Abstract
Adenosine deaminases that act on RNA [adenosine deaminase, RNA specific (ADAR)] catalyze the site-specific conversion of adenosine to inosine in primary mRNA transcripts. These re-coding events affect coding potential, splice sites, and stability of mature mRNAs. ADAR is an essential gene, and studies in mouse, Caenorhabditis elegans, and Drosophila suggest that its primary function is to modify adult behavior by altering signaling components in the nervous system. By comparing the sequence of isogenic cDNAs to genomic DNA, we have identified and experimentally verified 27 new targets of Drosophila ADAR. Our analyses led us to identify new classes of genes whose transcripts are targets of ADAR, including components of the actin cytoskeleton and genes involved in ion homeostasis and signal transduction. Our results indicate that editing in Drosophila increases the diversity of the proteome, and does so in a manner that has direct functional consequences on protein function.
Keywords: adenosine deaminase, Drosophila, RNA editing, ADAR, nervous system, transcriptome
INTRODUCTION
RNA editing is a well-established mechanism in which precursor messenger RNA transcripts are subject to re-coding by the enzyme adenosine deaminase, RNA specific (ADAR) (Bass 2002). ADAR catalyzes the deamination of adenosine to inosine. Inosine mimics guanosine in its base-pairing properties, and the translational machinery interprets I as G. In this way, an A-to-I conversion in the mRNA can alter the genetic information that can lead to changes in mRNA splicing and stability as well as changes in protein function. ADAR is essential in all animals examined to date, and studies in mouse, Caenorhabditis elegans, and Drosophila suggest that the function of pre-mRNA editing is to modify adult behavior by altering signaling components in the nervous system (Higuchi et al. 2000; Palladino et al. 2000; Wang et al. 2000; Tonkin et al. 2002). Considerable progress has also been made in understanding the mechanism of action of the ADAR enzyme (Cho et al. 2003; Haudenschild et al. 2004; Athanasiadis et al. 2005; Macbeth et al. 2005). The molecular details of target site specificity of RNA editing are drawn from studies of the mammalian glutamate receptor gene, GluR-B. Receptor pre-mRNA forms a double-stranded stem structure by imperfect base-pairing between an exonic sequence, where the edit(s) occur, and a noncoding intronic element called the editing site complementary sequence (ECS) (Higuchi et al. 1993). Most targets, however, are found serendipitously when isolating genes from cDNA libraries. Computational identification of ADAR substrates is difficult, and little progress has been made in designing algorithms for predicting putative target transcripts. ADAR targets in the human genome have been identified and are located predominantly in embedded Alu sequences, suggesting a role of ADAR in mRNA stability (Kim et al. 2004; Levanon et al. 2004). Editing can also introduce new splice sites (Rueter et al. 1999), and editing that occurs in 5′- and 3′-untranslated regions (UTRs) presumably alters the stability, localization, or translatability of the target mRNA (Morse et al. 2002; Prasanth et al. 2005). Recently, Yang and coworkers described an additional role of RNA editing in the biogenesis of select miRNAs involved in mammalian hematopoiesis (Yang et al. 2006).
The majority of verified targets of ADAR in Drosophila melanogaster come from a comparative genomic approach examining a distinct subset of genes consisting of ion channels, G-protein-coupled receptors (GPCRs), and proteins involved in synaptic transmission (Hoopengardner et al. 2003). Using this approach, conservation of the putative ECS site within neighboring introns was not an informative predictor; however, exons with the highest evolutionary conservation within a gene seemed to be the best predictive tool to identify ADAR targets for this subset of genes. The prevalence of editing sites found within the coding portions of Drosophila genes, as well as the stage-specific “self-tuning” of ADAR, could be a principal mechanism of increasing neuronal protein diversity (Keegan et al. 2005).
Although directed approaches to identify ADAR targets have been employed successfully, we have taken a different approach based on a systematic analysis of the Drosophila Gene Collection (DGC; http://www.fruitfly.org/DGC). The DGC contains cDNAs representing 10,398 of the predicted 13,449 genes in D. melanogaster (Release 4.1). This collection originates from cDNA libraries derived from a variety of tissues and developmental stages (Rubin et al. 2000; Stapleton et al. 2002a,b). Utilizing high-quality sequence data from full-insert sequences of adult head cDNA clones, we have identified 27 new targets of ADAR, doubling the total to 55. The edited sites verified in our analysis are within coding regions and in the 3′ UTRs of these targets. Previous studies (Dutzler et al. 2002, 2003) on one of our targets (CG31116), a member of the ClC family of chloride channels, suggest a clear functional consequence of editing upon the gating properties of the ion channel. Some target genes, like the G-protein-coupled receptor bride-of-sevenless (boss) and the calcium-activated potassium channel small conductance calcium-activated potassium channel (SK), have clear roles in neuronal development and signaling, while other target genes, like spir, which is an actin nucleating factor, represent components of the cytoskeleton. The largest class of ADAR targets consists of novel genes that have not yet been assigned a functional attribute in the Gene Ontology (GO) database (Ashburner et al. 2000).
Transcriptional analyses of the human genome (Kapranov et al. 2002), the Arabidopsis genome (Yamada et al. 2003), and the Drosophila genome (Stolc et al. 2004) suggest that upward of 50% of the predicted noncoding genome is expressed. In Drosophila, noncoding transcription has been well characterized and includes microRNAs (Lai et al. 2003; Lai 2005; Leaman et al. 2005), noncoding RNAs (Tupy et al. 2005), and anti-sense RNAs (Misra et al. 2002). The complexity of eukaryotic transcriptomes continues to expand, and editing of pre-mRNA transcripts adds yet another layer of intricacy.
RESULTS AND DISCUSSION
Identifying putative targets of ADAR
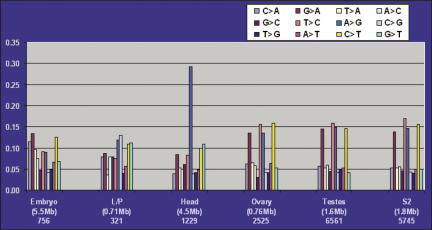
In order to identify putative new targets of ADAR, we took advantage of our full-insert sequenced gene collection. As part of our cDNA analysis pipeline, we aligned each high-quality cDNA sequence with the genome sequence using Sim4 (Florea et al. 1998) and recorded the base-pair discrepancies (see Materials and Methods). Figure 1 represents the results of tabulating all possible substitutions from ∼15 Mb of data from full-insert sequenced cDNAs. When substitutions are grouped by library and expressed as a percentage of the total, clones from the isogenic head library possess a threefold increase in the level of A-to-G transitions when compared to clones from the other two isogenic libraries. Moreover, the A > G transition frequency in the head libraries is the only type of substitution within the isogenic libraries that has a discernible frequency above background substitutions. This analysis led us to generate a list of clones with the highest likelihood of having an A-to-G substitution due to editing and reducing the possibility of strain-dependent polymorphisms. This approach identifies both authentic editing sites as well as errors introduced by the reverse transcriptase used to generate the cDNA libraries. To distinguish these events, further experimental validation of the sites is necessary and is described below.
FIGURE 1.
A-to-G transitions in head library cDNAs are 2.5–3-fold higher than all other types of base-pair substitutions within libraries derived from the same strain. Base-pair substitutions were identified and grouped by library and expressed as a percent of the total number of substitutions within each library. The embryo, larval/pupal (L/P), and head cDNA libraries are derived from the same isogenic strain used for delineating the genome sequence, while the ovary, testes, and the cell-line S2 libraries are not. Numbers below each library type correspond to the total number of base pairs (in parentheses, megabase pairs, Mb) and the total number of substitutions in each library.
The analysis resulted in a list of 198 putative editing targets. Most of the known edited genes in D. melanogaster are involved in rapid electrical and chemical neurotransmission (Stapleton et al. 2002a; Hoopengardner et al. 2003; Xia et al. 2005). This observation is, in part, due to the neurological phenotype associated with ADAR-null mutants coupled with the fact that genes of this type have been specifically evaluated for the presence of editing. Although our approach is not biased in this manner, we identified 11 of the known 28 targets of ADAR as putative targets and did not pursue them further. Additionally, 10 of the remaining 17 known target genes are represented in our collections, but do not possess the A-to-G transition, suggesting that since editing is <100% efficient, we captured the unedited mRNA in our cDNA collection. The remaining seven known targets do not have a representative EST or cDNA in our gene collection. The Sim4 alignment program encounters difficulties near the edges of sequence alignments, and putative targets with these alignment artifacts were removed from the list, further collapsing the set to a total of 108 clones containing 149 potential editing sites.
We interrogated these 149 putative editing sites experimentally by performing reverse-transcription–polymerase chain reaction (RT-PCR) followed by sequencing of the amplified products. RT-PCR was performed with total RNA isolated from adult heads of the isogenic strain. Amplicons ranged in size from ∼300 to 500 bp spanning the region(s) surrounding the potential editing site(s). Sequence traces from 27 of the 108 genes yielded 58 positions with either a mixed A and G signal (n = 51) or a pure G signal (n = 7), indicating the presence of edited transcripts (Fig. 2B; Supplemental Material, contact staple@fruitfly.org). We successfully amplified regions from all 108 putative targets and obtained high-quality sequencing data for all but one (CG7569). For targets whose chromatograms showed “weak” evidence of editing (predominant A at the putative editing site), we cloned the products from two independent RT-PCR reactions and sequenced 48 isolates per reaction. CG31116 is an example of this weak editing, where three of the four putative sites displayed almost no mixed signal. Cloning revealed these sites are edited with frequencies ranging from 4.5% to 10.4% (Supplemental Fig. S12 and table therein, contact staple@fruitfly.org). To rule out the unlikely possibility that the mix of A/G signals or the pure G signal seen in the RT-PCR was due to genome polymorphisms that have accumulated in the isogenic strain, we PCR-amplified and sequenced the same regions from genomic DNA isolated concomitantly with the RNA. None of the 58 sites is due to polymorphisms in the genomic DNA (Fig. 2B; Supplemental Material, contact staple@fruitfly.org). Similar analysis was performed on a subset of these 27 new targets using RNA isolated from ADAR-deficient animals, and all sites tested (n = 23) showed a pure A signal, indicating the unedited transcript (data not shown). Although this method of measuring the ratio of edited and unedited transcripts has been reported to be accurate to as low as 5% editing (Palladino et al. 2000), it cannot be used to determine the relative distribution of sites in cases where a transcript contains more than one edited site.
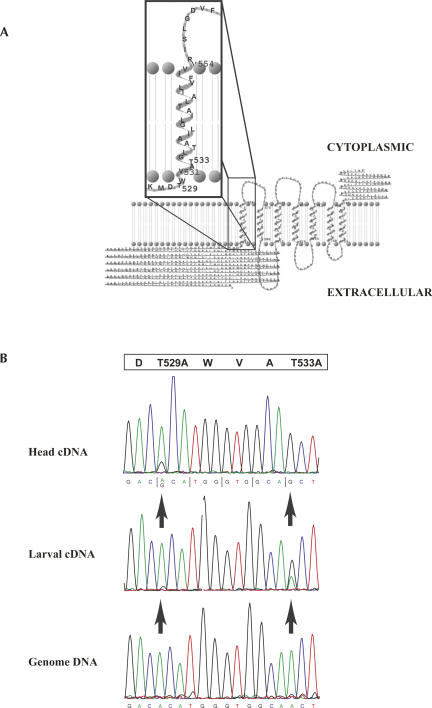
FIGURE 2.
Structure and editing of boss. (A) The transmembrane protein bride of sevenless (boss) is portrayed in the background and is predicted to pass through the lipid bilayer seven times with a large extracellular domain. An enlargement of the region found to be edited is in the boxed foreground. Threonines 529 and 533 lie at the junction between the extracellular and the first transmembrane domains, which, when edited by ADAR, are recoded to alanines. Representation of boss was visualized with TMRPres2D (Spyropoulos et al. 2004), a tool used for modeling transmembrane proteins. (B) Sequence chromatograms of the edited region of boss. The upper panel is a sequence trace from RT-PCR products of boss from wild-type animals that shows a mixed signal of A and G at threonine 529 when compared to the trace derived from the control genomic DNA (lower panel). Threonine 533 appears to be completely edited and displays a pure G signal in the wild-type trace when compared to the genomic trace. The larval chromatogram in the middle panel displays an intermediate level of editing at these two positions, suggesting that editing of boss is developmentally regulated. The open box at the top is the protein translation with the edited threonine codons indicated.
New targets of dADAR
The 27 new targets of ADAR are categorized into seven classes of genes, which are listed in Table 1. The seven gene classes include vesicular trafficking, ion homeostasis, signal transduction, ion channels, cytoskeletal components, an “other” class, and a novel or unknown class. For each target, the gene symbol, the cDNA representative that initially suggested editing, the protein, the amino acid residue(s), and 3′-UTR nucleotide(s) that are affected, along with the molecular function, if known, are listed.
TABLE 1.
New targets of ADAR editing
The vesicular traffic class of targets contains six genes, Rab26, Rlip, rab3-GEF, endophilin A (endoA), α-Adaptin, and sunday driver (syd). Small Ras-like G-proteins and their effectors are well-documented proteins that function in the intracellular transport of vesicles (Takai et al. 2001). Three of the six members fall into this category and are represented by the GTPase Rab26, the Ral GTPase activator protein Rlip, and the Rab3 guanyl-nucleotide exchange factor Rab3-GEF. The endoA and α-Adaptin genes encode proteins that are involved in the endocytic pathway: endoA promotes synaptic vesicle budding, and α-Adaptin is a subunit of the AP-2 complex (Guichet et al. 2002; Jafar-Nejad et al. 2002; Seto et al. 2002). The protein encoded by the syd gene was identified in a genetic screen in Drosophila for proteins involved in the axonal transport process (Bowman et al. 2000). Flies carrying mutations in syd reveal massive accumulations of various axonal membrane-bound cargoes, as well as axonal transport motors such as kinesin-I within the larval segmental nerves of multiple independent syd alleles. Syd is thought to function as a cargo adaptor in axonal transport by interacting directly with the kinesin light chain.
The ion homeostasis class consists of five genes involved in sodium and calcium ion exchange and calcium sequestration. Members of this group represent a new class of molecules that serve as substrates for ADAR and include Calphotin (Cpn), Nckx30C, and the related CG1090, Na pump α-subunit (Atpα), and CG32699. Atpα encodes a sodium:potassium-exchanging ATPase that undergoes a tyrosine-to-cysteine substitution due to editing. The Y390C substitution lies within the ATP hydrolase domain. The Atpα protein is predicted to contain 10 transmembrane-spanning segments and is detected in a subset of medial and lateral ventral cord neurons as well as in other larval and adult Drosophila tissues (Lebovitz et al. 1989). Animals carrying mutations in Atpα have behavioral abnormalities, reduced life span, and severe neuronal hyperexcitability along with the occurrence of age-dependent neurodegeneration (Palladino et al. 2003). The two related genes Nckx30C and CG1090 encode for proteins that function as potassium-dependent sodium–calcium exchangers. Nckx30C is critical for the rapid extrusion of calcium and is expressed in adult neurons and during ventral nerve cord development in the embryo (Haug-Collet et al. 1999). Cpn encodes a photoreceptor cell-specific calcium ion binding protein (Ballinger et al. 1993; Martin et al. 1993), and CG32699 is a gene that has an assigned GO functional attribute as having “calcium ion binding with acyltransferase activity.”
The signal transduction and ion channel groups contain two members each. The genes Mob1 and boss are grouped as signaling molecules, while the genes SK and CG31116 are ion channels. The G-protein-coupled receptor boss and the two ion channels are discussed in greater detail in the next section. Mob1 encodes a highly conserved protein shown to genetically and physically interact with Tricornered (trc), which is a member of the Drosophila Nuclear Dbf2 related (Ndr) subfamily of serine/threonine protein kinases (He et al. 2005). trc is required for the normal morphogenesis of polarized outgrowths such as epidermal hairs, sensory bristles, arista laterals, and sensory neuron dendrites (Geng et al. 2000; He and Adler 2002; Emoto et al. 2004). Furthermore, mutations in Mob1 were isolated in a behavioral screen for genes involved in long-term memory (Dubnau et al. 2003).
Another unique class of edited target genes is involved in actin nucleation and organization, represented by spire (spir), Ataxin 2 (Atx2), and CG32809. spir belongs to the posterior group of genes and is required for properly specifying the axis in the developing oocyte and embryo (Manseau and Schupbach 1989). spir mutants have a defect in microtubule plus-end orientation during oogenesis (Wellington et al. 1999). Recently, the gene product, Spir, has been shown to nucleate actin filaments in a unique way and may be a novel link between actin organization and intracellular vesicle transport (Kerkhoff et al. 2001; Quinlan et al. 2005). The amino acid change in Spir introduced by editing lies between two important conserved domains, the FYVE domain and the Spir box, which have been shown to be involved in its localization on intracellular membranes and possible association with Rab11, respectively. Whether this ADAR-modified residue in Spir alters its ability to nucleate actin filaments has yet to be shown. Another target of ADAR in this class, Atx2, is not implicated in actin nucleation or polymerization, but rather as a possible regulator of specific mRNAs that function to mediate the formation of actin filaments. Atx2 is the Drosophila homolog of the human polyglutamine disease gene SCA2, which is responsible for the dominantly inherited neurodegenerative disorder spinocerebellar ataxia type 2 (Huynh et al. 2000; Satterfield et al. 2002). In Drosophila, loss-of-function mutants of Atx2 suggest that its principal function is to regulate the formation or bundling of actin filaments by regulating a subset of RNA transcripts that encode mediators of actin filament formation. Studies of the SCA2 homolog, ATX-2, in C. elegans, demonstrate that it forms a complex with the poly(A)-binding protein PAB-1 and functions in translational regulation in the development of the germline (Ciosk et al. 2004). The last member in this class is CG32245 and is placed here only through its associated GO functional attribute of “structural constituent of cytoskeleton.”
There are two genes that comprise the “other” class, CG32809 and realtime (retm). CG32809 encodes a novel protein that is classified by GO as a proton-transporting ATPase. It also contains a partial Actin interacting protein 3 domain (AIP3, pfam03915) identified using the CD-Search engine at the National Center for Biotechnology Information (NCBI) (Marchler-Bauer and Bryant 2004). However, the K179R substitution due to editing of this transcript does not lie near the AIP3 domain. The gene retm has an assigned function as a phosphatidylinositol transporter whose protein contains SEC14 (cd00170), MSF1 (pfam04707), and CRAL_TRIO_N (pfam03765) domains. retm belongs to a novel family of mitochondrial proteins, is expressed in the developing midgut and central nervous system during embryogenesis, and is subcellularly localized to mitochondria (Dee and Moffat 2005).
The largest class of genes identified has no assigned function in the GO database and contains seven members (Table 1). Although this class does not have any associated GO attributes, mutations do exist for one of its members. Mutations in CG14616 result in lethality during the first and second instar larval stages (Drysdale and Crosby 2005).
Functional consequences of editing targets
Three notable targets of ADAR are the integral membrane-spanning proteins encoded by the genes boss, CG31116, and SK. The G-protein-coupled receptor boss is unique in that it acts as a ligand to determine the fate of the R7 photoreceptor cell in the Drosophila compound eye by a specific inductive interaction between the boss-bearing R8 photoreceptor neuron and the R7 precursor cell (Fortini et al. 1992; Hart et al. 1993). Interestingly, the edited sites convert threonines 529 and 533 to alanines, which lie at the junction of the large extracellular domain and the first transmembrane domain (Fig. 2A). At the larval stage, the T533 codon is a mix of signals, whereas in the adult, the T533 codon contains a pure G signal, indicating this is likely the predominant form of boss in the adult fly (Fig. 2B). Furthermore, this suggests that editing of boss at these sites is developmentally regulated. These substitutions may have an effect on the way in which the R8 photoreceptor cell presents boss to neighboring R7 cells, potentially modifying the action of boss at later stages in eye development.
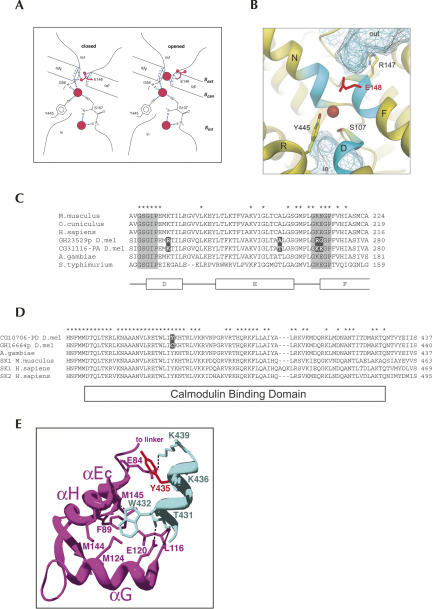
The gene CG31116 encodes a chloride channel belonging to the highly conserved ClC chloride channel family. In humans, mutations of ClC family members are associated with and underlying myotonia, Dent's disease, Bartter syndrome, osteoporosis, neurodegeneration, and possibly epilepsy (Jentsch et al. 2005b). Some ion channels, such as K+ channels, possess structurally distinct elements that perform the selective conduction of ions (filtering) and the actual opening or closing of the pore (gating) (Jiang et al. 2002). Crystallographic and electrophysiological studies of the anionic ClC channels from Escherichia coli and Salmonella typhimurium elegantly show that the processes of gating and filtering are closely tied (Dutzler et al. 2002) and that the side-chain carboxyl group of a glutamate residue serves as the gate of the pore lying within the selectivity filter (Fig. 3A,B). Obstructing the pore with its side chain closes the pore, while binding of a Cl− ion opens it. We have identified four edited sites, two of which affect residues in the selectivity filter (Fig. 3C). One of these residues is the highly conserved glutamate residue whose side chain acts as the anionic gate. We have found this glutamate codon to be edited in ∼10% of the adult head transcripts to encode a Gly residue, which interestingly is a residue that does not contain the carboxyl side chain capable of closing the ion pore and would be consistent with an open pore conformation. Electrophysiologic and crystallographic studies testing channel function support this observation. Converting the glutamate residue in the Torpedo ray ClC-0 channel to Ala, Gln, or Val causes the channel gate to stay open (Dutzler et al. 2003). Moreover, CG31116 is expressed in the ventral nerve cord, lateral cord glia, and the ventral midline, consistent with a neuronal function (Kearney et al. 2004).
FIGURE 3.
Structure and editing of ClC and SK ion channels. (A) Schematic of a ClC chloride channel conductance filter showing the closed and open conformations. The chloride ion is represented by red spheres, and the glutamate side chain is shown in red. Notice in the closed configuration the glutamate 148 side chain is occupying the ion binding site, while in the open configuration, it has moved out of the site and is replaced by a Cl− ion. Reprinted (with permission from AAAS © 2003) from Dutzler et al. (2003). (B) Ribbon structure of the ClC conductance filter depicted in the closed configuration with a chloride ion shown as a red sphere with the side chain of the edited glutamate 148 colored red. The amino acids at the ends of the α-helices, including the glutamate 148 at the end of helix F, are brought together near the membrane center to form the ion-binding site, shown in light blue. The vestibules toward the interior and exterior of the membrane are depicted as hatched domains. Reprinted (with permission from Macmillan Publishers Ltd. © 2002, http://www.nature.com/nature/index.html) from Dutzler et al. (2002). (C) Sequence alignment of the edited portion of ClC channels. Residues from the Cl− selectivity filter are represented by the gray shaded boxes, and identical residues are indicated with an asterisk. The four residues whose codons are edited in D. melanogaster are indicated by black boxes. The α-helices are depicted as open boxes below the alignment. The amino acid sequences and GenBank numbers are Mus musculus ClC2, NP_034030.1; Oryctolagus cuniculus ClC2, AAB05937.1; Homo sapiens ClC2, AAB34722.2; D. melanogaster GH23529p, AAM50193.1; D. melanogaster CG31116-PA, AAF54701.3; Anopheles gambiae, XP_312021.2; S. typhimurium, AE008704. The sequence alignments were performed with CLUSTALX (Thompson et al. 1997). (D) Sequence alignment of the Calmodulin binding domain of SK channels. The tyrosine 377 residue whose codon is edited in D. melanogaster is indicated by a black box. Identical residues are indicated with an asterisk, and the Calmodulin binding domain is depicted as an open box below the alignment. The amino acid sequences and GenBank numbers are D. melanogaster CG10706-PD, NP_726987.1; D. melanogaster GH16664p, AAM50183.1; A. gambiae, XP_314474.2; M. musculus SK1, AAK48900.1; H. sapiens SK1, AAH75037.3; H. sapiens SK2, AAP45946.1. The sequence alignments were performed with CLUSTALX (Thompson et al. 1997). (E) Ribbon structure of the rat SK2 calmodulin binding domain (colored in light blue) complexed with the apoCalmodulin C lobe (in purple). Notice that tyrosine 435 (colored in red) of SK2, which is the highly conserved tyrosine 377 in D. melanogaster, is involved in the interaction with apoCalmodulin. Reprinted (with permission from Elsevier © 2004) from Schumacher et al. (2004).
Another striking example of putative functional consequences of RNA editing on protein function is the transcript for the gene SK. This gene encodes a voltage-independent ion channel that is activated by submicromolar concentrations of intracellular calcium. These channels are high-affinity calcium sensors that transduce fluctuations in calcium concentrations into changes in membrane potential. SK channels are not gated by direct calcium binding, but are heteromeric complexes with calmodulin (CaM), where gating is mediated by binding of calcium to calmodulin and subsequent conformational alterations in the channel protein (Xia et al. 1998). The edited form of the SK transcript results in the tyrosine at position 377 being converted to a cysteine. This Y377C substitution is directly within the highly conserved calmodulin-binding domain (CaMBD) (Fig. 3D). The crystal structure of the CaMBD from rat SK2 complexed with apoCalmodulin reveals that the key region of CaMBD involved in apoCaM binding consists of 10 residues, which form an α-helix that binds only the C lobe of CaM (Schumacher et al. 2001). Interestingly, the structure suggests a van der Waals contact between the side chain of Y435 and residues in the E and H α-helix of CaM (Fig. 3E). Tyrosine 435 in the rat SK2 channel is the cognate tyrosine at position 377 that is edited in the Drosophila SK channel. Whether this Y377C substitution has a functional role in the ability of calmodulin to bind to the channel and therefore affect the transductive capacity of calmodulin remains to be tested.
Editing in D. melanogaster is thought to be a gene sparing strategy. That is, a different form of the same protein is used at different times in the animal's life cycle to perform similar functions and is facilitated by the differential activities of ADAR throughout development (Keegan et al. 2005). Our observations, including the example of the effect on Cl− gating, the possible developmental alteration of the activity of the boss ligand, and the other possible functional implications of editing in the 3′ UTRs, suggest an even greater role of editing by modulating specific protein function and transcript stability. It is very likely that more targets of editing exist in Drosophila because the bias of our sampling method was limited to the abundant mRNAs represented in our cDNA libraries coupled with the observation that editing is not 100% efficient. In summary, this analysis has identified 38 editing targets, 27 newly discovered, which doubles the total number of editing targets in D. melanogaster to 55.
MATERIALS AND METHODS
Identification of putative ADAR editing targets
We compared the nucleotide sequence of all full-length cDNA clones derived from libraries of adult head tissue to the Release 4 genome sequence and to the Release 4.1 computed gene models. Using Sim4 (Florea et al. 1998), we recorded all A>G substitutions, then manually inspected each clone for sequence quality discrepancies and Sim4 artifacts using Consed Autofinish (Gordon et al. 2001). This produced a curated list of 108 genes that were chosen as candidates for experimental testing.
Validation of targets
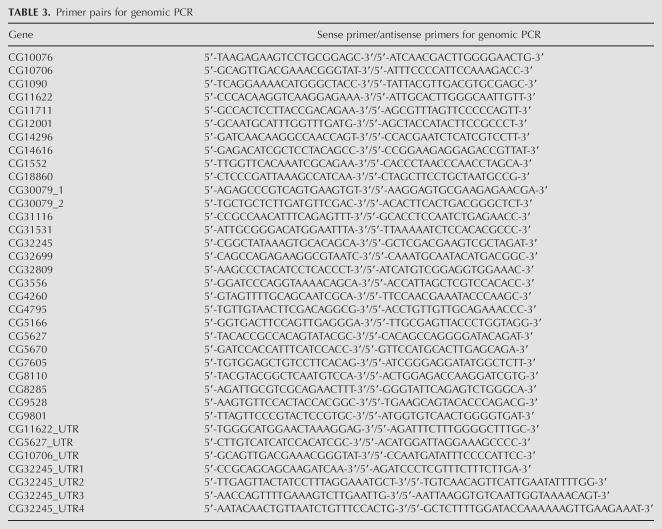
We used a similar method for RNA isolation and RT-PCR described previously (Stapleton et al. 2002a). RNA and genomic DNA were isolated at the same time using the Trizol reagent (Invitrogen) from adult heads from the isogenic strain y1; cn1 bw1 sp1. For the boss analysis, we isolated RNA from 0–24-h embryos, a mixed stage of larvae, and pupae from the isogenic strain. RT-PCR amplicons of the boss region from embryo and pupal RNA were negative, but positive for the larva RNA. RNA preps were digested with RNAase-free DNAase I (Roche) and tested by PCR to ensure the absence of contaminating genomic DNA. We designed gene-specific primers for the RT-PCR and the genomic PCR amplifications using Primer3 (Rozen and Skaletsky 2000) and the primer-picking feature in Consed Autofinish. The sense/antisense primer pair sequences for regions of the 27 validated genes are shown in Tables 2 and 3
TABLE 2.
Primer pairs for RT-PCR
TABLE 3.
Primer pairs for genomic PCR
Previously, we cloned PCR products from multiple RT reactions in order to evaluate the potential for editing. However, in this report, we directly sequenced the PCR products. For sequencing, PCR products were treated with 0.3 μL of Exonuclease I (USB), 0.5 μL of Shrimp Alkaline Phosphatase (Roche), and 0.5 μL of 1 M Tris (ph 8.0) in a final reaction volume of 75 μL and incubated for 45 min at 37°C and then heat-inactivated for 10 min at 65°C. The same sense and antisense primers used for RT-PCR and genomic PCR were used as sequencing primers in a 10-μL reaction containing 1 μL of ABI Big Dye III terminator mix (Applied Biosystems). Sequencing reactions were precipitated according to the manufacturer's protocol and loaded onto an ABI Prism 3730 DNA Analyzer. Sequence data were assembled and analyzed with the Phred/Phrap suite of programs and visualized with Consed. Chromatogram trace peaks at positions chosen for interrogation that displayed an unambiguous mixture of A and G signals or a pure G signal were recorded as being a positive target of ADAR. For the RT-PCR amplicons that were ambiguous or weak, we cloned PCR products from two independent RT-PCR reactions and picked 48 clones each for sequence analysis.
ACKNOWLEDGMENTS
We thank Gerry Rubin for providing support and encouragement; Ken Wan, Soo Park, and Richard Weiszmann for excellent technical support; and Roger Hoskins for critically reading and improving the manuscript. This work was supported by NIH grant HG002673 (to S.E.C.) and by the US Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.254306.
REFERENCES
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. The Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis, A., Placido, D., Maas, S., Brown, B.A., II, Lowenhaupt, K., Rich, A. The crystal structure of the Zβ domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 2005;351:496–507. doi: 10.1016/j.jmb.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Ballinger, D.G., Xue, N., Harshman, K.D. A Drosophila photoreceptor cell-specific protein, calphotin, binds calcium and contains a leucine zipper. Proc. Natl. Acad. Sci. 1993;90:1536–1540. doi: 10.1073/pnas.90.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, A.B., Kamal, A., Ritchings, B.W., Philp, A.V., McGrail, M., Gindhart, J.G., Goldstein, L.S. Kinesin-dependent axonal transport is mediated by the Sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- Cho, D.S., Yang, W., Lee, J.T., Shiekhattar, R., Murray, J.M., Nishikura, K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., DePalma, M., Priess, J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- Dee, C.T., Moffat, K.G. A novel family of mitochondrial proteins is represented by the Drosophila genes slmo, preli-like and real-time. Dev. Genes Evol. 2005;215:248–254. doi: 10.1007/s00427-005-0470-4. [DOI] [PubMed] [Google Scholar]
- Drysdale, R.A., Crosby, M.A. FlyBase: Genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau, J., Chiang, A.S., Grady, L., Barditch, J., Gossweiler, S., McNeil, J., Smith, P., Buldoc, F., Scott, R., Certa, U., et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Dutzler, R., Campbell, E.B., Cadene, M., Chait, B.T., MacKinnon, R. X-Ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- Dutzler, R., Campbell, E.B., MacKinnon, R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- Emoto, K., He, Y., Ye, B., Grueber, W.B., Adler, P.N., Jan, L.Y., Jan, Y.N. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Florea, L., Hartzell, G., Zhang, Z., Rubin, G.M., Miller, W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini, M.E., Simon, M.A., Rubin, G.M. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- Geng, W., He, B., Wang, M., Adler, P.N. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gaitan, M., Jackle, H. Role of Drosophila α-adaptin in presynaptic vesicle recycling. Cell. 1997;88:767–776. doi: 10.1016/s0092-8674(00)81923-6. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Desmarais, C., Green, P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichet, A., Wucherpfennig, T., Dudu, V., Etter, S., Wilsch-Brauniger, M., Hellwig, A., Gonzalez-Gaitan, M., Huttner, W.B., Schmidt, A.A. Essential role of endophilin A in synaptic vesicle budding at the Drosophila neuromuscular junction. EMBO J. 2002;21:1661–1672. doi: 10.1093/emboj/21.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, A.C., Kramer, H., Zipursky, S.L. Extracellular domain of the boss transmembrane ligand acts as an antagonist of the sev receptor. Nature. 1993;361:732–736. doi: 10.1038/361732a0. [DOI] [PubMed] [Google Scholar]
- Haudenschild, B.L., Maydanovych, O., Veliz, E.A., Macbeth, M.R., Bass, B.L., Beal, P.A. A transition state analogue for an RNA-editing reaction. J. Am. Chem. Soc. 2004;126:11213–11219. doi: 10.1021/ja0472073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug-Collet, K., Pearson, B., Webel, R., Szerencsei, R.T., Winkfein, R.J., Schnetkamp, P.P., Colley, N.J. Cloning and characterization of a potassium-dependent sodium/calcium exchanger in Drosophila . J. Cell Biol. 1999;147:659–670. doi: 10.1083/jcb.147.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B., Adler, P.N. The genetic control of arista lateral morphogenesis in Drosophila . Dev. Genes Evol. 2002;212:218–229. doi: 10.1007/s00427-002-0229-0. [DOI] [PubMed] [Google Scholar]
- He, Y., Emoto, K., Fang, X., Ren, N., Tian, X., Jan, Y.N., Adler, P.N. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell. 2005;16:4139–4152. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hoopengardner, B., Bhalla, T., Staber, C., Reenan, R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Huynh, D.P., Figueroa, K., Hoang, N., Pulst, S.M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad, H., Norga, K., Bellen, H. Numb: “Adapting” notch for endocytosis. Dev. Cell. 2002;3:155–156. doi: 10.1016/s1534-5807(02)00228-9. [DOI] [PubMed] [Google Scholar]
- Jentsch, T.J., Neagoe, I., Scheel, O. CLC chloride channels and transporters. Curr. Opin. Neurobiol. 2005a;15:319–325. doi: 10.1016/j.conb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Jentsch, T.J., Poet, M., Fuhrmann, J.C., Zdebik, A.A. Physiological functions of CLC Cl−channels gleaned from human genetic disease and mouse models. Annu. Rev. Physiol. 2005b;67:779–807. doi: 10.1146/annurev.physiol.67.032003.153245. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B.T., MacKinnon, R. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Jullien-Flores, V., Mahe, Y., Mirey, G., Leprince, C., Meunier-Bisceuil, B., Sorkin, A., Camonis, J.H. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: Involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 2000;113:2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., Gingeras, T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kearney, J.B., Wheeler, S.R., Estes, P., Parente, B., Crews, S.T. Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan, L.P., Brindle, J., Gallo, A., Leroy, A., Reenan, R.A., O'Connell, M.A. Tuning of RNA editing by ADAR is required in Drosophila . EMBO J. 2005;24:2183–2193. doi: 10.1038/sj.emboj.7600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff, E., Simpson, J.C., Leberfinger, C.B., Otto, I.M., Doerks, T., Bork, P., Rapp, U.R., Raabe, T., Pepperkok, R. The Spir actin organizers are involved in vesicle transport processes. Curr. Biol. 2001;11:1963–1968. doi: 10.1016/s0960-9822(01)00602-9. [DOI] [PubMed] [Google Scholar]
- Kim, D.D., Kim, T.T., Walsh, T., Kobayashi, Y., Matise, T.C., Buyske, S., Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, M., Hirschberg, B., Bond, C.T., Kinzie, J.M., Marrion, N.V., Maylie, J., Adelman, J.P. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- Kramer, H., Cagan, R.L., Zipursky, S.L. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. Whys and wherefores of miRNA-mediated regulation. Curr. Biol. 2005 miRNAs;15:R458–R460. doi: 10.1016/j.cub.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., Tomancak, P., Williams, R.W., Rubin, G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman, D., Chen, P.Y., Fak, J., Yalcin, A., Pearce, M., Unnerstall, U., Marks, D.S., Sander, C., Tuschl, T., Gaul, U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lebovitz, R.M., Takeyasu, K., Fambrough, D.M. Molecular characterization and expression of the (Na++K+)-ATPase α-subunit in Drosophila melanogaster . EMBO J. 1989;8:193–202. doi: 10.1002/j.1460-2075.1989.tb03364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Lloyd, T.E., Verstreken, P., Ostrin, E.J., Phillippi, A., Lichtarge, O., Bellen, H.J. A genome-wide search for synaptic vesicle cycle proteins in Drosophila . Neuron. 2000;26:45–50. doi: 10.1016/s0896-6273(00)81136-8. [DOI] [PubMed] [Google Scholar]
- Macbeth, M.R., Schubert, H.L., Vandemark, A.P., Lingam, A.T., Hill, C.P., Bass, B.L. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau, L.J., Schupbach, T. cappuccino and spire: Two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes & Dev. 1989;3:1437–1452. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J.H., Benzer, S., Rudnicka, M., Miller, C.A. Calphotin: A Drosophila photoreceptor cell calcium-binding protein. Proc. Natl. Acad. Sci. 1993;90:1531–1535. doi: 10.1073/pnas.90.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 2002;3:RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino, M.J., Keegan, L.P., O'Connell, M.A., Reenan, R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- Palladino, M.J., Bower, J.E., Kreber, R., Ganetzky, B. Neural dysfunction and neurodegeneration in Drosophila Na+/K+ATPase α subunit mutants. J. Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K.V., Prasanth, S.G., Xuan, Z., Hearn, S., Freier, S.M., Bennett, C.F., Zhang, M.Q., Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Quinlan, M.E., Heuser, J.E., Kerkhoff, E., Mullins, R.D. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Reinke, R., Zipursky, S.L. Cell–cell interaction in the Drosophila retina: The bride of sevenless gene is required in photoreceptor cell R8 for R7 cell development. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- Rozen, S., Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M., Harvey, D.A. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Satterfield, T.F., Jackson, S.M., Pallanck, L.J. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 2002;162:1687–1702. doi: 10.1093/genetics/162.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A., Rivard, A.F., Bachinger, H.P., Adelman, J.P. Structure of the gating domain of a Ca2+-activated K+channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- Schumacher, M.A., Crum, M., Miller, M. Crystal structure of apocalmodulin and an apocamodulin/SK2 CaMBD complex: Mechanism of Ca2+-activated SK channel grating. Structure. 2004;12:849–860. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Seto, E.S., Bellen, H.J., Lloyd, T.E. When cell biology meets development: Endocytic regulation of signaling pathways. Genes & Dev. 2002;16:1314–1336. doi: 10.1101/gad.989602. [DOI] [PubMed] [Google Scholar]
- Spyropoulos, I.C., Liakopoulos, T.D., Bagos, P.G., Hamodrakas, S.J. TMRPres2D: High quality visual representation of transmembrane protein models. Bioinformatics. 2004;20:3258–3260. doi: 10.1093/bioinformatics/bth358. [DOI] [PubMed] [Google Scholar]
- Stapleton, M., Carlson, J., Brokstein, P., Yu, C., Champe, M., George, R., Guarin, H., Kronmiller, B., Pacleb, J., Park, S., et al. A Drosophila full-length cDNA resource. Genome Biol. 2002a;3:RESEARCH0080. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002b;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M.F., Rifkin, S.A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P.E., et al. A gene expression map for the euchromatic genome of Drosophila melanogaster . Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., Matozaki, T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A., Saccomanno, L., Morse, D.P., Brodigan, T., Krause, M., Bass, B.L. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans . EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupy, J.L., Bailey, A.M., Dailey, G., Evans-Holm, M., Siebel, C.W., Misra, S., Celniker, S.E., Rubin, G.M. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster . Proc. Natl. Acad. Sci. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken, P., Kjaerulff, O., Lloyd, T.E., Atkinson, R., Zhou, Y., Meinertzhagen, I.A., Bellen, H.J. Endophilin mutations block clathrin-mediated endocytosis but not neurotransmitter release. Cell. 2002;109:101–112. doi: 10.1016/s0092-8674(02)00688-8. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Khillan, J., Gadue, P., Nishikura, K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Webel, R., Haug-Collet, K., Pearson, B., Szerencsei, R.T., Winkfein, R.J., Schnetkamp, P.P., Colley, N.J. Potassium-dependent sodium-calcium exchange through the eye of the fly. Ann. N.Y. Acad. Sci. 2002;976:300–314. doi: 10.1111/j.1749-6632.2002.tb04753.x. [DOI] [PubMed] [Google Scholar]
- Wellington, A., Emmons, S., James, B., Calley, J., Grover, M., Tolias, P., Manseau, L. Spire contains actin binding domains and is related to ascidian posterior end mark-5. Development. 1999;126:5267–5274. doi: 10.1242/dev.126.23.5267. [DOI] [PubMed] [Google Scholar]
- Xia, X.M., Fakler, B., Rivard, A., Wayman, G., Johnson-Pais, T., Keen, J.E., Ishii, T., Hirschberg, B., Bond, C.T., Lutsenko, S., et al. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature. 1998;395:503–507. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- Xia, S., Yang, J., Su, Y., Qian, J., Ma, E., Haddad, G.G. Identification of new targets of Drosophila pre-mRNA adenosine deaminase. Physiol. Genomics. 2005;20:195–202. doi: 10.1152/physiolgenomics.00093.2003. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yang, W., Chendrimada, T.P., Wang, Q., Higuchi, M., Seeburg, P.H., Shiekhattar, R., Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshie, S., Imai, A., Nashida, T., Shimomura, H. Expression, characterization, and localization of Rab26, a low molecular weight GTP-binding protein, in the rat parotid gland. Histochem. Cell Biol. 2000;113:259–263. doi: 10.1007/s004180000130. [DOI] [PubMed] [Google Scholar]