Abstract
Specific RNA recognition of proteins containing the double-strand RNA-binding domain (dsRBD) is essential for several biological pathways such as ADAR-mediated adenosine deamination, localization of RNAs by Staufen, or RNA cleavage by RNAse III. Structural analysis has demonstrated the lack of base-specific interactions of dsRBDs with either a perfect RNA duplex or an RNA hairpin. We therefore asked whether in vitro selections performed in parallel with individual dsRBDs could yield RNAs that are specifically recognized by the dsRBD on which they were selected . To this end, SELEX experiments were performed using either the second dsRBD of the RNA-editing enzyme ADAR1 or the second dsRBD of Xlrbpa, a homolog of TRBP that is involved in RISC formation. Several RNA families with high binding capacities for dsRBDs were isolated from either SELEX experiment, but no discrimination of these RNAs by different dsRBDs could be detected. The selected RNAs are highly structured, and binding regions map to two neighboring stem–loops that presumably form stacked helices and are interrupted by mismatches and bulges. Despite the lack of selective binding of SELEX RNAs to individual dsRBDS, selected RNAs can efficiently interfere with RNA editing in vivo.
Keywords: SELEX, dsRBD, RNA editing, ADAR1, Xlrbpa
INTRODUCTION
The double-stranded RNA-binding domain (dsRBD) is a motif common to a variety of RNA-binding proteins with diverse functions found in organisms ranging from Escherichia coli to man (St Johnston et al. 1992). The functions of dsRBD-bearing proteins range from RNA transport to RNA processing to RNA chaperonin activity (Hitti et al. 1998).
The dsRBD is ∼70 amino acids in length and has a typical secondary structure of two α-helices intervened by three β-sheets, which have an anti-parallel orientation with the α-helices being packed on one surface. Important amino acids for the interaction with the A-form helix of a double-stranded RNA were revealed in both biochemical and spectroscopical studies (Bycroft et al. 1995; Kharrat et al. 1995; Ryter and Schultz 1998; Ramos et al. 2000; Wu et al. 2004). Analysis of dsRBDs complexed with dsRNA indicate the absence of direct contacts between amino acid side chains and characteristic groups of bases, which would allow sequence-specific recognition (Ryter and Schultz 1998; Ramos et al. 2000). The major structural motif of the RNA that is recognized by dsRBDs seems to be its double strandedness, together with a slight distortion and bending that leads to an opening of the comparatively narrow major groove and a deviation of the number of bases per helical turn. The dsRBD contacts two neighboring minor grooves and spans across a central major groove, in this way covering 16 base pairs (bp). Most contacts between the dsRBD and the dsRNA are made with the sugar backbone, and by these means the exact width of the A-form RNA helix is measured, giving an explanation for the exclusive recognition of the RNA double helix. Interestingly, in the case of dsRBD #3 of Staufen and the dsRBD of Rnt1p, the first α-helix interacts specifically with bases of a terminal loop in a stem–loop substrate and seems to recognize the structural feature of tetra- or pentaloops (Ramos et al. 2000; Wu et al. 2004).
In general, two binding modes reflecting the function of the protein can be distinguished. While some dsRBD proteins are able to target their substrates specifically, others bind rather promiscuously. Adenosine deaminases that act on RNA (ADARs) can be used to describe this ambiguous behavior. ADARs convert adenosines to inosines by hydrolytic deamination (Bass 2002). There is a limited but increasing number of RNA substrates that are modified at individual sites in a very specific manner (Burns et al. 1997; Higuchi et al. 2000; Levanon et al. 2005). Editing in these substrates frequently leads to recoding of the original genomic sequence and as a consequence can lead to changes in the amino acid composition and properties of the resulting proteins. In contrast, so-called hyperedited substrates are deaminated to a high extent, but unspecifically. These targets form extended RNA duplexes with few mismatches or bulges and are frequently found in viral RNAs. Hyperedited substrates are deaminated until up to 50% of all adenosines are targeted, leading to partial unwinding of the dsRNA (Wong et al. 1989; Murphy et al. 1991; Kumar and Carmichael 1997). Thus ADARs seemingly can interact both selectively and nonselectively with their substrate RNAs.
Specific target recognition was studied in two different dsRBD-containing proteins, Rnt1p and ADAR2. Rnt1p is an RNase III-type endonuclease that contains a dsRBD at its amino terminus and functions as a key component of Saccharomyces cerevisiae rRNA-processing machinery. It processes RNA stem–loops in a sequence-independent manner but requires a characteristic tetra-loop with the consensus AGNN to position itself and cleaves 14–16 bp away from the loop (Chanfreau et al. 2000; Chanfreau 2003). The in-solution structure of Rnt1p with a 14 bp hairpin capped with an AGAA tetra-loop (snR47 precursor) shows that the first α-helix of the dsRBD rests in the fold of the tetra-loop and the adjacent base pairs, which together form a minor groove-binding pocket. The dsRBD specifically recognizes its target via this unique structure (Wu et al. 2004).
Human ADAR2, like its homologs in other vertebrates, contains two dsRBDs and a C-terminally located deaminase domain. The dsRBDs of ADAR2 are able to bind selectively close to the edited site on the Q/R site of GluR-B pre-mRNA. In contrast, PKR, which also contains two dsRBDs, shows a different binding pattern to this editing site. It was therefore proposed that different proteins obtain an intrinsic binding selectivity due to their individual set of dsRBDs (Stephens et al. 2004).
However, the role of dsRBDs in specific target recognition is still a matter of controversy. Again, ADAR provides a set of contradictive evidence to this discussion. If, for instance, the deaminase domains of ADAR1 and ADAR2 are exchanged, the resulting chimeric enzymes predominantly retain the substrate specificity determined by the deaminase domain (Wong et al. 2001). On the other hand, the exchange of dsRBDs of ADAR1 with the ones from PKR leads to a dramatic reduction in substrate recognition of the chimeric protein (Liu et al. 2000). Similarly, in Xenopus laevis oocytes it could be shown that Xenopus ADAR1 localizes to transcriptionally active lampbrush chromosomes and binds to the majority of nascent transcripts. Deleting, duplicating, or triplicating some dsRBDs of this protein leads to a clear change in ADAR1 distribution, causing a specific enrichment of the altered protein at a subset of transcripts (Doyle and Jantsch 2003), thus suggesting a crucial role of dsRBDs in substrate recognition.
There is a growing number of biological pathways in which dsRBD-containing proteins are involved, ranging from RNA editing to RNA interference to RNA localization. We therefore wanted to determine the recognition motifs of individual dsRBDs. To compare binding properties of different dsRBDs we performed SELEX experiments with the second dsRBDs of xlADAR1 and Xlrbpa, respectively. Previously, SELEX experiments had been performed on neighboring dsRBDs of PKR. However, these experiments were restricted to a single protein and were performed under different conditions (Bevilacqua et al. 1998; Zheng and Bevilacqua 2004). Our data indicate that short, stacked helices are the preferred binding substrate for dsRBDs. Moreover, RNAs obtained from SELEX with different dsRBDs can be bound promiscuously by other dsRBDs. Finally, we can show that the SELEX RNAs can interfere with an in vivo function of a dsRBD-containing protein, ADAR-mediated editing.
RESULTS
Selection for RNA aptamers that bind to dsRBD 2 of xlADAR1 and Xlrbpa
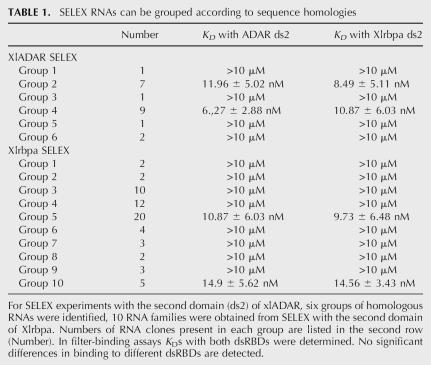
To identify RNA aptamers with binding specificity for a particular dsRBD we performed two in vitro RNA selection experiments. On the one hand, the second dsRBD of Xlrbpa, a homolog of human TRBP2 [TAR (HIV) RNA binding protein 2] was chosen (Eckmann and Jantsch 1997). Xlrbpa contains three dsRBDs and associates in an hnRNP-like manner with the majority of newly synthesized RNAs. Moreover, Xlrbpa exhibits RNA chaperonin activity (Hitti et al. 1998). The second dsRBD chosen for these experiments has the strongest RNA-binding activity in vitro and displays no apparent binding preference (Brooks et al. 1998). On the other hand, the second dsRBD of Xenopus (xl) ADAR1, which also showed the strongest in vitro RNA-binding activity in this protein, was chosen. Since ADARs are capable of selectively deaminating particular sites, it was hoped that this domain would contribute to sequence-specific binding. His-tagged versions of the proteins were purified from recombinant E. coli and used in SELEX experiments. The randomized RNA library was a gift of M. Famulok (Klug and Famulok 1994) and contained a 74-base-long randomized region, which is flanked by the primer binding sites, one with the T7-promoter. The library had an initial complexity of 1015 different molecules. Increase in binding affinity over the number of SELEX cycles was monitored by measuring the ratio between bound and unbound fraction of radiolabeled RNA (not shown). After the 10th cycle no further increase in affinity could be detected and therefore the cDNA of the 10th to 12th cycle were used for cloning into pGEM-T easy (Promega) and subsequent sequencing. The resulting sequences were aligned according to sequence homologies. For the xlADAR SELEX experiment six different groups of clones were obtained and for Xlrbpa SELEX 10 groups of clones wer obtained (Table 1). Clones obtained from the Xlrbpa SELEX in the 11th cycle are labeled as “kl” while clones from the 12th cycle are referred to as “Xl” clones. Similarly, clones from the ADAR SELEX are referred to as “Dr” clones with the number before the clone indicating the SELEX cycle from which they were isolated. Within each group high sequence homology was observed, with only a few, individual base differences observed among the members of a group. None of the single base exchanges led to a change of the predicted folding pattern of the RNAs. The lack of sequence homologies between groups suggested that no common primary nucleotide sequence but rather secondary structures might be the recognized feature of the RNAs.
TABLE 1.
SELEX RNAs can be grouped according to sequence homologies
Nonetheless, prior to further analysis and to limit the number of clone groups under investigation, we measured the strength of binding of all clone groups to dsRBDs. To do so, two clones of each group were picked that showed the least similarity to each other. Due to the high degree of homogeneity within each group of clones even the most divergent clones only differed in a few nucleotide positions. Using filter-binding assays the K Ds of the RNAs when bound to either protein domain were determined. The dissociation constant for the best binders ranged from 8 to 15 nM. Weak binders (K Ds > 10 μM) were not used for further investigations (Table 1). Two groups of RNA clones from each SELEX experiment displaying the lowest K Ds were used for further experiments (xlADAR SELEX: groups 2 and 4; Xlrbpa SELEX: groups 5 and 10). The four groups contained 42 of the 93 initially selected RNA clones. In all cases, binding of the RNA to either of the two protein domains was in a comparable range (Table 1; Fig. 2B, see below). Therefore, no obvious discrimination of RNAs derived from the two different SELEX experiments by different dsRBDs could be detected. Thus, secondary-structure predictions of RNAs representing groups that showed a lower K D were performed. While no primary sequence homology could be detected between clones of different groups, a structurally related folding pattern was predicted for clones representing each of the four groups. Clones of all groups were seemingly composed of three stems arranged around a central junction, designated as stem–loops I–III. Stem–loops I and II are short and contain few mismatches and bulges, whereas stem III is longer, containing several mismatches and bulges (Fig. 1). Apart from the tripartite stem–loop structure, no other structural motif could be identified as a potential recognition site for dsRBDs.
FIGURE 2.
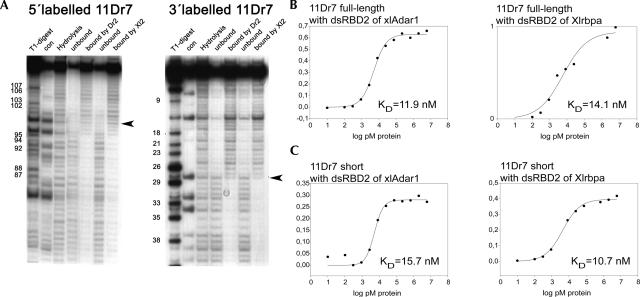
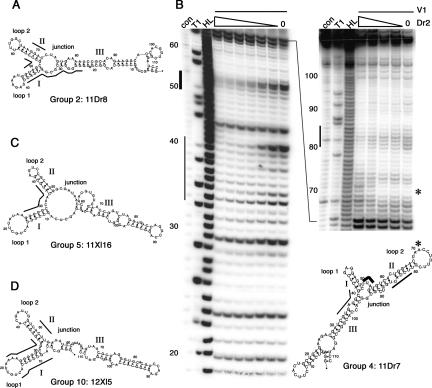
Boundary experiments reveal minimal binding sites, and shortened RNA retains binding affinity. (A) 5′ and 3′ labeled RNA from clone 11Dr7 was partially hydrolyzed. Fractions bound and unbound to the dsRBD2 of xlADAR (Dr2) or Xlrbpa (Xl2) were loaded side by side on denaturing polyacrylamide gels. (Hydrolysis) represents partially hydrolyzed RNA ladders. Denaturing (T1 digests) give the positions of G residues, (con) lane shows untreated RNA samples. The regions necessary for binding are depicted in Figure 1 as boxed nucleotides in the alignment. (B) Curves of representative filter-binding experiments of clone 11Dr7 of the group 4 of the XlADAR SELEX performed with dsRBD2 of ADAR (left) and dsRBD2 of Xlrbpa (right). Affinities to both domains are comparable. (C) Curve of representative filter-binding experiments of shortened clone11Dr7. Also the minimal RNA sequence of this clone retains binding affinity to both domains.
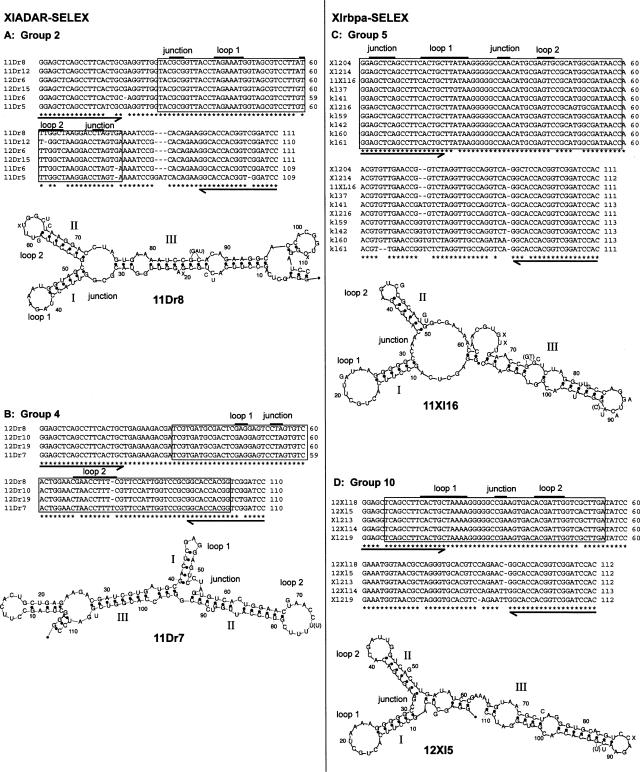
FIGURE 1.
Sequence alignments and folding predictions for best-binding RNA groups. From each SELEX experiment, two groups with the highest binding affinities were picked and used to determine binding sites. Groups 2(A) and 4 (B) were selected with ds2 of XlADAR, groups 5 (C) and 10 (D) with ds2 of Xlrbpa. Clones selected on ADAR have a Dr in their name while clones selected on Xlrbpa are designated as Xl or kl clones. Boxed regions correspond to minimal binding sites determined in boundary experiments. Arrows underneath alignments represent fixed primer sequences used to amplify the randomized cDNA pool. In groups 5 and 10 selected on ds2 of Xlrbpa the 5′ primer sequence participates in the formation of stem I in the binding site. Mismatches in the alignment are indicated. Mfold was used for folding predictions for representative clones. Roman numbers I, II, and III represent numbering of stem structures; loops and bulges are depicted in the figures and are also indicated above the alignments. Mismatches occurring in the alignments are also shown in the folding prediction next to the affected positions; deletions are indicated by an X and insertions are shown in brackets.
Binding sites on SELEX RNAs are not discriminated by different dsRBDs
A stem–loop structure containing 12 bp of dsRNA or 16 bp of a perfect RNA duplex can satisfy the binding requirements of a dsRBD (Ryter and Schultz 1998; Ramos et al. 2000). Folding predictions of RNAs originating from the four tightly binding groups of clones suggested the presence of more than one binding site of this length in these RNAs. Therefore, the minimal sequence required for binding by the dsRBD was determined in boundary experiments. Partially hydrolyzed RNAs labeled at either their 5′ or 3′ ends were incubated with one of the two dsRBDs. RNAs bound to the dsRBD were recovered by filtration through nitrocellulose while unbound RNAs were recovered from the flow-through. Bound and unbound RNA fractions were loaded alongside on denaturing polyacrylamide gels. These experiments showed that the minimal sequences necessary for dsRBD binding are identical for both dsRBDs used in these experiments (Fig. 2A). The selected RNAs not only bound with similar K Ds to the two different dsRBDs used in these experiments but also to the same regions. For RNAs derived from either group 2 or group 4, both selected on the xlADAR dsRBD, the minimal binding regions were located in the center of the RNA while flanking regions seemed dispensable for binding (Fig. 1, boxed region). In contrast, RNAs derived from either group 5 or group 10 that were selected with the Xlrbpa dsRBD all required their 5′-most region containing the fixed primer region for efficient binding. Mfold preditions indicate that the minimal RNA folds into a stem–loop that contains the primer sequence as one strand of the duplex (Fig. 1, boxed region; Mathews et al. 1999; Zuker 2003). This phenomenon can be explained by statistical means. The fixed primer region reduces the number of possible sequence variations necessary to generate double-stranded regions. Therefore, the SELEX starting pool contains more molecules capable of forming a double-stranded region including the fixed primer sequence. This overrepresentation can lead to an evolutionary advantage during the selection procedure.
Following the data of the boundary experiments, shortened clones expressing the minimal RNAs predicted to be essential for dsRBD binding were constructed, representing each of the four clone groups. Folding predictions suggested identical structures of this region in full-length and minimal clones; both full-length and shortened RNAs show conservation of two neighboring stem–loops. Using filter-binding assays we compared the K Ds of minimal and full-length RNAs. Surprisingly, only one group showed a comparable 10 nM binding constant for full-length and minimal constructs (Fig. 2C). In the case of the other groups, shortening of the RNA was detrimental to binding. Seemingly, stem III, although dispensable for binding, is necessary for folding and stability of the central loop of these structures. Along these lines it is also noteworthy that in all K D-measuring experiments also for full-length clones, the fraction of bound RNA never approached 100%. Instead, maximum binding ranged between 30% and 90%. A likely explanation for this finding could be an unfolded or, alternatively, another stable folding of the RNA, which is not recognized by the dsRBDs.
In silico foldings predict the energetically most favorable folding patterns for the clones investigated with a DG ranging from −37 to −40 kcal/mol. Foldings of the minimal regions determined by boundary experiments start to change at a DDG of 2.4–2.5 kcal/mol. Therefore, it is possible that these alternative structures account for the fraction of unbound RNA.
Chemical probing verifies folding prediction
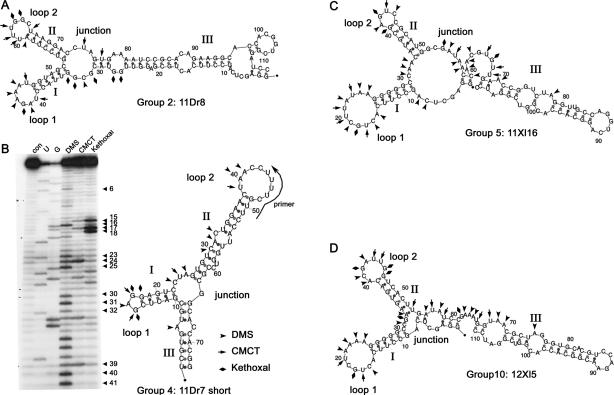
Although secondary-structure predictions are becoming increasingly accurate, they still need to be verified by chemical probing. We therefore performed structure mappings with the remaining four best-binding groups from both SELEX experiments. Three to five independent experiments were performed to determine sites that are consistently accessible to the modifying agents. These experiments clearly supported the energetically most favorable folding predictions of Mfold. Alternative, energetically less favorable foldings were also considered but did not fit our experimental data. RNA 11Dr8 of xlADAR group 2 contains two neighboring stem–loops. The shorter stem I contains 6 bp with a hepta-loop; stem II contains 7 bp capped with a loop of 6 bases. The dispensable third stem contains the two primer sequences (Fig. 3A). The second group represented by clone 11Dr7 short of clone group 4 is very similar in its conformation. Here stem I consists of 5 bp capped by a tri-loop, the longer stem II consists of 12 bp, is interrupted by a mismatch and a bulge, and is capped by a loop of 10 bases. The boundary experiment (see Fig. 2) showed that not only stem–loops I and II but also additional base pairs of stem III are required for binding (cf. Figs. 1B and 3B). In the case of clone 11Dr7 from this group the minimal RNA also did fold stably, showed the same K D as the full-length RNA, and could therefore be confirmed by chemical probing (Fig. 3B).
FIGURE 3.
Chemical structure mappings verify folding prediction RNA. (A) 11Dr8, (B) 11Dr7 short, (C) 11Xl16, and (D) 12Xl5 were subjected to chemical modifications with DMS, CMCT, and kethoxal, and were transcribed reversely together with two sequencing reactions. Three to five experiments were done independently. Sites that were consistently accessible to modification and thus represent unpaired regions were marked on the secondary-structure prediction plot by different symbols, as indicated. A representative gel of the mapping of RNA 11Dr7 short is given in B. The primer used for RT reaches into loop 2. The folding predictions for the full-length sequence of 11Dr7 (not shown) could be verified independently.
The clones of the groups selected with the second dsRBD of Xlrbpa are similar in structure to the ones selected on dsRBD2 of ADAR1. Clones of both group 5, represented by clone 11Xl16, and group 10, represented by clone 12Xl5, contain two neighboring stem–loops resembling those of the clones selected on the dsRBD of ADAR1. However, in the clones originating from the Xlrbpa SELEX, stem I is partially composed of the primer region, which reaches into the 10- or 11-nt loops. In both groups of clones the primer region is base paired with a track of five Gs, which form two non-Watson–Crick base pairs with U. The loop region differs significantly with the exception of the nucleotides originating from the fixed primer region. Stem II is seven (group 10), or, in the case of group 5, six nucleotides long and capped by a tetra-loop (group 5) or in case of group 10 by a hepta-loop. The two stems are separated by a junction, which is followed by stem III with several bulges (Fig. 3C,D). This stem III was not essential for binding in boundary experiments. However, deletion of the stem leads to a dramatic reduction in binding. Mfold analysis predicts identical foldings of the minimal region for full-length and shortened constructs. Nonetheless, alternative foldings that differ from the energetically most favorable one by <1 kcal/mol are also predicted for the shortened clones. It therefore seems likely that stem III is required for a stable folding of the RNA.
The two neighboring RNA stem–loops serve as a binding platform for dsRBDs
None of the individual stem–loops of our SELEX clones could satisfy the published minimal binding length for dsRBDs of at least 13, if not 16 bp (Ryter and Schultz 1998; Ramos et al. 2000). Therefore, to investigate whether two neighboring stem–loops could form stacked helices we tested whether adjacent stem–loops are protected by a dsRBD using a nuclease protection assay. 5′ labeled RNA was incubated with purified dsRBD and treated with the double-strand-specific ribonuclease V1. RNase V1 recognizes any 4–6 nucleotide (nt) segments of backbone with an approximately helical conformation. Therefore the RNA does not have to be entirely double stranded but has to follow the helical turn of an A-form helix (Lowman and Draper 1986). For clones of group 2 of the xlADAR SELEX, bases of both stems I and II are protected. The region, which shows protection from V1 digest, lies between the two terminal loops and covers 12 bases (11Dr8; Fig. 4A). In the case of group 4 of the xlADAR SELEX, V1 footprinting could confirm contacts to stem III, confirming the results of boundary experiments (11Dr7; Fig. 4B). Additionally, protection of stem II and parts of the very short stem I could be observed. It seems likely that protection of stem I is not due to direct dsRBD binding but rather it is protected by the bulky dsRBD, binding to the adjacent stem II. Also, this stem of 4 bp is rather short to be properly recognized and cleaved by nuclease V1. Stems II and III potentially form a stacked helix that is interrupted by stem I, which makes a distortion in the helical structure. Here we find a protection of up to 19 nt.
FIGURE 4.
Protection from RNase V1 digestion by dsRBD on neighboring stem–loops. Addition of dsRBD protein prevents the double-stranded RNA-specific ribonuclease from cleavage on SELEX clones. (A,C,D) show the protection of RNA clones 11Dr8, 11Xl16, and 12Xl5, each representing SELEX groups 2, 5, and 10, respectively. The dsRBD added correspond to the ones these RNAs were selected on. Protected regions are marked by bars alongside the predicted folding pattern. (B) Protection of 11Dr7, a representative of SELEX group 4, by increasing amounts of ds2 of XlADAR (100; 33.3; 11.1; 3.7; 1.2 ng/μL final concentrations for the short run gel on the left-hand side; 100; 20; 4; 0.8 ng/μL final concentrations for the long-run gel on the right). Different regions that are protected are shown in differently weighted lines along the stems of the RNA. Upon addition of increasing amounts of dsRBD protein, loop 2 seems to adopt a helical structure as RNase cleavage can occur in the loop. These binding-dependent cleavage sites are marked by an asterisk next to the gel and in the folding prediction.
The protection pattern for the two groups of the Xlrbpa SELEX is very similar to group 2 of the xlADAR SELEX. Again, the two neighboring stem–loops form a contiguous helix that is interrupted by a junction in the middle, which does not seem to influence the helical structure severely and provides the binding platform for the dsRBD (Fig. 4C,D). In all experiments we find clear protection on the predicted stems but cannot distinguish the helical surfaces to which dsRBDs bind. Interestingly, regions appearing unpaired without dsRBDs become more helical with increasing amounts of dsRBD protein. It seems that the dsRBD forces bases at the edge of loops and bulges into a helical structure, making it a target for V1 recognition (asterisk in Fig. 4A). Most interestingly, these experiments indicate that the dsRBD can interact with two adjacent helices that are separated by an unstructured region even when each of the individual helices is too short to allow a dsRBD to bind.
Interestingly, the general folding pattern of three helical structures fused by a large multiway junction was also predicted for all other groups of clones binding the dsRBD less tightly. The maximum number of unpaired nucleotides fusing two adjacent helices was found in 11Xl16, where three unpaired nucleotides link helixes I and II, suggesting that larger unpaired regions might disrupt binding of the dsRBD to two adjacent helices.
SELEX RNA can interfere with in vivo editing activity of X. laevis oocytes
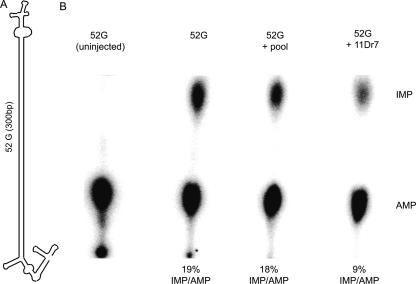
DsRNAs injected into oocytes are subjected to a high extent of editing due to the activity of xlADAR (Bass and Weintraub 1988). We asked whether coinjection of one of our selected RNAs with a known edited target could reduce the extent of editing by blocking or sequestering endogenous ADAR1. As a substrate we used a known hyperedited RNA named 52G (F55A4.9), isolated in a screen for new ADAR targets in C. elegans (Morse et al. 2002). The 810-nt-long 52G-RNA consists of a nearly perfect RNA duplex of 300 bp and is flanked by two structured ends. This helical structure provides several potential ADAR1 binding sites, and up to 74 adenosines that can be edited (B. Bass, pers. comm.). Oocyte nuclei were injected with radioactively labeled RNA either alone, together with our unlabeled SELEX RNA 11Dr7, or with the initial, randomized SELEX pool in an equimolar ratio. The SELEX RNA on its own was also tested whether it was a target for editing. However, no evidence for editing could be detected for the SELEX RNA on thin layer chromatography (data not shown). By adding equimolar amounts of SELEX RNAs (clone 11Dr7) a clear reduction of 52G editing could be observed (Fig. 5). The extent of reduction in editing varied from 30% to 50%, depending on the experiment. The initial randomized SELEX pool, in contrast, had no effect on 52G editing. Nuclease protection assays indicate that 11Dr7 RNA provides a single binding site for a dsRBD (see above). In contrast, the base-paired region of the 52G RNA offers several binding sites. Therefore, equimolar injection of 52G with SELEX RNA provides a great excess of binding sites on the 52G RNA. It therefore appears likely that the SELEX RNA shows a higher affinity to ADAR in vivo and is therefore able to reduce editing activity significantly.
FIGURE 5.
SELEX RNA interferes with editing in vivo. (A) Schematic picture of 52G RNA, a noncoding RNA containing numerous ADAR editing sites that was identified in C. elegans (Morse and Bass 1999). This abundantly expressed 800-nt RNA folds into an intramolecular duplex of ∼300 bp, flanked by terminal structures. (B) Radioactively labeled 52G RNA was injected either in the presence or absence of SELEX RNA 11Dr7 or the initial randomized SELEX pool into X. laevis oocyte nuclei. Injected RNAs were isolated and submitted to P1 digestion and nucleotides were separated on TLC plates. The ratio of inosines is significantly reduced upon coinjection of equimolar amounts of SELEX RNA 11Dr7. On average, editing levels are reduced 30%–50% upon coinjection of 11Dr7.
DISCUSSION
Mimicking coevolution of RNAs with dsRBDs by applying SELEX
DsRBDs are involved in the recognition of double-stranded RNAs in a wide range of biological processes. In some cases, dsRBD-containing proteins are able to mediate specific RNA targeting while in other cases dsRBDs interact with RNAs rather promiscuously. Rnt1p, for instance, is involved in processing of rRNAs, snRNAs, and snoRNAs, by targeting stem–loops of 12–15 bp preferentially capped with an AGNN tetra-loop. The in-solution structure of the dsRBD of Rnt1p with a stem–loop of its substrate snR47 precursor shows that the tetra-loop and the first helical turn of the RNA fold into a specific conformation that provides a binding pocket. The first α-helix of Rnt1p fits exactly into this pocket. The first α-helix of the dsRBD folds to the dsRBD core in a slightly different angle than other published dsRBDs, which suggests that dsRBDs probably coevolved with their targets in a structure-specific manner (Wu et al. 2004). We tried to determine if we could imitate this evolution by applying a SELEX with two different dsRBDs. The second dsRBDs of either xlADAR1 or Xlrbpa, two proteins of different RNA-binding specificities, were chosen. These dsRBDs proved to be the strongest binders in the respective proteins as judged by Northwestern assays, and thus seemed the ideal candidates for initial target recognition (Brooks et al. 1998). Interestingly, our results indicate that binding sites on the RNA clones obtained by SELEX show structural similarities, but share no common motif or sequence homology. Two neighboring stem–loops, which are interrupted by bulges, are capped by differently sized loops and provide the binding platform for the dsRBD. The best-binding RNAs are bound by both dsRBDs with a comparable strength in the 8–15 nM range.
We have also analyzed binding of the dsRBDs to a perfect dsRNA oligonucleotide 25 bases in length. This oligo bound either dsRBD with a K D of 2–10 nM (data not shown). Furthermore, this dsRNA efficiently competed with the binding of SELEX RNAs. It may therefore seem surprising that no perfect double-stranded RNAs were isolated during the SELEX procedure. However, perfect dsRNAs are rather stable and might therefore become underrepresented during reverse transcription. The RNAs isolated by us, in contrast, consist of multiple, short helical regions that are interrupted by bulges. The structure of these RNAs will therefore be more efficiently converted to cDNA during reverse transcription.
SELEX RNAs contain two neighboring stems for binding
A comparison of RNA-folding structures obtained in our experiments with those from previous experiments on other dsRBD-containing proteins reveals surprisingly little overlap. Bevilacqua et al. (1998) used the two dsRBDs of PKR, a protein kinase that can be activated by dsRNAs for a similar SELEX experiment. The binding site on selected RNAs mapped to a single stem–loop of 14 bp with a characteristic AG mismatch. These selected RNAs not only bound tightly, but were also able to activate PKR.
A comparison of the structures assumed by the selected RNAs obtained in our experiments to known dsRBD targets shows some similarities: Three out of the four best-binding RNA families show two neighboring stem–loops, which are interrupted by a junction. The two stacked stems would provide a binding site of 12–13 bp, which is in agreement with the published stem–loop structures found in RNAs that bind to the dsRBDs of Staufen and Rnt1p. Ramos et al. (2000) showed that a stem–loop longer than 12 bp is ideal for binding a dsRBD in Staufen. Similarly, for the dsRBD of Rnt1p it could be shown that 12–15 bp are required for binding and recognition (Wu et al. 2004). Currently, we have no direct evidence that the loops capping the stems are involved in the interaction with the dsRBD. Nonetheless, since the double-stranded regions on the RNA are shorter than 16 bp, a participation of the loops in the interaction with dsRBDs seems likely.
Importance of mismatches for recognition
Clones of group 4 of the xlADAR SELEX form a slightly different structure. Here the structure that is protected by a dsRBD from V1 digestion spans up to 20 bp (Fig. 4A). The protected region in 11Dr7 harbors two mismatched regions and two bulges. Interestingly, the two mismatches contain cytosine residues, somewhat resembling the R/G site of the pre-mRNA of a B-subunit of the glutamate receptor (Lomeli et al. 1994). It is thus possible that the second dsRBD of ADAR1 has the ambiguous ability to either recognize stem–loops in a Staufen-like fashion or, alternatively, to bind duplex RNA-containing mismatches, reflecting the binding of Xlrbpa to two stacked helices in the crystal structure (Ryter and Schultz 1998; Ramos et al. 2000). Evidence supporting this assumption comes from two studies determining the interaction of ADAR2 with its substrate. The R/G site of GluR-B is located in a stem–loop structure of 70 nt that is interrupted by three mismatches. One of these mismatches (A–C) contains the adenosine that can be deaminated by either ADAR1 or 2. Stefl et al. (2005) used NMR-chemical shift perturbation studies to investigate the interaction of the two dsRBDs of rat ADAR2 with the R/G stem–loop. They find selectivity for either of the two dsRBDs of ADAR2 for defined locations on the RNA. DsRBD1 interacts with the stem–loop structure comparable with the Staufen dsRBD. DsRBD2, in contrast, occupies the site opposite of the edited A, making specific contacts with the two Cs in the mismatches. Using shortened fragments of the RNA containing the mismatch region without the loop, they show that both dsRBDs are able to interact with the shortened fragment. Conversely, the loop region can only be bound by dsRBD1 (Stefl and Allain 2005; Stefl et al. 2005). A similar observation was obtained by Stephens et al. (2004) with the Q/R site in the same pre-mRNA. The Q/R site is located in a duplex RNA of 30 bp with two mismatches, where the deaminated A is 5′ of these mismatches. For both dsRBDs, binding sites could be mapped to different regions of mismatches.
Sequence variation does not interfere with folding of the RNAs
Analyzing the location of sequence variation occurring in each of our SELEX groups, we can show that these do not interfere with folding. In most cases the variations occur in loops or lead to a wobble base pair instead of a Watson–Crick base pair. Other mutations create mismatches in stems due to a rearrangement of the base-pairing nucleotides, but leaving the overall stem–loop structure intact. Taken together, the occurrence of sequence variation supports the folding predictions but also argues for a discrimination of specific structures rather than primary sequence by dsRBDs.
A junction and large loops form the SELEX RNAs
We can only speculate on the influence of the junctions separating the neighboring RNA stems. Bulges are generally able to introduce bends into A-form helices. Zacharias and Hagerman (1995) show that an increasing number of adenosines (1–6) cause a stable bending of the helix from 15° to 90°. However, it is not clear whether the junctions that join the helices in the SELEX RNAs behave as bulges. In our experiments junctions of different sizes can be found in the selected RNAs. It would therefore be interesting to test if they introduce a specific angle or give flexibility to the structure that is necessary for dsRBD binding.
The structures of large unpaired regions in the junctions and central bulges might not be as open as depicted in the folding predictions. Our structure mappings clearly showed that the involved nucleotides are accessible for chemical modification. However, some base interactions might form since loops and bulges of eight or more unpaired nucleotides are very rare (for review, see Burkard et al. 1999).
SELEX RNAs interfere with editing
The ability of one of the SELEX RNAs to interfere with in vivo editing reactions clearly demonstrates that the RNAs isolated in vitro can bind dsRBDs with high affinity also in vivo. Xenopus oocytes contain a high level of adenosine deaminase activity (Bass et al. 1994). As our SELEX RNA proved not to be edited, its binding to one or even to all dsRBDs of ADAR1 seemingly inhibits target recognition dramatically. Our SELEX RNA just provides one binding site for one dsRBD in contrast to several binding sites on the 300-bp-long stem of 52G. This, in turn, suggests a higher affinity of 11Dr7 to dsRBDs than the in vivo substrate 52G.
Binding specificity of dsRBD-containing proteins
In our SELEX experiments we chose those dsRBDs that have the strongest binding to dsRNA in vitro. We rationalized that initial RNA recognition would presumably occur between the most avid RNA-binding dsRBD and the substrate and that this initial contact might lead to a discrimination of different targets. To further substantiate this point we used a dsRBD of an enzyme capable of specific target recognition on the one hand and an unspecific binder on the other hand. Yet, we find no binding specificity for different SELEX groups. This suggests that the initial contacts with RNAs are tight but unspecific.
As most dsRBD proteins contain a set of motifs, combinations of dsRBDs may contribute to substrate specificity by arranging the protein on specific tertiary structures of the RNA target.
MATERIALS AND METHODS
Expression and purification of the second dsRBD from xlAdar1 and Xlrbpa
The second dsRBD of xlADAR1 and Xlrbpa, respectively, was cloned into pRSET-C, which adds a 6xHis-tag N-terminally to the dsRBD sequence (Brooks et al. 1998). Plasmids were transformed into E. coli BL21(DE3). Fifty milliliter cultures were inoculated with single colonies, and protein expression was induced by addition of 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) for 4 h. After cell lysis by sonication, fusion proteins were purified over Ni2+-NTA agarose (Quiagen) according to the manufacturer's protocols. Fractions containing protein were pooled and dialyzed against 0.05% trifluoro acetic acid in ddH2O overnight at 4°C. The dialyzed proteins were lyophilized for storage.
Selection of RNAs binding dsRNA-binding domains
A randomized DNA pool (estimated 1015 different molecules) was provided by M. Famulok from the Ludwig-Maximilians-University of Munich (Klug and Famulok 1994). The DNA pool consisted of a randomized region of 74 nt flanked by primer regions, one of which contained a T7 promotor for in vitro transcription (M38.27 5′-TCTAATACGACTCACTATAGGAGCTCAGCCTTCACTGC, M20.106 5′-GTGGATCCGACCGTGGTGCC). The PCR-amplified DNA pool was in vitro transcribed. Six hundred micrograms of DNA pool were incubated with 10 mM rNTPs and 5000 units of T7 RNA polymerase in 40 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 50 mM NaCl, 2 mM spermidine, 30 mM DTT, in a 5 mL reaction volume. The DNA template was removed by digestion with 500 units of DNase I (RNase-free, Roche Applied Science) for 1 h. The RNA was gel purified on a denaturing polyacrylamide gel, eluted from the gel by diffusion in 500 mM NH4-acetate, 0.2% SDS, and 10 mM EDTA, and precipitated.
Before every selection step, the RNA pool (in 100 mM NaCl, 50 mM KCl, 10 mM HEPES at pH 7.4, 1 mM MgCl2, 1 mM dithiothreitol) was again treated with DNase I for 1 h, heated to 70°C for 5 min, left at room temperature for 10 min, and preselected by filtering through presoaked nitrocellulose, to eliminate RNAs with affinities for nitrocellulose. The remaining RNA pool was incubated with 5 μg of protein and 10 μg yeast t-RNA (Sigma) for 10 min at room temperature and filtered through nitrocellulose. The RNA bound to the filter was recovered by adding 500 μL Trizol (Invitrogen, Life Technologies) and vortexed at 60°C for 5 min. After mixing with 100 μL chloroform the phases were separated by centrifugation. The RNA was ethanol precipitated with glycogen added as a tracer. The recovered RNA was reverse transcribed at 42°C with superscript-RTase (Life Technologies). The reverse transcribed DNA was PCR amplified and the PCR product served as a template for the next in vitro transcription. For in vitro transcriptions for the following SELEX cycles, 1 μg of DNA, 1 mM rNTPs, 10 μCi of α-32P-rATP, and 100 units of T7 Polymerase were used.
After every selection step the fraction of total bound RNA versus unbound RNA was determined by scintillation counting. From the 10th cycle onward, no further increase of binding affinity was observed; therefore these cycles were used for cloning into pGEM-T easy (Promega), plasmid preparation, and sequencing. The resulting sequences were aligned according to sequence homologies.
Filter-binding assay
A dilution series of either dsRBD in 1× binding buffer (1 mM MgSO4, 20 mM KCl, 10 mM HEPES at pH 7.8, 1 mM dithiothreitol, 250 μg/mL BSA) with concentrations ranging from 6000 nM to 100 pM (in 1:3 dilution steps), as well as binding buffer alone, were pipetted into a 96-well microtiter plate. An equal volume of 5′-labeled RNA in binding buffer was added to each well and incubated for 20 min at room temperature. The reaction was applied to a Bio-Dot Micro filtration apparatus (Biorad), filtered through nitrocellulose, and washed twice with wash buffer (1 mM MgSO4, 20 mM KCl, 10 mM HEPES at pH 7.8, 1 mM dithiothreitol). The dried membrane was exposed to a PhosphorImager screen. The data were analyzed using Quantity One (Biorad). The data points were fit to a sigmoid curve (Sigma-Plot) and the point of inflection was used to determine the K D according to the formula used by Bevilacqua (Bevilacqua and Cech 1996).
Boundary experiments
3′ and 5′ radioactively labeled RNAs together with 1 μg of t-RNA were partially hydrolyzed in 40 mM NaHCO3 by heating for 2 min to 95°C. After neutralization of the reaction with acidic acid the RNA ladder was precipitated. These different sized RNAs in 20 μL 1× binding buffer were incubated with 1 μg of the second dsRBD of xlADAR1 and Xlrbpa for 20 min at room temperature. The fraction of RNA that was bound by the proteins was retained by filtering it through nitrocellulose and washed with washing buffer. The unbound fraction and the wash fraction were collected, pooled, and precipitated. The bound fraction was recovered with Trizol. Bound and unbound fractions were loaded side by side together with a hydrolysis ladder and a denaturing T1 digest on denaturing polyacrylamide gels. The minimal RNAs were cloned, transcribed in vitro, and tested in filter-binding assays to determine whether they had retained their binding capacity.
Structure mapping
To verify structures, which were predicted by the RNA and DNA folding and hybridization software Mfold (Mathews et al. 1999; Zuker 2003), RNAs were chemically modified to distinguish between unpaired and paired regions. Unpaired nucleotides in structured RNAs are accessible for reactive chemicals like dimethyl sulfate (DMS), kethoxal or 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT). DMS preferentially methylates A and C, kethoxal reacts with Gs, and CMCT binds to U and G. Aliquots of 10 pmol RNA were denatured in 30 μL 1× binding buffer for 2 min at 95°C and renatured for 10 min at room temperature.
Fifty micrograms of kethoxal were added and incubated for 15 min at room temperature. Adding 2 μl 0.5 M K-borate and 4 μL of 0.2 M EDTA stopped the reaction and the RNA was precipitated. After centrifugation the pellet was resuspended in 30 mM K-borate and phenol extracted. It is necessary to add borate, as the adduct is only stable in its presence.
DMS (in ethanol) was added to the reaction to a final concentration of 0.7% and incubated for 10 min at room temperature. The reaction was stopped by adding 15 μL of STOP-Mix B (1 M Tris at pH 7.5, 1 M (β-Mmercaptoethanol, 0.1 M EDTA), precipitated, and phenol extracted.
CMCT (1.3 μg in 15 μL 1× binding buffer) was added and incubated for 10 min at room temperature. The reaction was stopped by adding 14 mM EDTA and precipitated.
All modified RNAs were phenol extracted and precipitated before primer extension.
We hybridized 0.3 pmol of 32P-5′-labeled primer to 0.4 pmol of RNA template in 10 μL of hybridization buffer (25 mM Tris at pH 8.4, 30 mM KCl) for 30 min at 42°C.
After the annealing step, 10 μL extension mixture (200 mM Tris at pH 8.4, 20 mM MgCl2, 20 mM dithiothreitol, 500 μM dNTPs, 0.4 units AMV reverse transcriptase) were added to the modified and also, as a control, to unmodified RNAs. Sequencing reactions were performed in parallel using 200 μM ddNTPs in the same buffer. The reactions were stopped by precipitation after 45 min of incubation at 42°C. Reactions were loaded on 10% denaturing acrylamide gels. The gels were fixed, dried, and exposed to PhosphorImager screens.
Nuclease protection assay
RNase V1 recognizes any 4–6-nt segments of polynucleotide backbone with an approximately helical conformation and cleaves leaving 5′phosphates (Lowman and Draper 1986).
For the partial digest with RNase V1, RNAs were 5′ labeled with T4 polynucleotide kinase and γ-32P-ATP. Approximately 20 fmol (corresponding to 50,000 cpm) of these RNAs were centrifuged, washed, dried, and resuspended in structure buffer (100 mM Tris at pH 7, 1 M KCl, 100 mM MgCl2). After annealing, 1 μL of t-RNA (10 mg/mL) was added, followed by the addition of 1 μL of decreasing protein concentrations, starting with 1 μg/μL and diluting it in 1 in 3 dilution steps (1μg/μL; 333, 111, 37, 12, 4 ng/μL). To guarantee binding of the protein the samples were incubated at room temperature for 15 min.
Then 0.002 units of RNase V1 were added and left at room temperature for a further 10 min (Ambion). The reaction was stopped and precipitated by adding 25 μL of provided STOP buffer. Samples were loaded side by side together with a hydrolysis ladder and a denaturing T1 digest of the RNA on denaturing RNA gels.
In vivo editing assay
The hyperediting substrate 52G (F55A4.9) cloned into pGem3zf(+) (a gift from B.L. Bass; see Morse et al. 2002) was linearized with HindIII. The cut vector was used for in vitro transcription with T7-RNA polymerase to give rise to an 810-nt-long RNA. For injection into X. laevis oocyte nuclei, ∼0.5 fmol of α-32P-ATP radioactively labeled RNA (∼10,000 cpm) was used per single oocyte. For coinjections, equimolar amounts of unlabeled SELEX RNAs were used. After 90 min oocytes were hand enucleated and the nuclei were homogenized. The lysate was proteinase K digested, and the RNA was isolated by phenol extraction and precipitation with ethanol. The isolated RNA was heat denatured (in 25 mM Tris at pH 7.5, 0.1 mM ZnCl2) and placed on ice before adding P1 nuclease. The resulting 5′ mononucleotides were separated on cellulose TLC plates with saturated (NH4)2SO4:0.1 M sodium acetate (pH 6):isopropanol (79:19:2 by volume). The dried TLC plates were exposed to PhosphorImager screens and resulting spots were quantified using Quantity One (Biorad).
ACKNOWLEDGMENTS
The authors thank Brenda Bass, University of Utah, for supplying them with a clone encoding the C. elegans substrate 52G. Michael Famulok, University of Munich, provided the initial pool of randomized sequences used for SELEX. We also thank Edward Hitti, Roman Brooks, and Andrea Stahlmann for help with the SELEX procedure. This work was supported by Austrian Science Foundation grant numbers P13534 and SFB 17-16 to M.F.J.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.125506.
REFERENCES
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L., Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bass, B.L., Hurst, S.R., Singer, J.D. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 1994;4:301–314. doi: 10.1016/s0960-9822(00)00069-5. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Cech, T.R. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., George, C.X., Samuel, C.E., Cech, T.R. Binding of the protein kinase PKR to RNAs with secondary structure defects: Role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–6316. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- Brooks, R., Eckmann, C.R., Jantsch, M.F. The double-stranded RNA-binding domains of Xenopus laevis ADAR1 exhibit different RNA-binding behaviors. FEBS Lett. 1998;434:121–126. doi: 10.1016/s0014-5793(98)00963-6. [DOI] [PubMed] [Google Scholar]
- Burkard, M.E., Turner, D.H., Tinoco, I., Jr. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. The interactions that shape RNA structure In The RNA world (eds. R.F. Gesteland et al.) pp. 233–264. pp. [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Bycroft, M., Grunert, S., Murzin, A.G., Proctor, M., St Johnston, D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5 [published erratum appears in EMBO J 1995 Sep 1;14(17):4385] EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G. Conservation of RNase III processing pathways and specificity in hemiascomycetes. Eukaryot. Cell. 2003;2:901–909. doi: 10.1128/EC.2.5.901-909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Buckle, M., Jacquier, A. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. 2000;97:3142–3147. doi: 10.1073/pnas.070043997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, M., Jantsch, M.F. Distinct in vivo roles for double-stranded RNA-binding domains of the Xenopus RNA-editing enzyme ADAR1 in chromosomal targeting. J. Cell Biol. 2003;161:309–319. doi: 10.1083/jcb.200301034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann, C.R., Jantsch, M.F. Xlrbpa, a double-stranded RNA-binding protein associated with ribosomes and heterogeneous nuclear RNPs. J. Cell Biol. 1997;138:239–253. doi: 10.1083/jcb.138.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hitti, E., Neunteufl, A., Jantsch, M.F. The double-stranded RNA-binding protein X1rbpa promotes RNA strand annealing. Nucleic Acids Res. 1998;26:4382–4388. doi: 10.1093/nar/26.19.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharrat, A., Macias, M.J., Gibson, T.J., Nilges, M., Pastore, A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug, S.J., Famulok, M. All you wanted to know about SELEX. Mol. Biol. Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- Kumar, M., Carmichael, G.G. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y., Hallegger, M., Kinar, Y., Shemesh, R., Djinovic-Carugo, K., Rechavi, G., Jantsch, M.F., Eisenberg, E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Lei, M., Samuel, C.E. Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR. Proc. Natl. Acad. Sci. 2000;97:12541–12546. doi: 10.1073/pnas.97.23.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., Seeburg, P.H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Lowman, H.B., Draper, D.E. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Morse, D.P., Bass, B.L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D.G., Dimock, K., Kang, C.Y. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology. 1991;181:760–763. doi: 10.1016/0042-6822(91)90913-v. [DOI] [PubMed] [Google Scholar]
- Ramos, A., Grunert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St Johnston, D., Varani, G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter, J.M., Schultz, S.C. Molecular basis of double-stranded RNA-protein interactions: sStructure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston, D., Brown, N.H., Gall, J.G., Jantsch, M. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl, R., Allain, F.H. A novel RNA pentaloop fold involved in targeting ADAR2. RNA. 2005;11:592–597. doi: 10.1261/rna.7276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl, R., Skrisovska, L., Xu, M., Emeson, R.B., Allain, F.H. Resonance assignments of the double-stranded RNA-binding of adenosine deaminase acting on RNA 2 (ADAR2) J. Biomol. NMR. 2005;31:71–72. doi: 10.1007/s10858-004-6058-x. [DOI] [PubMed] [Google Scholar]
- Stephens, O.M., Haudenschild, B.L., Beal, P.A. The binding selectivity of ADAR2’s dsRBMs contributes to RNA-editing selectivity. Chem. Biol. 2004;11:1239–1250. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Wong, S.K., Sato, S., Lazinski, D.W. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, T.C., Ayata, M., Hirano, A., Yoshikawa, Y., Tsuruoka, H., Yamanouchi, K. Generalized and localized biased hypermutation affecting the matrix gene of a measles virus strain that causes subacute sclerosing panencephalitis. J. Virol. 1989;63:5464–5468. doi: 10.1128/jvi.63.12.5464-5468.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Henras, A., Chanfreau, G., Feigon, J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, M., Hagerman, P.J. Bulge-induced bends in RNA: Quantification by transient electric birefringence. J. Mol. Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- Zheng, X., Bevilacqua, P.C. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]