Abstract
Over 100 different chemical types of modifications have been identified in thousands of sites in tRNAs, rRNAs, mRNAs, small nuclear RNAs, and other RNAs. Some modifications are highly conserved, while others are more specialized. They include methylation of bases and the ribose backbone, rotation, and reduction of uridine, base deamination, elaborate addition of ring structures, carbohydrate moieties, and more. We have developed a systematic approach to detect and quantify the extent of known RNA modifications. The method is based on the enzymatic ligation of oligonucleotides using the modified or unmodified RNA as the template. The efficiency of ligation is very sensitive to the presence and the type of modifications. First, two oligo pairs for each type of modification are identified. One pair greatly prefers ligation using the unmodified RNA template over the modified RNA template or vice versa. The other pair has equal reactivity with unmodified and modified RNA. Second, separate ligations with each of the two oligo pairs and the total RNA mixture are performed to detect the presence or absence of modifications. Multiple modification sites can be examined in the same ligation reaction. The feasibility of this method is demonstrated for three 2′O-methyl modification sites in yeast rRNA.
Keywords: RNA modification, DNA ligase, 2′O-methylation, ribosomal RNA, snoRNA
INTRODUCTION
Biological RNAs undergo extensive post-transcriptional modifications consisting of numerous chemical types at many sites. More than 100 chemical types of post-transcriptional RNA modifications have been identified to date (http://medlib.med.utah.edu/RNAmods/; Rozenski et al. 1999). The most extensive modifications occur in tRNAs and rRNAs, although modifications are also present in spliceosomal RNAs (snRNA), small nucleolar RNAs (snoRNA), mRNAs, and micro-RNAs (Maxwell and Fournier 1995; Ebhardt et al. 2005; Yu et al. 2005). A 75–95 nucleotide tRNA contains on average seven to eight modifications, some of which are conserved in all tRNAs, while others are present among just a few tRNAs. rRNAs also contain a large number of modifications, from 36 in Escherichia coli to >210 in humans (Bjork 1995; Kiss 2001; Decatur and Fournier 2003). Some modifications, in particular, those in the anticodon loop of tRNA, have well-defined functions (Agris 2004). However, the functions of the vast majority of RNA modifications remain unclear.
Three general methods have been used to locate and characterize RNA modifications in a biological sample: thin-layer or liquid chromatography, mass spectrometry, and enzymatic detection by reverse transcriptase or RNase H (Andachi et al. 1989; Kowalak et al. 1993; Yu et al. 1997; Qiu and McCloskey 1999; Zhao and Yu 2004). To a large extent, the chromatographic and mass-spectrometric methods usually involve purification of the RNA molecule of interest, followed by partial or complete nuclease digestion. Differences in the chromatographic mobility or mass then allow detection of modified nucleotides. No purification is needed for the enzymatic detection method, which relies on reverse transcriptase to stop at the modified nucleotide. The RNase H method is limited to a few modifications and involves a two-step procedure: labeling the RNA of interest followed by primer extension either before or after RNase H cleavage. Though powerful and successful, these methods are difficult to scale up to allow simultaneous studies of many modifications. Furthermore, quantitative comparison of modifications between two samples is challenging because a single data point is obtained in most cases. Each biological sample can have varying reactivity, so it is not straightforward to compare unambiguously the extent of modifications in two samples using a single data point.
tRNAs or rRNAs are not always fully modified. Lack of modification can be derived from either incomplete modification of new RNA transcripts or reversal at selected sites of previously modified RNA. Incomplete modification can be due to low availability of co-substrates (e.g., S-adenosyl-methionine for methylation), regulated concentrations or activities of modification enzymes, or imbalances in the speed of RNA folding and the modification reaction (e.g., Bjork 1995; Lazaro et al. 1996; Zhong et al. 1999). Lack of rRNA modifications can lead to antibiotic resistance in bacteria (Weisblum 1995; Vila-Sanjurjo et al. 1999; Gaynor and Mankin 2003; Kehrenberg et al. 2005).
About half of the 100 modification types are chemically reversible. For example, the highly conserved 1-methyl-A in eukaryotic tRNAs may be demethylated in an enzyme-catalyzed reaction, akin to the demethylation of deoxy-A bases in DNA repair. The universally conserved DNA repair enzyme AlkB has been shown to repair methylated RNA under physiological conditions, including possible demethylation of 1-methyl-A (Aas et al. 2003; Ougland et al. 2004). In a few demonstrated cases, tRNA missing particular modifications can perform a distinct function, e.g., promotion of ribosomal frameshift (Bjork et al. 1989; Carlson et al. 1999). These considerations underscore the need for a rapid and reliable method to study the quantitative extent of RNA modifications.
This work describes a new, systematic method to detect and quantify RNA modifications. The method is based on the sensitivity of an enzymatic ligation using both modified and unmodified RNA in the total RNA mixture as a template pair. First, a collection of oligonucleotide pairs is tested on a model RNA oligonucleotide containing a different modification type to find at least one pair that distinguishes unmodified from modified (or modified from unmodified) RNA, and one other pair that reacts equally well with the unmodified and the modified RNA. The former oligo pair discriminates the unmodified from modified RNA (hence, the D-oligo pair). The latter oligo pair calibrates the total amount of unmodified and modified RNA (hence, the nondiscriminating or N-oligo pair). Second, D- and N-oligos are designed to enable ligation at specific sites in a biological RNA. The method allows simultaneous characterization of multiple modification sites without purification from a total RNA mixture. Furthermore, the availability of two data points using D- and N-oligos permits the comparison of the relative extent of modifications between two samples. We describe the D- and N-oligo pairs for seven modification types and demonstrate the feasibility of this method by examining three rRNA modification sites in yeast.
RESULTS AND DISCUSSION
Method description
Every modification in a biological RNA is characterized by its chemical type and its location. A ligation-based approach is applied to detect and quantify the extent of modification at each site (Fig. 1). The method relies on T4 DNA ligase joining two oligonucleotides using the RNA as the template. The specificity for interrogating a given modification site is provided by the hybridizing sequences of these two oligonucleotides. The extent of modification is characterized by the efficiency of the ligase reaction that is dependent on the presence or absence of modifications.
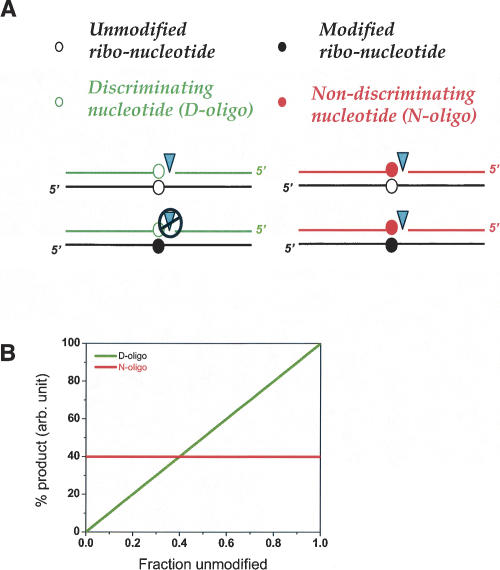
FIGURE 1.
Outline of the ligation method to study RNA modifications. (A) The RNA (black line) containing an unmodified or modified nucleotide (open or filled black circles) is hybridized with two oligonucleotides. These oligonucleotides are ligated by the T4 DNA ligase using this RNA as a template (triangle). The D- and N-oligos have different nucleotide compositions (green or red circles) at the ligation junction or different ligation sites. (B) For the D-oligo (green line), ligation proceeds very efficiently when the RNA is unmodified, but very poorly when the RNA is modified (or the other way around). For the N-oligo (red line), ligation has a constant yield regardless of the status of RNA modification. The amount of D-oligo ligation product corresponds to the amount of unmodified over modified RNA template (or modified over unmodified). The amount of N-oligo product corresponds to the sum of the unmodified and modified RNA.
Two oligo pairs are needed for each modification type (e.g., 2′Ome-U or Um). In the ligation reaction, one oligo pair discriminates unmodified over modified RNA (or modified over unmodified RNA), so that the ligation occurs with significantly different efficiency when the RNA is unmodified (or modified). This discriminating oligo pair is designated as the D-oligo (Fig. 1). The ligation efficiency of the other oligo pair has little preference for the presence or the absence of modification. This nondiscriminating oligo pair is designated as the N-oligo. The ligation reaction using the D-oligo pair provides a ratio of unmodified/modified (or modified/unmodified). The reaction using the N-oligo provides the amount of unmodified plus modified RNA and also normalizes the varying efficiencies of ligation when using biological samples isolated from diverse sources (Fig. 1). These two data points allow quantitative correlation of the extent of modification at each site.
The identification of the D- and N-oligos for each modification type is carried out experimentally by screening the ligation efficiency of a collection of oligonucleotide pairs using model 30-mer RNA templates (Table 1). The model RNA sequence is designed to have minimal amount of secondary structure when the central nucleotide 15 is uridine to expedite the screening of D- and N-oligos. For this study, the model RNAs contain the modified nucleotide at this 15th position (2′Ome-A, C, G, or U; 3-methyl-U, 5,6-dihydro-U, and inosine) and the corresponding unmodified nucleotide (A, C, G, or U, as appropriate). Two configurations are designed for the oligonucleotide substrates by placing the ligation junction either 3′ (set I) or 5′ (set II) to the modified nucleotide in the model RNA (Fig. 2A). Here, anchor refers to a constant oligo substrate, and floater refers to oligo substrates containing variations at their 5′ (set I) or 3′ end (set II). All other positions of these ligation substrates are made of deoxy residues (Table 1).
TABLE 1.
Sets I and II floaters and anchors used in this work
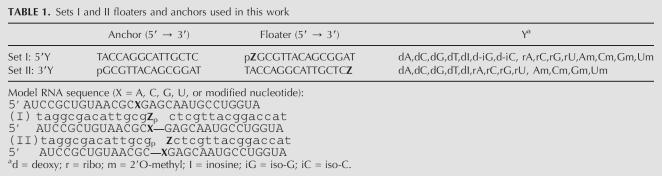
FIGURE 2.
Finding D- and N-oligos for each modification type. (A) Two sets of oligo pairs with the ligation junction 5′ (set I) or 3′ (set II) to the modification site. Black = model 30-mer RNA containing unmodified (open) or modified (filled circles) nucleotide. Blue = floater and anchor oligonucleotides as ligation substrates. (B) A total of 15 set I pairs and 13 set II pairs are tested for 2′O-methyl-U, 3-methyl-U and 5,6-dihydro-U modifications. I-dA is the only sufficiently discriminating D-oligo among these 28 oligo pairs, whereas II-rA, II-Am, II-rG, and II-Gm can be used as N-oligos. (C) Ligation using the designated D-oligo (I-dA floater) to discriminate unmodified U from 2′Ome-U, 3-methyl-U and 5,6-dihydro-U (left, >10-fold). Ligation using a designated N-oligo (II-rA floater) shows <1.3-fold variation between the unmodified U and the other modifications (right).
We first describe our results on obtaining D- and N-oligos for seven modification types using this model 30-mer RNA template. To demonstrate the feasibility of this approach for biological samples, we then apply these D- and N-oligos to detect the loss of 2′O-methyl modifications at two sites in yeast rRNA.
Finding D- and N-oligos for seven modification types
T4 DNA ligase can join two DNA oligonucleotides using an RNA template (Kleppe et al. 1970; Nilsson et al. 2000). We therefore adopted an empirical approach to screen for D- and N-oligos for seven modification types. Fifteen set I floaters and 13 set II floaters were used in this screen because of their commercial availability. In these sets of floaters, the nucleotide at the ligation junction was allowed to vary in a background containing deoxynucleotides complementary to the template (set I = 5′ pZGCGTTACAGCGGAT; set II = 5′ TACCAGGCATTGCTCZ, Z = variable nucleotide; see Table 1). This small number of floaters was sufficient to provide D- and N-oligo “solutions” to these modification types, although we are not limited to these floaters. Moreover, the variable nucleotide needs not be confined to the ligation junction. Even within the limits of commercial availability, the oligo-pair collection could be readily expanded. For example, expansion of the oligo-pair collection by threefold (from 28 to 84) could be made simply by changing the 3′ deoxy nucleotide of the set I anchor or the 5′ deoxy nucleotide of the set II anchor to a ribo or a 2′Ome nucleotide. For the unmodified RNA, sets I and II anchors containing the corresponding terminal ribo or 2′Ome nucleotide show distinct ligation efficiencies as the deoxy anchors (data not shown).
When the ligation junction is placed 3′ to the modified nucleotide (anchor I and floaters I), the amount of product is very sensitive to the base identity and backbone composition of the floater 5′ nucleotide (Fig. 2B, left). When the model 30-mer RNA contains an unmodified U at the designated 15th position, significant amount of products (15%–20%) can be obtained when the corresponding floaters have 5′ dA or deoxy-Inosine (I-dA or I-dI), but not when the corresponding floaters have 5′ dG, dC, or dT (<4%). This result apparently reflects the Watson–Crick and wobble base pairing of rU–dA and rU–dI. However, little ligation product (<2%) is obtained when the floaters contain 5′ rA or 2′O-methyl-A (mA) even though rU–dA, rU–rA, and rU–mA pairs are all expected to have an A-form-like helical conformation. This result shows that T4 DNA ligase exhibits significant specificity for the backbone constitution and presumably conformation of the base-pair 3′ to the ligation junction.
When the model 30-mer RNA contains a backbone modification of 2′O-methyl-U, the ligation efficiency compared to the unmodified RNA is reduced by at least 10-fold (from 20% to 1%) using the I-dA floater (Fig. 2C, left), and by approximately threefold (from 15% to 5%) using the I-deoxy-Inosine floater. The ligation efficiencies are very similar using the I-dG floater. Applying an arbitrary cutoff of 10-fold in differential ligation efficiency between the unmodified and the 2′O-methyl modified U templates, I-dA floater is therefore selected as an acceptable discriminating oligo (D-oligo) for the 2′O-methyl-U modification.
When the model 30-mer RNA contains base-pair disrupting modifications of 3-methyl-U and 5,6-dihydro-U, the largest reductions in the ligation efficiencies are again observed using the I-dA floater. Therefore, the I-dA floater is also acceptable as the discriminating oligo (D-oligo) for these two base modifications (Fig. 2C, left).
When the ligation junction is placed 5′ to the modified nucleotide (anchor II and floaters II), the ligation reaction has a markedly different sensitivity to base and backbone compositions of the floater 3′ nucleotide compared to set I oligo pairs (Fig. 2B, right). Very efficient ligation is observed for floaters containing 3′ Am, Gm, rA, or rG (II-Am, II-Gm, II-rA, or II-rG). These four floaters also give <1.5-fold discrimination when the model RNA contains the same modified nucleotides, 2′O-methyl-U, 3-methyl-U, and 5,6-dihydro-U (Fig. 2C, right). These results show that all four floaters are acceptable as the nondiscriminating oligo (N-oligo) for these modifications.
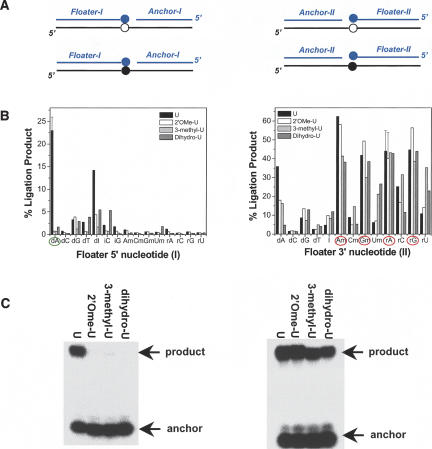
To further confirm that the correct choices for D- and N-oligos are made on the basis of the above results, ligations were performed using defined mixtures of unmodified and modified model RNAs (Fig. 3). In the cases of D-oligos, the amount of product increases linearly from very low (x = 0 corresponds to 100% modified RNA) to very high (x = 1 corresponds to 100% unmodified RNA). Under these conditions, the ligation efficiency differs by ∼50-fold when the RNA template is either unmodified or all 2′O-methyl-U or 2′O-methyl-A. In the cases of N-oligos, the amount of products varies by <1.5-fold among all mixtures. These results confirm that the selection of the D- and N-oligos based on the initial screen (Fig. 2) is indeed correct.
FIGURE 3.
The calibration curves for modifications. (A) Ligation reactions using a defined mixture of unmodified and 2′Ome-modified model 30-mer RNA templates. (B) The percent product from gel analysis is plotted against the fraction of unmodified RNA.
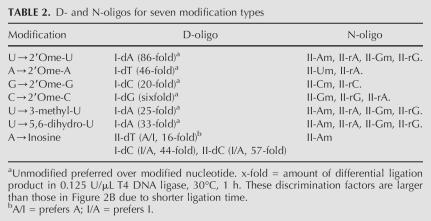
Applying this same two-stage procedure, initial screening followed by analysis using defined unmodified/modified model 30-mer RNA mixtures, we obtained D- and N-oligos for seven modifications (Table 2). While a single D-oligo was found for the 2′O-methyl, 3-methyl-U, and 5,6-dihydro-U modifications among the 28 oligo-pairs tested, multiple D-oligos were identified for A → Inosine modification. In most cases, more than one N-oligo can be obtained.
TABLE 2.
D- and N-oligos for seven modification types
The identity of the most effective D- and N-oligos can be rationally explained for some, but not for all, modifications. For the 2′O-methyls, the Watson–Crick base-pairing nucleotide in its deoxy form is preferred for the unmodified RNA, suggesting that the ligation efficiency is sensitive to the backbone geometry only in the context of Watson–Crick base pairs. The ability of Watson–Crick base pairing clearly explains the preference of unmodified U over 3-methyl-U and 5,6-dihydro-U. However, the N-oligos for these same modifications cannot base pair either. Multiple D-oligos are found for the inosine modification, all of which can be explained by the base-pairing preference to either A or Inosine. The N-oligo for A → I modification is far less sensitive to the base-pairing abilities of the unmodified and the modified RNA.
Detection of loss of 2′O-methylation in yeast rRNA
To demonstrate the application of this method for analysis of biological samples, we next used it to detect the loss of 2′O-methylations at two sites in yeast rRNA. The 18S and the 25S rRNA in Saccharomyces cerevisiae contain 55 2′O-methyl modifications, and all but one are guided by 42 C/D box snoRNAs complexed with distinct proteins; a protein-only enzyme catalyzes one 2′O-methylation (Lowe and Eddy 1999; Decatur and Fournier 2003; Lapeyre and Purushothaman 2004; Davis and Ares 2006). We used two mutant yeast strains. One does not express snR53 that normally guides the 2′O-methylation of A796 in 18S rRNA. The other contains snR67 with a point mutation in the guide element that disrupts the 2′O-methylation of G2618 in 25S rRNA (equivalent to E. coli Gm2251). In the wild-type sample, which contains 2′O-methyl modification at both sites, we expect very low ligation yields for D-oligo reactions at these two sites. In contrast, the mutant samples should show much higher ligation efficiency for their respective D-oligo products.
As controls, we tested ligation at two additional sites. A617 in 18S rRNA is expected to be equally 2′O-methylated in the wild-type and the two mutant yeast samples, so that its D-oligo pair should have similarly low ligation yields in all samples. A “pseudo” D-oligo pair was used for G2811 in 25S rRNA (“G2811”). This oligo pair has a ligation junction one-nucleotide away from the supposed D-oligo pair for the 2′O-methylated G2811, so that this “D-oligo” pair should have a similarly high ligation yield in all samples.
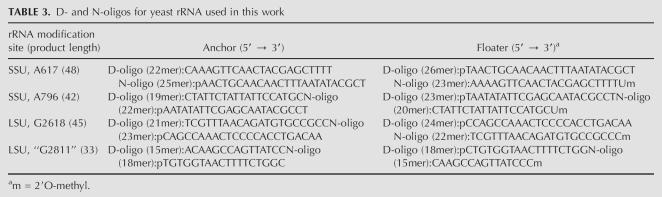
To enable analysis of the four modification sites in the same reaction, we designed the D- and N-oligo pairs to produce ligation products of different lengths (Table 3; 48 for A617, 45 for G2618, 42 for A796, and 33 for G2811). Because every biological RNA sample may contain DNA ligase inhibitors, the model 30-mer RNA was included at a known concentration in all ligation reactions with the yeast samples as overall ligation and loading control. Figure 4 shows the ligation results using yeast total RNA mixtures. Without yeast RNA, only the ligation product derived from the 30-mer RNA standard is observed. The N-oligo mixture gives five reaction products at the expected length when yeast RNA is included (Fig. 4, right). The four products (as well as those from the D-oligo reactions) derived from yeast RNA indeed correspond to our design as confirmed by ligation reactions containing each N- or D-oligo pair in separate reactions (data not shown).
TABLE 3.
D- and N-oligos for yeast rRNA used in this work
FIGURE 4.
Detection of the loss of 2′O-methylation in yeast rRNA. (A) Ligation reactions carried out using yeast total RNA from the wild-type or snoRNA mutant strains using D- or N-oligos. “Standard” corresponds to the product derived from a known amount of the model 30-mer RNA added to the ligation mixture. ΔsnR53 corresponds to a loss of 2′O-methylation at A796 in 18S rRNA, and ΔsnR67 corresponds to a loss of 2′O-methylation at G2618 in 25S rRNA. In this panel, the ligation reaction was performed for 120 min. (B) Relative amount of ligation product at the A796 or G2618 site for the snoRNA mutant strains over the wild-type strain. The decreased ratio at the latest time (120 min) is due to increased amount of product in the wild-type strain, as the amount of product in the mutant strains approaches saturation.
The D-oligo reactions work as designed where the loss of A796 and G2618 modification is easily detected in the corresponding snoRNA deficient strain (Fig. 4, left). At this high ligase concentration (0.5 U/μL), the relative amount of ligation product is 30–40-fold higher in the deletion strains over the wild type at an early time point (15 min), and then decreases to 8–10-fold at the latest time point shown (120 min). This decrease in the relative amount of ligation product at the later time is primarily due to the increased yield in the wild-type sample, as the yield in the snoRNA deletion strains reaches an endpoint. This result shows a robust time window during which the discrimination between the unmodified and modified rRNA is high (i.e., eightfold or more).
For the D-oligos, the absolute amount of ligation product derived from the A796 site is almost 25-fold higher than the amount derived from the G2618 site (Fig. 4, left). This result may be explained by two factors: the secondary structure of the rRNA around the modification site, and the rRNA sequence at the ligation junction. A796 is in a region of 18S rRNA with little secondary structure and the ligation occurs at A796U797. G2618 is in a highly structured region of 25S rRNA and the ligation occurs at G2618G2619. The secondary structure of rRNA clearly influences the hybridization efficiency of D- (and N-) oligos as an 11-mer in place of the 24-mer D-oligo for the G2618 site with a similar melting temperature, as the D-oligo for A796 failed to produce any detectable ligation product (data not shown). However, the G2618 site is accessible for hybridization to a significant extent as demonstrated by the N-oligo reaction at this site (Fig. 4, right). Clearly, different rRNA sequences at the ligation junction affect ligation yield, possibly through subtle structural differences of base pairs in the transition state of the DNA ligase reaction.
SUMMARY AND CONCLUSIONS
We have developed a systematic, ligation-based approach to study RNA modifications. As a semiempirical approach, this method can be applied for any modification type as long as a corresponding model RNA oligonucleotide can be made to allow identification of D- and N-oligos for that type. A brief survey of the literature (Q. Dai, J.A. Piccirilli, and T. Pan, unpubl.) shows that 84 modified nucleotides among a total of ∼100 natural modification types are either commercially available for RNA synthesis (12/84), or have been synthesized (72/84). Additionally, using only commercially available phosphoramidites (e.g., 2′F-A, 2′βOH-A, 8-oxo-A, 2′amino-A), additional floaters and anchors can be synthesized to expand the oligonucleotide collection for screening from 28 here to 720 (20 floaters and 18 anchors for sets I and II = 20 × 18 × 2).
The sensitivity of the ligation method for biological samples strongly depends on the structural and sequence context of the modification site. Examples in this work (Fig. 4) show that for a “good” rRNA site (A796 in 18S), a few tens of nanograms of total RNA would be sufficient for analysis. For a “poor” rRNA site (G2618 in 25S), 1 μg of total RNA is needed. The detectable changes in the modification fraction depend on the differential ligation yield between fully modified and fully unmodified RNA. When the D-oligo reaction differential between unmodified and modified RNA is 10-fold, a change from 100% to 90% modified RNA would result in a twofold increase in the ligation yield (1 + 0.1 × 10 = 2, so 1 → 2). However, this 10-fold difference only results in a 0.9-fold decrease in the ligation yield upon a change in unmodified RNA from 100% to 90% (10–0.1 × 10 = 9, so 10 → 9). When the reaction differential between unmodified and modified RNA is 50-fold, a change from 100% to 90% unmodified RNA would result in a twofold decrease in the ligation yield (10–0.1 × 50 = 5, so 10 → 5).
A major advantage of this approach is its ease for adaptation for high throughput analysis of RNA modifications. We show here (Fig. 4) that four distant sites in the ribosomal RNA sequence can be simultaneously analyzed by gel electrophoresis. Future work will apply this method for microarray analysis of, e.g., all 2′O-methylation sites in yeast (55 sites) or human rRNA (107 sites) during cell growth, differentiation, and environmental adaptation.
MATERIALS AND METHODS
RNA and DNA oligonucleotides
RNA oligonucleotides with or without modifications were purchased from Dharmacon Research, de-protected, and purified according to manufacturer's protocols. All DNA oligonucleotides (anchors and floaters) were purchased from Integrated DNA Technologies. When needed, RNA or DNA oligonucleotides were purified on denaturing polyacrylamide gels containing 7 M urea.
Ligation reaction using a model 30-mer RNA
The screening step was carried out with 0.15 μM model 30-mer RNA with or without modifications, 0.5 μM of floater oligonucleotides (15 set I and 13 set II floaters), and 0.38 μM of 5′ 32P-labeled anchor oligonucleotide in 66 mM Tris-HCl, pH 7.6, 6.6 mM MgCl2, 10 mM DTT, 66 μM ATP, 15% DMSO, and 0.125 U/μL T4 DNA ligase (USB Inc.). All components were mixed and incubated at 30°C for 2 h, and the ligation product separated on denaturing polyacrylamide gels.
The ligation reaction using a defined mixture of modified and unmodified model 30-mer RNA was carried out under the same conditions as above. The reaction was carried out at 30°C for 1 h.
Ligation reaction using total yeast RNA
The two snoRNA-mutant strains are derived from the wild-type YS602 strain (MAT:α ade2-101 his3-11,15 trp1Δ901 ura3-52 leu2-3,112). The snR67/snR53 region was disrupted by insertion of the TRP1 gene, and in the case of the strain not expressing snR53, snR67 expression was restored with the snR67 gene and its upstream sequence on a CEN-based yeast shuttle vector carrying the LEU2 marker. In the case of the strain not guiding the P-loop 2'-O-methylation Gm2618, the restoring vector harbors the entire snR67/snR53 region; however, the guide for the P-loop site has a substitution at the nucleotide that corresponds to the one pairing with the modified site, resulting in disruption of 2'-O-methylation.
Total yeast RNA from wild-type and mutant strains was isolated using standard protocols described previously (Schattner et al. 2004). To remove impurities that interfered with the T4 DNA ligase reaction, total RNA was mixed with an equal volume of 50 mM KOAc/200 mM KCl, extracted with phenol/CHCl3 (1:1), and precipitated with ethanol. The RNA samples were dissolved in water at ∼2 μg/μL.
The anchor and floater oligonucleotides were first hybridized with yeast total RNA in 20 mM TrisHCl, pH 7.5, 50 mM NaCl, 0.4 μg/μL yeast RNA, 0.5 μM floater, and 0.4 μM anchor oligonucleotides targeting the rRNA. The hybridization mixture also contained 10 nM of the model 30-mer RNA, its 60 nM floater, and 30 nM 32P-labeled anchor as the control for ligation efficiency and loading. To accommodate the varying melting temperatures of the floater and anchor oligos targeting the rRNA, hybridization was carried out first at 55°C for 1 h followed by another hour at 44°C.
Following hybridization, the ligation reaction was performed upon the addition of a 2× ligation mixture containing 132 mM TrisHCl, pH 7.5, 13.2 mM MgCl2, 20 mM DTT, 132 μM ATP, 30% DMSO, and 1 U/μL T4 DNA ligase. The ligation proceeded at 37°C from 15 to 120 min. In order to remove excessive background of the unreacted, 32P-labeled oligos (the 32P-label is always in the middle of the ligation product), 5 μL aliquots of the ligation mixture were treated with 0.1 U/μL calf-intestine alkaline phosphatase (from Boehringer-Mannheim), 0.1 U/μL RNase H (from Epicenter Technologies), and 42 μM Na-pyrophosphate at 37°C for 10 min. The reaction was quenched with the addition of an equal volume of 9 M urea/50 mM EDTA. The mixture was boiled for 2 min and rapidly cooled on ice prior to its loading on denaturing polyacrylamide gels containing 7 M urea.
ACKNOWLEDGMENTS
This work was supported by the NIH (R21GM73747 to T.P. and J.A.P., and GM19351 to M.J.F.), Chicago BioMedical Consortium, and a graduate fellowship from the Burrough Wellcome Fund Interfaces ID 1001774 (M.S.). We thank Xue-hai Liang and Dorota Piekna-Przybylska in the M.J.F. laboratory for helpful advice. We also thank Dr. A. Mankin and Dr. Yi-Tao Yu for insightful discussions, and the reviewers for comments.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.208906.
REFERENCES
- Aas, P.A., Otterlei, M., Falnes, P.O., Vagbo, C.B., Skorpen, F., Akbari, M., Sundheim, O., Bjoras, M., Slupphaug, G., Seeberg, E., et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi, Y., Yamao, F., Muto, A., Osawa, S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 1989;209:37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Bjork, G.R. Biosynthesis and function of modified nucleosides. In: Soll D., RajBhandary U., editors. tRNA: Structure, biosynthesis, and function. ASM Press; Washington, DC: 1995. pp. 165–206. [Google Scholar]
- Bjork, G.R., Wikstrom, P.M., Bystrom, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- Carlson, B.A., Kwon, S.Y., Chamorro, M., Oroszlan, S., Hatfield, D.L., Lee, B.J. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- Davis, C.A., Ares, M., Jr. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W.A., Fournier, M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Ebhardt, H.A., Thi, E.P., Wang, M.B., Unrau, P.J. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, M., Mankin, A.S. Macrolide antibiotics: Binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 2003;3:949–961. doi: 10.2174/1568026033452159. [DOI] [PubMed] [Google Scholar]
- Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L.H., Vester, B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe, K., Van de Sande, J.H., Khorana, H.G. Polynucleotide ligase-catalyzed joining of deoxyribo-oligonucleotides on ribopolynucleotide templates and of ribo-oligonucleotides on deoxyribopolynucleotide templates. Proc. Natl. Acad. Sci. 1970;67:68–73. doi: 10.1073/pnas.67.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak, J.A., Pomerantz, S.C., Crain, P.F., McCloskey, J.A. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre, B., Purushothaman, S.K. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Lazaro, E., Rodriguez-Fonseca, C., Porse, B., Urena, D., Garrett, R.A., Ballesta, J.P. A sparsomycin-resistant mutant of Halobacterium salinarium lacks a modification at nucleotide U2603 in the peptidyl transferase centre of 23 S rRNA. J. Mol. Biol. 1996;261:231–238. doi: 10.1006/jmbi.1996.0455. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M., Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- Maxwell, E.S., Fournier, M.J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Nilsson, M., Barbany, G., Antson, D.O., Gertow, K., Landegren, U. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nature Biotechnol. 2000;18:791–793. doi: 10.1038/77367. [DOI] [PubMed] [Google Scholar]
- Ougland, R., Zhang, C.M., Liiv, A., Johansen, R.F., Seeberg, E., Hou, Y.M., Remme, J., Falnes, P.O. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Qiu, F., McCloskey, J.A. Selective detection of ribose-methylated nucleotides in RNA by a mass spectrometry-based method. Nucleic Acids Res. 1999;27:e20. doi: 10.1093/nar/27.18.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., McCloskey, J.A. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner, P., Decatur, W.A., Davis, C.A., Ares, M., Jr., Fournier, M.J., Lowe, T.M. Genome-wide searching for pseudouridylation guide snoRNAs: Analysis of the Saccharomyces cerevisiae genome . Nucleic Acids Res. 2004;32:4281–4296. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Sanjurjo, A., Squires, C.L., Dahlberg, A.E. Isolation of kasugamycin resistant mutants in the 16 S ribosomal RNA of Escherichia coli . J. Mol. Biol. 1999;293:1–8. doi: 10.1006/jmbi.1999.3160. [DOI] [PubMed] [Google Scholar]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R.W., Steward, R., Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., Steitz, J.A. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Yu, Y.T. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, P., Cao, Z., Hammond, R., Chen, Y., Beyer, J., Shortridge, V.D., Phan, L.Y., Pratt, S., Capobianco, J., Reich, K.A., et al. Induction of ribosome methylation in MLS-resistant Streptococcus pneumoniae by macrolides and ketolides. Microb. Drug Resist. 1999;5:183–188. doi: 10.1089/mdr.1999.5.183. [DOI] [PubMed] [Google Scholar]