Abstract
Double-stranded RNA (dsRNA) can induce post-transcriptional gene silencing in a wide variety of organisms. Commonly, inverted repeats are used to produce dsRNA to silence genes of interest. However, cloning inverted repeats still remains a rate-limiting step for widely applying this technique. Here we describe a pGEM-T-based vector, pGEM-WIZ, designed to produce inverted repeats for any Drosophila gene. pGEM-WIZ has a high efficiency in assembling inverted repeats and the repeats in this vector are stable in regular Escherichia coli strains. Furthermore, we have developed a method for rapid selection of clones with an inverted repeat based on size and relative copy number of the vector with or without an insert. This method further eases the cloning process. The inverted repeat cassette assembled in pGEM-WIZ can be easily transferred to commonly available expression vectors suitable for stably expressing inverted repeats in vitro and in vivo.
Keywords: RNA interference, inverted repeat, cloning, Drosophila
INTRODUCTION
Since double-stranded RNA (dsRNA) was first discovered as the trigger of gene silencing in Caenorhabditis elegans (Fire et al. 1998), RNA interference (RNAi) has become a standard tool for studying the function of genes in situ and in a broad array of experimental organisms. For example, it has been shown that introduction of dsRNA into Drosophila embryos by injection can silence specific target genes (Kennerdell and Carthew 1998; Misquitta and Paterson 1999). However, down regulation of gene expression by injecting dsRNA is transient. A single injection may not provide a sufficient window of functional depletion for some genes that encode proteins with long half-lives, and silencing effects cannot be inherited to the next generation. In addition, effects mediated by this transient introduction of dsRNA are variable.
To overcome these problems, methods have been developed to use vectors that stably express dsRNA in cells or transgenic animals. For such vector-based RNAi, dsRNA can be produced by either bidirectional transcription or by transcription of an inverted repeat or “hairpin” sequence. Vectors carrying inverted repeats for RNAi are more commonly used, presumably due to their higher efficiency in making dsRNA (e.g., Fortier and Belote 2000; Kennerdell and Carthew 2000; Lam and Thummel 2000; Martinek and Young 2000). However, difficulty in cloning an inverted repeat has hindered using this approach. Most bacteria do not tolerate plasmids with an inverted repeat. To overcome this cloning difficulty, an RNAi vector, pWIZ, has been developed for generating transgenes carrying an inverted repeat (Lee and Carthew 2003). In this vector, a functional intron of the Drosophila white gene was introduced as a spacer to separate the repeats, whereby the stability of an inverted repeat sequence in bacteria was enhanced. When the inverted repeat in pWIZ is expressed, the corresponding transcript forms loopless hairpin RNA after splicing. In addition, pWIZ derives from pUAST (Brand and Perrimon 1993). The combination of the Gal4/UAS modular expression system enables hairpin RNA to conditionally silence gene activity in Drosophila in a variety of tissues at any specific developmental stage.
To use pWIZ, a single PCR fragment is inserted on each side of the white intron sequentially, and the vector with an inverted repeat is identified by restriction mapping (Lee and Carthew 2003). However, due to the large size of pWIZ (9.1 kb) in relation to an insert (∼500–700 bp) and limited unique restriction sites, in some cases restriction bands are difficult to interpret. Thus, restriction mapping alone fails to resolve the ambiguity, necessitating DNA sequencing. In addition, for reasons that are not entirely clear, bacteria poorly tolerate an inverted repeat in pWIZ. To deter DNA recombination in bacteria, the recombination-deficient Escherichia coli strains such as the SURE strain (Stratagene) are often used to propagate pWIZ containing an inverted repeat. Typically, selection of a clone with the repeat involves testing a large number of clones. Thus, screening for an appropriate clone in pWIZ can prove time consuming and laborious. Furthermore, we have experienced variable success with pWIZ. Attempts to clone inverted repeats for a number of genes using pWIZ have repeatedly proven unsuccessful in a variety of E. coli strains.
To enhance the efficiency in cloning inverted repeats for RNAi, we have constructed an all-purpose vector, pGEM-WIZ, for assembling the repeat for any Drosophila gene. pGEM-WIZ has proven much more efficient than pWIZ. Some inverted repeats that previously had failed to be cloned in pWIZ were assembled efficiently in pGEM-WIZ. The inverted repeats in pGEM-WIZ are stable in common E. coli strains. In addition, we have developed a method for fast selection of clones with the repeat. This method can further shorten the time for identifying the correct clones and reduce the labor involved in generating RNAi vectors.
RESULTS AND DISCUSSION
Use of pGEM-WIZ for cloning inverted repeats
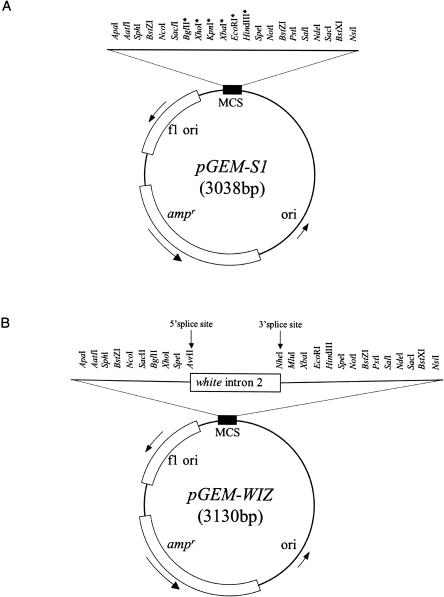
We have generated a multifunctional vector, pGEM-WIZ, to assemble inverted repeats targeting any Drosophila genes by shuttling intron 2 of the Drosophila gene white from pWIZ into a cloning vector, pGEM-S1 (Fig. 1A,B; see also Materials and Methods). As pGEM-S1, pGEM-WIZ uses pGEM-T (Promega) as its backbone. Like other pUC derivatives, pGEM-WIZ lacks the rop gene that is involved in controlling copy number. As a result, pGEM-WIZ replicates to a much higher copy number than do other plasmids that carry a ColE1 origin (Sambrook et al. 1989). In addition, pGEM-WIZ carries the white intron as a spacer to enhance the stability of inverted repeat sequences in bacteria. The vector contains AvrII and NheI restriction sites flanking the intron that retain the consensus sequences for 5′- and 3′-splice sites, respectively (Fig. 1B).
FIGURE 1.
Schematic representation of the pGEM-S1 and pGEM-WIZ vectors. (A) pGEM-S1 was constructed by circularizing pGEM-T (Promega). As a result, an additional six unique sites are present in pGEM-S1, marked by asterisks (*). (B) pGEM-WIZ was constructed by shuttling the intron 2 of the Drosophila gene white from pWIZ into pGEM-S1. The white intron contains consensus splicing sites: 5′-splice site (AG↑GTRAGT) and 3′-splice site (TTTYYYYTNCAG↑RT), where ↑ stands for the splice site; R for A or G; Y for C or T; and N for any base. MCS=multiple cloning sites.
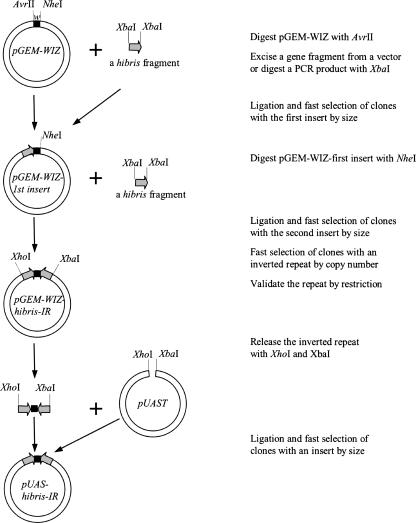
To assemble an inverted repeat into pGEM-WIZ, a DNA fragment corresponding to the gene of interest was inserted into AvrII and NheI sites sequentially in opposite orientations (Fig. 2). Note that XbaI, NheI, SpeI, and AvrII have compatible cohesive ends so that a fragment bearing any of these restriction sites can be ligated into pGEM-WIZ at an AvrII or NheI site. As an example, to generate an inverted repeat for the Drosophila gene hibris, a 504-nucleotide (nt) fragment was amplified by PCR (Bao and Cagan 2005) and cloned into pGEM-T-easy (Promega). The verified fragment was released by XbaI from a single clone and inserted into pGEM-WIZ at AvrII and NheI sites sequentially, as shown in Figure 2. After the repeat is assembled in pGEM-WIZ, it can be easily shuttled to common expression vectors such as pUAST or CaSpeR-hs (see below). When the whole inverted repeat cassette is transferred to a Drosophila expression vector, the same fragments that are inserted twice at AvrII and NheI sites in opposite orientations within the cassette will be spliced as two exons arranged in a head–head or tail–tail fashion, leading to a loopless hairpin RNA that serves for RNA interference.
FIGURE 2.
Experimental strategy for cloning inverted repeats using pGEM-WIZ. For clarity, assembling an inverted repeat for the Drosophila gene hibris is given as an example. Please note, to form a functional inverted repeat, the two fragments can be arranged in either a head–head or a tail–tail fashion. w=intron 2 of the white gene.
The use of the white intron in pWIZ has proven to be a significant advance in stabilizing inverted repeat sequences in bacteria. Some inverted repeats that previously had failed to be cloned were able to be assembled in pWIZ (Lee and Carthew 2003). However, we have experienced variable success in cloning inverted repeats using pWIZ. When simultaneously generating 10 different inverted repeats, three were successfully completed, but the remaining seven proved problematic in pWIZ (R.I. Johnson, pers. comm.). As an example, out of 194 clones examined, no single inverted repeat in pWIZ has been obtained when generating an RNAi construct against the Drosophila locus CG2970 using a variety of E. coli strains. This construct together with the remaining six was later successfully generated using the pGEM-WIZ strategy described here. Regularly, two to four out of 10 colonies picked contained an inverted repeat when using pGEM-WIZ (see below). In contrast, among the three RNAi constructs successfully generated in pWIZ, on average 36 clones were tested to find one containing the repeat. Thus, pGEM-WIZ significantly enhances the efficiency of assembling inverted repeats. It is also noteworthy that inverted repeats are stable in pGEM-WIZ in all the common E. coli strains tested so far including TOP10 or DH5α (Invitrogen). Furthermore, compared with pWIZ, pGEM-WIZ has a smaller size (3.1 kb), which can be used to one's advantage to screen inserts (see below). Multiple cloning sites present in pGEM-WIZ provide an easy access to determine the orientation of inserts and shuttle the inverted repeat cassette. The method we describe in this paper, however, requires an extra step in order to shuttle the repeat to an expression vector such as pUAST. The ligation of an inverted repeat to an expression vector is normally very efficient. For example, when an inverted repeat is ligated to pUAST, normally >80% of the clones contained a correct insert (see below). Thus, despite an additional cloning step, the pGEM-WIZ provides a versatile and all-purpose platform for cloning a variety of inverted repeats for RNA interference.
As for pWIZ (Lee and Carthew 2003), the same considerations apply when utilizing pGEM-WIZ: (1) Preferably, exclude fragments that contain XbaI sites. If a fragment with an XbaI site is required, it should not have an additional NheI, SpeI, or AvrII site. (2) The template fragment should not contain either a 5′ or 3′ consensus splice site. (3) Add either an XbaI, NheI, SpeI, or AvrII site to the end of the PCR primers; select a site that is absent from the target fragment. (4) The DNA fragment should be 500–700 bp. Although the fragment can be a PCR product or a verified fragment restriction-isolated from pGEM-T-easy (Promega), the latter showed more consistent results.
Fast screening of inverted repeats
Screening for inserts
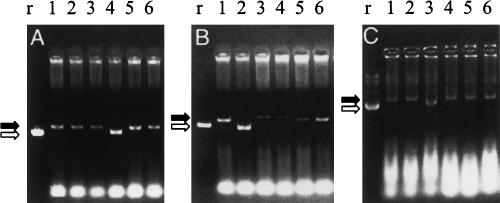
The smaller size of pGEM-WIZ (3.1 kb) is advantageous for selecting clones. A clone with an insert of 500–700 bp in pGEM-WIZ can be unambiguously distinguished from an empty vector based on size alone through examining the mobility of superhelical DNA (Fig. 3A). As an example, after the hibris fragment was inserted into the AvrII site in pGEM-WIZ, supercoiled DNA from the correct clones migrated more slowly than the one from an empty vector (Fig. 3A). Similarly, clones containing a second insert can be distinguished from the ones with a single insert based on the mobility of superhelical DNA. One round of screening, from harvesting cells to results, required ∼50 min.
FIGURE 3.
Rapid selection for clones with an inverted repeat based on size and relative amount of plasmid DNA. Selection of an inverted repeat for the Drosophila gene hibris is shown here as an example. A total of 40 μL of plasmid DNA was isolated from 0.8 mL of a 4 mL overnight culture by a modified STET-lysozyme method (see Materials and Methods). Four microliters of DNA samples were loaded on a 1% agarose gel containing 0.5 μg/mL ethidium bromide. Fill arrows point to the clones with an insert that run at a position corresponding to a higher molecular weight, while open arrows point to the control vectors. (A) A 504-bp fragment verified by sequencing was inserted into pGEM-WIZ at an AvrII site. The clones (1,2,3,5,6) with an insert were unambiguously identified by size compared with the control pGEM-WIZ alone (r). (B) The same fragment was inserted into the NheI site of pGEM-WIZ that contains the first insert. The clones with double inserts were selected by size (1,3,4,5,6). Among them, the ones with significantly weaker intensity of the bands (3,4,5) were identified to contain an inverted repeat. pGEM-WIZ with one insert was used as a control (r). (C) The whole inverted repeat was excised from pGEM-WIZ with XhoI/XbaI and ligated into pUAST. Clones with an inverted repeat were identified by size (1,2,4,5,6). pUAST was used as a control (r). All selected clones have been confirmed by restriction mapping.
Screening for orientations
We observed that the presence of an inverted repeat in a vector negatively affects plasmid replication in E. coli. Without exception, pGEM-WIZ with an inverted repeat propagates in a significantly lower copy number than the same vector with a direct repeat (two inserts in the same orientation). Actually, this can also be used to one's advantage. When using the same culture conditions and same quantity of cells, a clone containing an inverted repeat is easily separated from the ones with a direct repeat by comparing the relative amount of plasmid DNA, as revealed by staining intensity on the gel. As an example, to select clones with an inverted repeat for hibris, miniprep samples were directly loaded onto the gel and analyzed by electrophoresis. Clones with an inverted repeat were easily identified by size and the intensity of the bands (Fig. 3B). Typically, two to four out of 10 colonies picked contained an inverted repeat in pGEM-WIZ. Again, from harvesting cells to identifying the correct clones required ∼50 min. The inverted repeats were then further validated by restriction analysis.
Use of inverted repeats for RNA interference
Inverted repeats assembled in pGEM-WIZ can be excised and shuttled into other vectors to target RNA in vitro or in vivo. Multiple restriction sites present in pGEM-WIZ make it convenient to excise an inverted repeat assembled in this vector. As an example, pAc5.1/V5-HisA and CaSpeR-hs are commonly used vectors for tissue cell culture. To shuttle an inverted repeat into pAc5.1/V5-HisA, the repeat can be excised from pGEM-WIZ by XhoI/XbaI or SpeI. To clone an inverted repeat into CaSpeR-hs, BglII/XbaI, BglII/NotI, or SpeI can serve the purpose. To generate RNAi for transgenic animals, an inverted repeat can be excised from pGEM-WIZ by XhoI/XbaI, BglII/XbaI, BglII/NotI, or SpeI and inserted into pUAST or other suitable vectors (Table 1). As an example, the inverted repeat for hibris (hibris-IR) was excised from pGEM-WIZ using XhoI/XbaI and inserted into XhoI/XbaI-digested pUAST. Five out of six colonies contain an inverted repeat (Fig. 3C). Importantly, the transgene carrying the inverted repeat for hibris made by this method was functional (Bao and Cagan 2005). Transferring an inverted repeat into common expression vectors is summarized in Table 1.
TABLE 1.
RNA interference delivered by some common vectors
In summary, pGEM-WIZ inherits the advantage of using a functional intron as a spacer to enhance the stability of inverted repeat sequences in bacteria but exhibits a much higher efficiency in assembling inverted repeats than pWIZ. The repeats in pGEM-WIZ are stable in all common E. coli strains tested so far. In addition, pGEM-WIZ provides a versatile and all-purpose platform for cloning a variety of inverted repeats for RNA interference. When pGEM-WIZ is coupled with a rapid screening method based on the size and amount of plasmid DNA, the cloning process can be further eased, and screening a large number of clones is practical even when encountering difficult inverted repeats. Although pGEM-WIZ was designed for RNAi in Drosophila, it can be easily modified to assemble inverted repeats for RNAi in other organisms. For example, by replacing the white intron in pGEM-WIZ with an intron of a murine gene flanked by consensus splice sites, the vector can be used to assemble an inverted repeat for RNAi in mice.
MATERIALS AND METHODS
Construction of pGEM-S1
The vector pGEM-S1 derives from pGEM-T (Promega). It was constructed by circularizing pGEM-T using a synthesized fragment comprised of two oligonucleotides: gem-lk1122-f (AGATCTCTCGAGGGTACCTCTAGAGAATTCAAGCTTA) and gem-lk1122-r (AAGCTTGAATTCTCTAGAGGTACCCTCGAGAGATCTA). pGEM-S1 contains additional unique restriction sites: BglII, XhoI, KpnI, XbaI, EcoRI, and HindIII (Fig. 1A). A high copy number and multiple cloning sites improve the utility of pGEM-S1 as a cloning vector. For sequencing, M13 primers were used: M13-forward (CGCCAGGGTTTTCCCAGTCACGAC) and M13-reverse (TCACACAGGAAACAGCTATGAC).
Construction of pGEM-WIZ for assembling inverted repeats
An XhoI/XbaI fragment containing the 74-nt intron 2 of the Drosophila gene white was excised from pWIZ (Lee and Carthew 2003) and inserted into XhoI/XbaI-digested pGEM-S1. The resulting vector, pGEM-WIZ, contains the white intron 2 flanked by ApaI, AatII, SphI, NcoI, SacII, BglII, XhoI, and AvrII on the 5′ side and by NheI, XbaI, EcoRI, HindIII, NotI, PstI, HincII, SalI, and NdeI on the 3′ side (Fig. 1B). The sequences of both pGEM-S1 and pGEM-WIZ have been deposited in GenBank (accession numbers: DQ917670 and DQ917671).
Selection of clones by electrophoresis
To select clones, plasmid DNA from a small fraction of overnight cultures was extracted by miniprep using a modified STET-lysozyme method (Holmes and Quigley 1981). In brief, 0.8 mL of a 4 mL overnight culture inoculated using a single colony was pelleted, resuspended in 38 μL of STET solution (8% sucrose, 50 mM Tris at pH 8.0, 50 mM EDTA, and 5% Triton X-100), and further lysed with the addition of 2 μL of lysozyme stock solution (20 mg/mL in water). The lysate was mixed by inversion, boiled for 3 min, and centrifuged to remove proteins and cell debris. DNA was recovered in the supernatant.
DNA samples from miniprep were directly analyzed on a 1% agarose gel containing 0.5 μg/mL ethidium bromide by standard DNA electrophoresis. When the samples were run on a minigel (8.5 × 6.0 cm) at 90 V in the GIBCO BRL horizontal gel electrophoresis apparatus Horizon 85 (Life Technologies, Inc.), 35 min was sufficient to resolve the size difference between clones with an insert and the ones without by examining the mobility of superhelical DNA.
ACKNOWLEDGMENTS
We are grateful to Ruth Johnson for the statistics on pWIZ and helpful discussions. We thank Julia Cordero, Marcos Vidal, and Chunlai Wu for discussions and invaluable suggestions. We also thank Catherine Collins, Midori Seppa, and Stephen Warner for comments on the manuscript, and David Larson for help in preparing the figures. This work is supported by the National Institute of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.258406.
REFERENCES
- Bao, S., Cagan, R. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev. Cell. 2005;8:925–935. doi: 10.1016/j.devcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fortier, E., Belote, J.M. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila . Genesis. 2000;26:240–244. doi: 10.1002/(sici)1526-968x(200004)26:4<240::aid-gene40>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Holmes, D.S., Quigley, M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R., Carthew, R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R., Carthew, R.W. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Lam, G., Thummel, C.S. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila . Curr. Biol. 2000;10:957–963. doi: 10.1016/s0960-9822(00)00631-x. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Carthew, R.W. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Martinek, S., Young, M.W. Specific genetic interference with behavioral rhythms in Drosophila by expression of inverted repeats. Genetics. 2000;156:1717–1725. doi: 10.1093/genetics/156.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta, L., Paterson, B.M. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): A role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]