deletion (Fig. 8B). In contrast, other measurable perturbations (e.g., in G4, G6, C8+ H42 protons) or lack of perturbation (in G29) are common to both ΔC25 and C27A substitutions. As can be seen in the model of the C27A RNA (Fig. 8C), the G7 imino, G12 imino, and C8+ amino protons are closest to the C27A base substitution. Although exchange rates of cytidine amino protons are complicated by intermediate rotations around the C4–N4 bond, examination of the structure of C27A RNA reveals that the C8+ H41 is more exposed to solvent than in the wild-type RNA by virtue of the rearrangements in the junction region (see Fig. 5); this is compatible with the solvent exchange data. Unfortunately, a comparison of kex values for C8+ H3 proton could not be obtained since G30 and C8+ are overlapped in the C27A RNA; however, the solvent exchange rate for C8+ in the wild-type and ΔC25 RNAs is two- to threefold greater than internal Watson–Crick base-paired regions, and in fact, more comparable to terminal base pairs. This is in contrast to previous findings with the PEMV-1 pseudoknot (Nixon et al. 2002b). In any case, the data suggest that the C27A substitution specifically perturbs the kinetic stability of the triple-helical stack at the helical junction region of this RNA.
DISCUSSION
The solution structure of the C27A ScYLV mRNA pseudoknot represents the first high-resolution structure of a mutant luteoviral pseudoknot with significantly reduced frameshift stimulatory activity. The overall structure of the mutant RNA is essentially identical to the wild-type pseudoknot, despite distinct helical junction architectures (Figs. 5,6). This, in turn, leads to an ≈1.5 kcal·mol−1 reduction in global stability at 37°C of the C27A ScYLV RNA as measured by thermal denaturation experiments over a wide range of pH values (Cornish et al. 2005; Cornish and Giedroc 2006). More rapid imino proton solvent exchange rates (Fig. 8) localized to the two triple base pairs that straddle the helical junction in the C27A versus wild-type ScYLV RNAs (see Fig. 6) may also be reporting on these differences in global stability.
In previous work, several structures of variant mouse mammary tumor virus (MMTV) gag-pro pseudoknots were solved by solution NMR methods (Shen and Tinoco 1995; Chen et al. 1996; Kang et al. 1996; Kang and Tinoco 1997). Of the five structures determined, three with the highest frameshifting efficiencies (VPK, U13C, and APK A27G) possessed an intercalated adenosine (A14) between S1 and S2; this appeared to cause S1 to bend toward the major groove of S2, creating an apparent interhelical bend angle of ≈90°. The weakly functional mutant ΔA14U13C RNA, which lacks the wedged adenosine at the junction, appeared not to adopt a bent conformation. Another poorly functional mutant, APK, was found to adopt a bent conformation apparently opposite of that found in the VPK, U13C, and APK A27G RNAs because it contained 2 nt between S1 and S2. This suggested that a single unpaired and intercalated adenosine coupled with a specifically bent global conformation collaborate to mediate efficient frameshift stimulation in the MMTV gag-pro system.
A subsequent study that investigated the functional activities of chimeric pseudoknots revealed that in pseudoknots with short L2s (≤8 nt), an adenosine in the 3′-most position of L2 (analogous to A27 in C27A ScYLV RNA) was clearly an important determinant for efficient frameshift stimulation, more so than the wedged adenosine (Liphardt et al. 1999). However, even the functional importance of a 3′ L2 adenosine decreases as the length of L2 is increased. Functional and structural studies with simian retrovirus-1 (SRV-1) and human endogenous retrovirus-K10 (HERV-K10) gag-pro pseudoknots then confirmed that pseudoknots with coaxially stacked helical stems could stimulate high levels of −1 frameshifting (Michiels et al. 2001; Wang et al. 2002). Finally, our comparative studies of ScYLV and BWYV pseudoknots show that a 3′ L2 adenosine is in fact inhibitory, relative to cytidine, in the luteoviral RNA structural context (Cornish et al. 2005), a result contrary to simple predictions from previous work (Liphardt et al. 1999; Su et al. 1999). These disparate findings, taken together, indicate that the impact of specific interactions at the helical junction are strongly context dependent, and structural/functional rules that emerge from investigation of a specific frameshift signal may not be generally applicable to systems. Indeed, there may be multiple mechanistic paths the ribosome can take to achieve high levels of −1 frameshifting since antisense oligonucleotides capable of base pairing to a site just downstream of the slip site have recently been shown to induce significant levels of frameshifting in vitro (Howard et al. 2004).
What then drives −1 frameshift stimulation by H-type RNA pseudoknots? Given the fact that the lowest energy global structures of the wild-type and C27A ScYLV RNAs are virtually identical, we consider the possibility remote that global structure and/or interhelical bend angle are primary determinants for frameshift stimulation, as has been argued previously for the MMTV gag-pro system (Chen et al. 1996). More recent structural studies of the HIV-1 gag-pol frameshift signal reveal an extended hairpin containing two helical elements separated by a three-purine bulge that creates a 60° interhelical bend angle (Gaudin et al. 2005; Staple and Butcher 2005). While the possibility was raised that the magnitude of the interhelical bend angle might somehow influence levels of frameshift stimulation, perhaps by functioning as a “positioning element” for the elongating ribosome (Staple and Butcher 2005), the mechanistic importance of the lower stem in the HIV-1 signal is likely distinct from the upper stem S1 of H-type pseudoknots since it sequesters the linker between the 3′ edge of the slip site and the upper stem–loop element. Distinct electrostatics may also potentially play a functional role in wild-type versus C27A RNAs, given the small differences in backbone geometry that are apparent in the average structures of these RNAs (Fig. 7); this possibility requires further refinement of the backbone dihedral angles in these RNAs.
Rather than global RNA structure, then, it seems more likely that the helical junction region in pseudoknots functions as a kinetic barrier to the unwinding mediated by the mechanical force of the elongating ribosome while the decoding center is positioned over the slip site. The relative impact of the helical junction region in maintaining significant frameshifting efficiency is expected to be more pronounced in those frameshifting pseudoknots that have very short S2 stems, like the luteoviral mRNAs studies here. Small perturbations in helical junction architecture would, in turn, effect significant perturbations in the kinetics or thermodynamics of unfolding relevant to translocation. Although the structural origin of the reduced thermodynamic stability of the C27A versus wild-type ScYLV RNAs (Cornish et al. 2005) is difficult to directly assess, distinct base stacking and/or hydrogen bonding interactions at the helical junction are likely to be involved. Inspection of the triple base-pair stack that crosses the helical junction (Fig. 6) reveals that the base stacking appears slightly poorer for the C27A versus wild-type ScYLV RNAs (Cornish and Giedroc 2006). Another possibility is that the proposed C27 N4-H4•••C15 O4′ hydrogen bond in the wild-type RNA (Fig. 5E) may make a more favorable contribution to the local stability of the helical junction than the corresponding A27 N6-H6•••C8+ O2 hydrogen bond in the ScYLV C27A RNA (Fig. 5F). Note that the analogous substitution in the BWYV RNA (A25C) also increases frameshifting levels by threefold (Cornish et al. 2005). It seems likely that these distinct hydrogen bonding interactions are largely responsible for the global differences in stability between the wild-type and C27A ScYLV RNAs (Cornish et al. 2005). An elucidation of the distinct pairwise coupling free energies between hydrogen bonds that straddle the helical junction (C27A·C14 versus A27·C14 with C8+·C12) in the wild-type versus C27A ScYLV pseudoknots is consistent with this idea (Cornish and Giedroc 2006).
Our structural and dynamic data argue in favor of a mechanical model of luteoviral mRNA pseudoknot-stimulated −1 frameshifting recently proposed on the basis of a structural model of a ribosome-frameshifting mRNA complex thought to be an intermediate derived from cryo-EM reconstructions (Namy et al. 2006). In this model, upon encountering the pseudoknot, the small subunit stalls during translocation with eIF2 bound, inducing tension in the mRNA that bends the P-site tRNA in the 3′ direction. This tension disrupts the codon–anticodon interaction, allowing the P-site tRNA to relax and repair in the new −1 frame. If two mRNA pseudoknots, e.g., wild-type and C27A ScYLV RNAs, possess distinct abilities to resist these mechanical forces, they will have vastly different functional activities, in the absence of pronounced structural differences in the ground state. Mechanical force-induced unwinding experiments with luteoviral RNAs studied here promise to provide additional mechanistic insight into this complex process.
MATERIALS AND METHODS
RNA preparation
The C27A (Fig. 1A) and ΔC25 ScYLV RNAs were prepared by run-off transcription using SP6 RNA polymerase and purified as described previously (Cornish et al. 2005). The final NMR buffer was 10 mM potassium phosphate, 100 mM KCl, and 5 mM MgCl2 (pH 6.0). The RNA concentration was 2.7 mM and 1.2 mM for the C27A and ΔC25 RNAs, respectively. For residual dipolar coupling experiments, ∼15 mg/mL Pf1 phage (ASLA Biotech Ltd.) was added, producing a 2H2O splitting of 15.4 Hz.
NMR spectroscopy
All NMR experiments were performed on a Varian Inova 500 MHz spectrometer at the Biomolecular NMR Laboratory, Texas A&M University. Homonuclear 1H-1H experiments included short (τmix = 60 msec) and long (τmix = 280 msec) mixing time D2O-NOESY spectra and 60-, 200-, 280-, and 320-msec NOESY spectra acquired in 10% D2O/90% H2O. A TOCSY spectrum acquired in 100% D2O was used to obtain ribose and pyrimidine H5/H6 resonance assignments, facilitated by a natural abundance 1H-13C HSQC spectrum. Imino proton resonance assignments were made on the basis of an analysis of imino/amino proton connectivities observed in a long-mixing-time (τm = 320 msec) NOESY spectrum. NMR data were processed using nmrPipe and analyzed using Sparky (Goddard and Kneller 2002; Delaglio et al. 1995). For measurement of 1DCH residual dipolar couplings, a sensitivity- and gradient-enhanced TROSY experiment was run (Weigelt 1998). For each subspectrum, 192 scans were recorded with 1024 and 128 complex points in t2 and t1, respectively. The total acquisition time for each experiment was ∼24 h.
NMR restraints
A total of 242 NOE-derived distance restraints were obtained from the NOESY experiments as follows. NOEs from the 60-msec D2O and 10% D2O/90% H2O NOESY were categorized as strong (2.5 ± 0.5 Å), medium (3.3 ± 0.7 Å), weak (4.0 ± 1.0 Å), and very weak (5.0 ± 1.0 Å) distance restraints essentially as described (Cornish et al. 2005; Theimer et al. 2005), with weaker NOEs from longer mixing time NOESY experiments (τm200, 280, or 320 msec) catagorized as very weak restraints (5.0 ± 1.0 Å). Incorporation of looser bounds on the weaker NOEs (5.0 ± 2.0 Å) revealed that RMSD values obtained for the overall structure (2.52 ± 0.60 Å, n = 16) as well as S1, S2, and L1 nucleotides fell well within the range of values given in Table 1 (data not shown). Seventy-six NOE-type and base planarity restraints were also used to constrain the Watson–Crick base pairs and the trans Watson–Crick/Hoogsteen base pair formed by C8+·G12. The N1 to 2′-OH distances for the cis Watson–Crick/sugar-edge pairs involving A21, A22, A24, and A27 were constrained with a square well potential of 2.8 ± 0.3 Å (Cornish et al. 2005). Although omitting these restraints does not significantly alter the structure as found previously for the wild-type RNA (Cornish et al. 2005), they were primarily used to facilitate the convergence of the calculated structure bundle (see below) since the total number of NOE restraints was low. Loose sugar and backbone dihedral angle restraints (±60°) were used for S1 and S2, analogous to that used to determine the wild-type RNA structure (Cornish et al. 2005). Sixty-three 1DCH base and ribose residual dipolar couplings were used as restraints during refinement.
Structure calculations
Solution structure calculations were performed in two stages using XPLOR-NIH 2.10. In the first stage, 100 structures were generated from random coordinates using simulated annealing as previously described (Nixon et al. 2002b). All restraints excluding the residual dipolar couplings were used in this step. In the second stage of refinement, 20 lowest energy structures were chosen from the pool of 100 structures and refined against the experimental residual dipolar coupling restraints. Eighteen picoseconds of molecular dynamics were performed while cooling the system from 1000 K to 100 K and incrementally increasing the force constant for the residual dipolar couplings from 0.005 to 1 kcal·mol−1·Hz−2 while holding the force constant for the dihedral angle and NOE restraints constant at 100 kcal·mol−1·deg−2 and 200 kcal·mol-1·Å−2, respectively. One thousand steps of conjugate gradient restrained energy minimization followed this molecular dynamics run. A grid search estimated D a = 12.55 Hz and R = 0.54. The 20 lowest energy structures and an average structure have been deposited in the PDB as 2AP0 and 2AP5, respectively.
In a separate series of structure calculations, the 20 lowest energy structures from the wild-type ScYLV pseudoknot bundle (1YG3) were used as a starting point for refinement, with C27 first replaced with an adenosine in each structure using Insight II (Accelrys Inc.). Forty picoseconds of molecular dynamics were performed with the temperature decreasing from 1000 K to 100 K and the force constant for the dihedral angle and NOE restraints held constant at 10 kcal·mol−1·deg−2 and 200 kcal·mol-1·Å−2, respectively. Following 200 steps of conjugate gradient restrained energy minimization, the subsequent RDC-based stage of refinement was identical to that described above. Structures generated in this way gave similar statistics, and a pairwise comparison of the two average minimized structures revealed that they were indistinguishable (1.15 Å global RMSD, excluding extra helical nucleotides G9, A13, and C25; data not shown).
A pairwise global RMSD between the average wild-type (PDB accession code 1YG4) and C27A (2AP5) RNAs (excluding nucleotides G9, A13, C25, and C27/A27) is 1.98 Å, well within the range of pairwise RMSDs obtained for individual structure bundles (1YG3 and 2AP0, respectively). Thus, these data suggest that global structures are statistically indistinguishable at this level of refinement.
Saturation transfer solvent exchange experiments
The rates of exchange of imino protons with solvent protons were measured with RNA samples (0.8–1.5 mM) dissolved in 90% H2O/10% D2O on a 500-MHz Varian INOVA spectrometer operating at 10°C using a penta probe. The water proton resonance was selectively inverted using a Gaussian 180° pulse (5.3 msec) with exchange delay times ranging from 2 to 2000 msec (with 30–60 increments per experiment) (Coman and Russu 2004). A weak gradient (0.1 G/cm) was applied during the exchange delay following water inversion to prevent the effects of radiation damping, with a second Gaussian pulse (2.8 msec) applied at the end of the exchange delay to bring the water magnetization back to the z axis. Exchangeable proton resonances were detected using a gradient-enhanced spin echo sequence. There was a 6-sec recycle delay between transients, with 256 transients acquired at each exchange delay increment. The resulting spectral array was processed with 1-Hz line broadening and the resonance intensities quantified using a spectral deconvolution routine in NUTS (Acorn NMR, Inc.). The observed imino proton resonance intensities were fitted to the following equation (Dhavan et al. 1999) using a nonlinear least-squares fitting routine in Kaleidagraph (Synergy Software):
where I t is the imino proton intensity at exchange delay time t, I 0 is the initial intensity with no solvent saturation delay, E is the efficiency of water inversion (assumed equal to −2 for an inversion efficiency of 100%) (Dhavan et al. 1999), R1w is the longitudinal relaxation rate of water protons (measured in a separate inversion recovery experiment), and R1i is the sum of the imino proton longitudinal relaxation rate and the imino proton exchange rate, kex.
ACKNOWLEDGMENTS
We thank Dr. Jerry Tsai (TAMU) for the use of his computers for structure calculations. We acknowledge support from the NIH (AI040187) and the Texas Higher Education Coordinating Board Advanced Research Program (010361-0278-1999, 010366-0172-2001). P.V.C. was supported in part by an NIH Chemistry–Biology Interface Training Grant (T32 GM008523). S.N.S was supported in part by an NIH Molecular Biophysics Training Grant (T32 GM065088).
Footnotes
Article published online ahead of print. Article and publication date at http://www.rnajournal.org/cgi/doi/10.1261/rna.199006.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Baranov, P.V., Gesteland, R.F., Atkins, J.F. Recoding: Translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Chen, X., Chamorro, M., Lee, S.I., Shen, L.X., Hines, J.V., Tinoco, I., Jr., Varmus, H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: Nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Kang, H., Shen, L.X., Chamorro, M., Varmus, H.E., Tinoco, I., Jr. A characteristic bent conformation of RNA pseudoknots promotes −1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Cochella, L., Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coman, D., Russu, I.M. Site-resolved stabilization of a DNA triple helix by magnesium ions. Nucleic Acids Res. 2004;32:878–883. doi: 10.1093/nar/gkh228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish, P.V., Giedroc, D.P. Pairwise coupling analysis of helical junction hydrogen bonding interactions in luteoviral RNA pseudoknots. Biochemistry. 2006;45:11162–11171. doi: 10.1021/bi060430n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish, P.V., Hennig, M., Giedroc, D.P. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish, P.V., Giedroc, D.P., Hennig, M. Dissecting non-canonical interactions in frameshift-stimulating mRNA pseudoknots. J. Biomol. NMR. 2006;35:209–223. doi: 10.1007/s10858-006-9033-x. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. DeLano Scientific; San Carlos, CA: 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- Dhavan, G.M., Lapham, J., Yang, S., Crothers, D.M. Decreased imino proton exchange and base-pair opening in the IHF–DNA complex measured by NMR. J. Mol. Biol. 1999;288:659–671. doi: 10.1006/jmbi.1999.2690. [DOI] [PubMed] [Google Scholar]
- Egli, M., Minasov, G., Su, L., Rich, A. Metal ions and flexibility in a viral RNA pseudoknot at atomic resolution. Proc. Natl. Acad. Sci. 2002;99:4302–4307. doi: 10.1073/pnas.062055599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J., Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Gaudin, C., Mazauric, M.H., Traikia, M., Guittet, E., Yoshizawa, S., Fourmy, D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- Giedroc, D.P., Theimer, C.A., Nixon, P.L. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc, D.P., Cornish, P.V., Hennig, M. Detection of scalar couplings involving 2′-hydroxyl protons across hydrogen bonds in a frameshifting mRNA pseudoknot. J. Am. Chem. Soc. 2003;125:4676–4677. doi: 10.1021/ja029286t. [DOI] [PubMed] [Google Scholar]
- Goddard, T.D., Kneller, D.G. University of California; San Francisco: 2002. SPARKY 3. [Google Scholar]
- Gueron, M., Kochoyan, M., Leroy, J.L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328:89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- Harger, J.W., Meskauskas, A., Dinman, J.D. An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- Howard, M.T., Gesteland, R.F., Atkins, J.F. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H., Tinoco, I., Jr. A mutant RNA pseudoknot that promotes ribosomal frameshifting in mouse mammary tumor virus. Nucleic Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H., Hines, J.V., Tinoco, I., Jr. Conformation of a non-frameshifting RNA pseudoknot from mouse mammary tumor virus. J. Mol. Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- Kim, Y.G., Su, L., Maas, S., O'Neill, A., Rich, A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl. Acad. Sci. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.G., Maas, S., Wang, S.C., Rich, A. Mutational study reveals that tertiary interactions are conserved in ribosomal frameshifting pseudoknots of two luteoviruses. RNA. 2000;6:1157–1165. doi: 10.1017/s1355838200000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzycki, J.A. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B., Stombaugh, J., Westhof, E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt, J., Napthine, S., Kontos, H., Brierley, I. Evidence for an RNA pseudoknot loop–helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Matsugami, A., Katahira, M., Uesugi, S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J. Mol. Biol. 2002;322:955–970. doi: 10.1016/s0022-2836(02)00876-8. [DOI] [PubMed] [Google Scholar]
- Michiels, P.J., Versleijen, A.A., Verlaan, P.W., Pleij, C.W., Hilbers, C.W., Heus, H.A. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy, O., Rousset, J.P., Napthine, S., Brierley, I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Namy, O., Moran, S.J., Stuart, D.I., Gilbert, R.J., Brierley, I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, P.L., Giedroc, D.P. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- Nixon, P.L., Cornish, P.V., Suram, S.V., Giedroc, D.P. Thermodynamic analysis of conserved loop–stem interactions in P1-P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry. 2002a;41:10665–10674. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- Nixon, P., Rangan, A., Kim, Y., Rich, A., Hoffman, D., Hennig, M., Giedroc, D. Solution structure of a luteoviral p1–p2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002b;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Orlova, M., Yueh, A., Leung, J., Goff, S.P. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell. 2003;115:319–331. doi: 10.1016/s0092-8674(03)00805-5. [DOI] [PubMed] [Google Scholar]
- Pallan, P.S., Marshall, W.S., Harp, J., Jewett, F.C., 3rd, Wawrzak, Z., Brown, B.A., 2nd, Rich, A., Egli, M. Crystal structure of a luteoviral RNA pseudoknot and model for a minimal ribosomal frameshifting motif. Biochemistry. 2005;44:11315–11322. doi: 10.1021/bi051061i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant, E.P., Dinman, J.D. Torsional restraint: A new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant, E.P., Jacobs, K.L., Harger, J.W., Meskauskas, A., Jacobs, J.L., Baxter, J.L., Petrov, A.N., Dinman, J.D. The 9-A solution: How mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald, A., Zhu, W., Major, T.A., Roy, H., Palioura, S., Jahn, D., Whitman, W.B., Yates, J.R., 3rd, Ibba, M., Soll, D. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- Shen, L.X., Tinoco, I., Jr. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Snoussi, K., Leroy, J.L. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry. 2001;40:8898–8904. doi: 10.1021/bi010385d. [DOI] [PubMed] [Google Scholar]
- Staple, D.W., Butcher, S.E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Su, L., Chen, L., Egli, M., Berger, J.M., Rich, A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar, S., Hickerson, R.P., Noller, H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- ten Dam, E.B., Pleij, C.W., Bosch, L. RNA pseudoknots: Translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Giedroc, D.P. Equilibrium unfolding pathway of an H-type RNA pseudoknot which promotes programmed −1 ribosomal frameshifting. J. Mol. Biol. 1999;289:1283–1299. doi: 10.1006/jmbi.1999.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Giedroc, D.P. Contribution of the intercalated adenosine at the helical junction to the stability of the gag-pro frameshifting pseudoknot from mouse mammary tumor virus. RNA. 2000;6:409–421. doi: 10.1017/s1355838200992057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Blois, C.A., Feigon, J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Theobald-Dietrich, A., Giege, R., Rudinger-Thirion, J. Evidence for the existence in mRNAs of a hairpin element responsible for ribosome dependent pyrrolysine insertion into proteins. Biochimie. 2005;87:813–817. doi: 10.1016/j.biochi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Varnai, P., Canalia, M., Leroy, J.L. Opening mechanism of G.T/U pairs in DNA and RNA duplexes: A combined study of imino proton exchange and molecular dynamics simulation. J. Am. Chem. Soc. 2004;126:14659–14667. doi: 10.1021/ja0470721. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Wills, N.M., Du, Z., Rangan, A., Atkins, J.F., Gesteland, R.F., Hoffman, D.W. Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: A mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral gag-pro frameshift site. RNA. 2002;8:981–996. doi: 10.1017/s1355838202024044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt, J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 1998;120:10778–10779. [Google Scholar]
- Wilson, D.N., Nierhaus, K.H. The ribosome through the looking glass. Angew. Chem. Int. Ed. Engl. 2003;42:3464–3486. doi: 10.1002/anie.200200544. [DOI] [PubMed] [Google Scholar]
- Wilson, D.N., Blaha, G., Connell, S.R., Ivanov, P.V., Jenke, H., Stelzl, U., Teraoka, Y., Nierhaus, K.H. Protein synthesis at atomic resolution: Mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Pept. Sci. 2002;3:1–53. doi: 10.2174/1389203023380846. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons, W.M.J., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H., Noller, H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]